Abstract
We determined and annotated the complete 2.2-megabase genome sequence of Pyrobaculum aerophilum, a facultatively aerobic nitrate-reducing hyperthermophilic (Topt = 100°C) crenarchaeon. Clues were found suggesting explanations of the organism's surprising intolerance to sulfur, which may aid in the development of methods for genetic studies of the organism. Many interesting features worthy of further genetic studies were revealed. Whole genome computational analysis confirmed experiments showing that P. aerophilum (and perhaps all crenarchaea) lack 5′ untranslated regions in their mRNAs and thus appear not to use a ribosome-binding site (Shine–Dalgarno)-based mechanism for translation initiation at the 5′ end of transcripts. Inspection of the lengths and distribution of mononucleotide repeat-tracts revealed some interesting features. For instance, it was seen that mononucleotide repeat-tracts of Gs (or Cs) are highly unstable, a pattern expected for an organism deficient in mismatch repair. This result, together with an independent study on mutation rates, suggests a “mutator” phenotype.
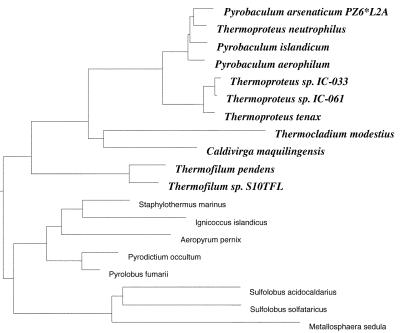
Pyrobaculum aerophilum is a hyperthermophilic (Tmax = 104°C, Topt = 100°C) and metabolically versatile member of the crenarchaea (Fig. 1), which are predominantly anaerobic respirers. Unlike most hyperthermophiles, P. aerophilum can withstand the presence of oxygen, growing efficiently in microaerobic conditions, thus making it relatively easy to work with in the laboratory. Unlike most of its phylogenetic neighbors, the growth of P. aerophilum is inhibited by the presence of elemental sulfur, but it grows well anaerobically using nitrate reduction (1). Here we have determined the complete genome sequence of P. aerophilum IM2, which was isolated from a boiling marine water hole at Maronti Beach, Italy (1). We obtained the sequence by a low coverage random shotgun sequencing strategy, with gap closure and resolution of ambiguities aided by the creation of a genomic fosmid map (2). We present an overview of the features and content of the genome, including a possible explanation of the organism's intolerance to sulfur and evidence of a possible lack of mismatch repair activity. Studies of the genus Pyrobaculum provide important opportunities for understanding the boundaries of life in extreme habitats. In a recent molecular sampling of a deep subsurface geothermal water pool, the only organisms detected were hyperthermophilic archaeal members closely related to Pyrobaculum (3).
Figure 1.
Small subunit rRNA-based phylogenetic tree of the crenarchaea. Constructed by Harald Huber, Regensburg University, Regensburg, Germany.
The P. aerophilum IM2 genome has 2,222,430 base pairs, 51% G + C. Two thousand five hundred and eight-seven protein-coding regions were designated, with an average length of 759 amino acids, covering 88% of the genome (Table 1). Circular and linear maps of genome features, including many features discussed below, are published as supporting information (Figs. 4 and 5) on the PNAS web site, www.pnas.org.
Table 1.
Chromosomal coding sequences
| Similar to | No. | Av bp | Av pI |
|---|---|---|---|
| Known proteins | 992 (38%) | 965 | 8.4 |
| Proteins of unknown function | 577 (22%) | 695 | 8.2 |
| Only within P. aerophilum genome | 302 (12%) | 693 | 8.8 |
| None | 716 (28%) | 553 | 8.5 |
| Total | 2,587 |
Methods
Fosmid and pUC18 libraries were constructed as described (2). Approximately 26,000 sequences were obtained from ≈14,000 pUC18 clones by using both Dye-terminator (18,000 sequences) and Dye-primer (8,000 sequences) chemistry on Applied Biosystems 373 and 377 sequencing machines. Twenty-three thousand four hundred and thirty-seven sequences were used to generate the final assembly, giving ≈4-fold coverage of the genome. Sequence assembly and editing were done by using phred/phrap/consed software (http://www.phrap.org). Sequence assembly was verified by comparison to sequence tagged sites of the published fosmid map (2) and PCR where necessary. After extensive manual inspection and resequencing, ≈3% of the genome was left covered by a single clone and ≈6% left unsequenced on both strands or with complementary sequencing chemistries.
Annotation was initially assisted by use of the magpie suite of software for automated genome interpretation (4). Coding regions were identified by using genemark (5), and exact start site predictions were refined with genemarks (6). fasta (7) and blast (8) programs were used to compare sequences against National Center for Biotechnology Information nonredundant protein and nucleotide databases. Predicted proteins were also compared against the pfam database of protein domain hidden Markov models (9) by using hmmer (http://hmmer.wustl.edu/). Where possible, the most likely major function of each predicted protein and its associated category was determined by manual inspection of results from the above analyses with emphasis placed on matches to characterized proteins or genes. Regions of synteny between the P. aerophilum genome and other completed genomes (at the protein-coding level) were identified and used to help specify functions and functional categories. Transfer RNAs were predicted by tRNAscan-SE (v. 1.21) (10) and manually refined by Todd Lowe (http://rna.wustl.edu/tRNAdb). Homologs of small nucleolar RNAs were identified by Todd Lowe (11). Repeat sequences were initially identified by using miropeats (12). Transmembrane helices were predicted for the T/A box families by using tmhmm v. 2.0 (13)
Replication and Repair
Mismatch Repair and Mononucleotide Runs.
Some of the most intriguing discoveries coming from archaeal genome sequencing projects are those pertaining to genes involved in DNA repair. The two major mechanisms for avoiding mutations during DNA replication are immediate editing of the growing strand by the DNA polymerase and detection and correction of mismatches soon after replication by the mismatch repair system (see reviews in refs. 14–16). Homologs of the Escherichia coli proteins involved in mismatch repair have been found in humans, and damage to them is involved in inherited predispositions to colon (HNPCC), endometrial, and ovarian cancer (17–20). However, homologs of the mismatch repair proteins have not been detected in the P. aerophilum genome, nor have they been found in any of the six published genomes of thermophilic archaea [Methanobacterium thermoautotrophicum and Archaeoglobus fulgidus each have a divergent MutS (MutS2) homolog without a MutL homolog]. It remains to be seen whether mismatch repair activities can be detected in these organisms. However, there are indications that P. aerophilum, at least, may be mismatch repair deficient.
During assembly and editing of the P. aerophilum genome, 10 regions were identified where high-quality sequences from different clones gave varying lengths of a mononucleotide run (Table 2). Further characterization of cultures grown from single colonies confirmed that these variable length mononucleotide runs were because of rapid fluctuations in the genome (M. M. Slupska, A. Conrad, L. Garibyan, S.T.F., J. Chiang, J. Pan, H. A. Nguyen, and J.H.M, unpublished work). Mononucleotide runs are particularly susceptible to strand slippage, and frequent frameshifting at such runs has been demonstrated in mismatch repair-deficient strains for both E. coli and yeast (21–23). An increased rate of variation of mononucleotide run lengths was also discovered during the genome sequencing project of Campylobacter jejuni (24). This genome has only a divergent MutS2 and no detectable MutL homolog. The data in Table 2 indicate that repeat-tracts are more unstable in runs of Gs (or Cs) than in runs of As (or Ts), as has been found in other systems (see, for instance, ref. 21). In C. jejuni, the repeat-tract sequences are proposed to have a function in producing variable surface structures that can be important for virulence and rapid adaptation to changing environments. It does not appear that the unstable mononucleotide runs in P. aerophilum have any obvious function, because they are distributed within tolerant regions, such as suspected pseudogenes and long intergenic regions. Rather, it seems that the repeat-tract instability is a consequence of the lack of a mismatch repair system. If P. aerophilum is indeed lacking mismatch repair, then an increased rate of frameshifting would be expected. During annotation, 32 genes were clearly found to have one or more frameshifts, many occurring at mononucleotide runs of from four to seven bases. Undetected frameshifts near the ends of genes, or in less conserved proteins, could double or triple this number.
Table 2.
Mononucleotide runs
| Genome position | Base | Lengths* | Context (intergenic region), bp† |
|---|---|---|---|
| 40977 | G | 11,15 | Intergenic (1,176) |
| 93514 | G | 11,11,11 | Intergenic (121) |
| 159685 | A | 9 | Intergenic (400) |
| 221366 | C | 11,12,12,16 | Intergenic (548) |
| 380761 | G | 12,13,14,14 | Intergenic (988) |
| 463202 | C | 14,15 | Within gene |
| 496647 | G | 11,11,13 | Within gene |
| 832639 | T | 8,10 | Intergenic (122) |
| 839489 | A | 9,9 | Intergenic (121) |
| 909549 | C | 7,8,8,9 | Within gene |
| 1028272 | T | 11,11,11 | Intergenic (253) |
| 1059643 | A | 9,9,9,9,9 | Intergenic (782) |
| 1110478 | C | 9,9,10,10,10,13 | Within gene |
| 1181593 | A | 9,9 | Intergenic (226) |
| 1217896 | C | 12 | Within gene |
| 1225895 | C | 12,15 | Intergenic (126) |
| 1293892 | A | 9,9,9,9,9 | Intergenic (85) |
| 1818426 | C | 11,12,15 | Intergenic (140) |
All mononucleotide runs of nine or more bases found in the P. aerophilum genome are listed.
Each of the lengths listed corresponds to sequences (one or more) from one genomic library clone.
The size of each intergenic region is given in brackets. The average size of intergenic regions in the genome is 121 bp.
A study of specific base substitution rates in P. aerophilum (M. M. Slupska, A. Conrad, L. Garibyan, S.T.F., J. Chiang, J. Pan, H. A. Nguyen, and J.H.M, unpublished work) reveals that transitions occur at the level expected of a mismatch repair deficient mutator. Although lack of a mismatch repair system is advantageous under certain selective conditions (see, for instance, ref. 25) and as a way of generating diversity and responding to changing environments (26–30), a permanent mutator lifestyle, in which the absence of the function generates high rates of mutation (a mutator phenotype), can have serious consequences as deleterious mutations accumulate (22). Other enzymes might partially compensate for the lack of a canonical mismatch repair system. Higher fidelity polymerases or editing functions might be used during replication, or additional as yet uncharacterized systems might operate. However, the repeat-tract instability observed here indicates that like C. jejuni (24), the high-temperature archaea such as P. aerophilum might be extraordinary examples of organisms that can survive as permanent mutators (we cannot at this stage rule out the possibility that the P. aerophilum species is not a mutator, but the IM2 isolate is a mutator variant). Studies with Mycobacterium smegmatis have also led to the idea that M. smegmatis and Mycobacterium tuberculosis might often have a mutator lifestyle (31).
Base Excision Repair.
Repair of damaged or altered bases often begins with excision of the altered base by DNA glycosylases and abasic-site endonucleases. The major enzyme in P. aerophilum cell free extracts responsible for removing uracil from DNA has been identified and characterized (32). It is similar in sequence to the recently characterized uracil DNA glycosylases of Thermotoga maritima and A. fulgidus (33). Most identified archaeal DNA glycosylases are in the MutY/Nth family, with every archaeal genome having from one to three homologs. P. aerophilum has three MutY/Nth homologs, two of which have been functionally characterized. Pa-MIG has been shown to have DNA glycosylase activity specific to U/G and T/G mismatches as well as an uncoupled AP lyase activity (34). The second characterized P. aerophilum DNA glycosylase (PaNth) recognizes and begins repair of pyrimidine adducts in a manner similar to E. coli endonuclease III (EcNth). It has DNA glycosylase/β-lyase activity with the modified pyrimidine base 5,6-dihydrothymine (DHT), which is enhanced when the DHT is paired with G (35).
Nucleotide Excision Repair (NER).
As with most other archaea (29), P. aerophilum has only a small number of homologs of the eukaryal NER enzymes (Rad25, Rad2, and possibly Rad3). Whether these function in NER (along with unidentified analogs of the numerous missing enzymes) is unknown.
Recombination.
P. aerophilum has two copies of the RecA/RAD51 recombinase homolog RadA. Otherwise, proteins involved in recombination have not been clearly identified. Evidence was found for recent events of homologous recombination with the discovery of two gene pairs that have partially recombined. Fig. 2 shows the alignments for two pairs of paralogs, a Cdc48 pair and a replication factor C small subunit pair. In both cases, most of the genes share 55–65% identity with their paralog, but both alignments have an anomalous region of 100% nucleotide identity covering hundreds of bases. In both cases, the sequence was confirmed by additional PCR and sequencing. A better understanding of how frequently such partial gene recombinations occur among prokaryotic lineages may help in interpreting results of phylogenetic analyses.
Figure 2.
Evidence for two recent events of homologous recombination. Nucleotide alignments from start codon to stop codon of (a) cell cycle protein Cdc48 paralogs PAE0696 and PAE3171 and (b) replication factor C small subunit paralogs PAE0734 and PAE1646. In each case, a section of several hundred nucleotides has become identical between the two paralogs.
DNA Polymerases.
In eukaryotes, multiple family B DNA polymerases are involved in DNA replication. Family B polymerases have been found in all archaea; however, the complement found in the crenarchaea differs from the complement found in the euryarchaea. The euryarchaea tend to have one family B DNA polymerase, plus a novel (possibly euryarchaea specific) heterodimeric DNA polymerase (36), although, on the basis of limited data, the crenarchaea, like the eukarya, seem to have multiple family B polymerases that may all play a role at the replication fork (37). The P. aerophilum genome sequence codes for three family B DNA polymerases, one of each of the B1, B2, and B3 subfamilies (38). The P. aerophilum B3 DNA polymerase shares 78% amino acid identity with a recently cloned and characterized DNA polymerase from the closely related Pyrobaculum islandicum. This P. islandicum enzyme was shown to have 3′ to 5′ exonuclease activity and, under suitable assay conditions for PCR, to amplify DNA fragments of up to 1,500 bp (37).
As with all archaea, the other replication factor homologs detected in the genome are more similar to their eukaryotic counterparts than bacterial. They include two copies of the sliding clamp processivity factor (proliferating cell nuclear antigen-like), one large subunit, and two copies of the small subunit of the clamp loading protein (replication factor C), DNA ligase, minichromosome maintenance protein, and a possible origin recognition protein (orc/cdc6). Single strand-binding protein (replication factor A) was not detected.
Cell Division.
Surprisingly, although FtsZ homologs are found in the euryarchaea and FtsZ ring structures have been visualized in Haloferax mediterranei (39), no homologs of FtsZ have been found within the crenarchaea, including P. aerophilum. A type of “snapping division” has been visualized for Thermoproteus tenax (40), which may be an example of a crenarchaeal FtsZ independent mechanism (41).
Transcription and Translation.
It has recently been shown experimentally and computationally that P. aerophilum mRNAs generally lack 5′ untranslated regions, and it was shown computationally that Aeropyrum pernix, the other crenarchaea with an available complete genome, probably also uses mainly leaderless transcripts (42). Thus the mechanism for translation initiation at the 5′ end of transcripts must be fundamentally different for these crenarchaea from the mechanism generally used by the characterized euryarchaea in which the ribosome binds to a Shine–Dalgarno site upstream of the translation start site. One might expect to find similarities to the eukaryotic translation initiation system in which the ribosome is first directed to a cap structure on the 5′ end of the mRNA. However, the genome in this case has not provided us clues as to the possible mechanism. The set of ribosomal proteins and translation initiation factors found in the genome is similar to those found in euryarchaeal genomes.
P. aerophilum has a similar set of translation factors as have been identified in other archaeal genomes. Aminoacyl tRNA synthetases have been identified for all amino acids except glutamine. Glu-tRNA (Gln) amidotransferase subunits are found; thus, as seen in other archaea and Gram-positive bacteria (43), glutamine tRNAs are probably amino-acylated with glutamic acid, followed by a transamidation to yield the glutaminyl-tRNA. Two apparent asparaginyl-tRNA synthetases are found, as well as two tyrosyl-tRNA synthetases. The C-terminal section of the canonical methionyl-tRNA synthetase is found as a separate ORF in the P. aerophilum genome, 1,334 bases upstream of the N-terminal gene.
No evidence was found for the use of the 21st amino acid selenocysteine. No matches were found to the set of genes coding for the proteins involved in the selenocysteine incorporation system. No selenocysteine tRNA was found, and no UGA codons were found in known selenocysteine positions.
Nonprotein Coding RNA.
After careful manual inspection of the results from the tRNAscan-SE program, 46 tRNAs and 1 pseudogene were identified (T. M. Lowe, personal communication). This corresponds to exactly one copy of each expected tRNA necessary to decode all 61 canonical codons. The most interesting feature of the P. aerophilum tRNAs is the prediction of large numbers of noncanonically placed introns (at least 11 of the 15 detected introns). This was also observed in the A. pernix genome (T. M. Lowe, personal communication) and is surprising in that it contrasts with the high consistency of intron placement among the vast majority of both archaeal and eukaryal tRNAs. Current tRNA annotations are available in the Genomic tRNA Database (T. Lowe, http://rna.wustl.edu/tRNAdb/).
Homologs of the eukaryal small nucleolar RNAs have been identified in archaea, with generally increasing numbers found in the genomes of organisms with higher growth temperatures (11). This may be because of an increased need for methylations of structural RNAs in higher-temperature environments (11). A record 53 homologs were identified in P. aerophilum by computational methods (T. M. Lowe, personal communication), most of which could be experimentally confirmed by primer extension (T. M. Lowe, A. Omer, L. Garibyan, S.T.F., J.H.M., and P. P. Dennis, unpublished work).
A 20-mer (ggcggcggcgtgggggtttt) inverted repeat was found surrounding the single rRNA operon, 51 and 15 bases upstream and downstream, respectively. The single 5S rRNA gene was not near the 16S-23S operon. Concurring with a previous report for P. aerophilum IM2 (44), a 713-bp intron was found within the 16S rRNA gene. Otherwise, introns were found within 15 tRNAs and not within any other genes.
Metabolism.
P. aerophilum is metabolically versatile, able to grow using organic and inorganic substrates for both aerobic and anaerobic respiration. Maximum cell densities are obtained with complex organics as substrates, although in the absence of organic material, P. aerophilum will grow chemolithoautotrophically using molecular hydrogen or thiosulfate (1). A set of genes for a complete citric acid cycle (TCA cycle) using 2-oxoacid/ferredoxin oxidoreductase was found.
Glyoxylate Cycle.
Consistent with P. aerophilum's growth on acetate (1), candidates for the key enzymes of the glyoxylate cycle, isocitrate lyase and malate synthase, have been found in the genome. The glyoxylate cycle is required to synthesize precursors for carbohydrates only when the carbon source is a C2 compound. To date, the only demonstrated occurrence of the glyoxylate cycle within the archaea is for a halophilic euryarchaeota, Haloferax volcanii (45). Interestingly, the H. volcanii malate synthase falls into a distinct class separate from the previously known sequence-based classes (A and G) (45). However, this class may not be typical of the archaea, as the P. aerophilum putative malate synthase falls clearly within class A, sharing 30% amino acid identity (blast Expect = e−23) with a yeast glyoxysomal malate synthase over the first two-thirds of the yeast protein length. Further, a database search revealed a putative malate synthase in the recently published Sulfolobus solfataricus genome (46), which shares 60% amino acid identity over the full length (813 amino acids) of the P. aerophilum protein.
2-Oxoacid Dehydrogenase.
Another metabolic similarity between P. aerophilum and H. volcanii is the presence of genes with highly significant homology to 2-oxoacid dehydrogenase multienzyme complexes. 2-Oxoacid dehydrogenases are used in eukarya, and most aerobic bacteria to convert pyruvate to acetyl-CoA, whereas archaea are generally known to use a simple pyruvate oxidoreductase (47). An operon containing genes homologous to 2-oxoacid dehydrogenase enzymes: E1 (2-oxoacid decarboxylase), E2 (dihydrolipoamide acetyltransferase), and E3 (dihydrolipoamide dehydrogenase) has been characterized for H. volcanii; however, the enzymatic activity was not detected in the organism (47). The P. aerophilum genome contains an apparent operon of not only the E1, E2, and E3 genes but also genes for lipoic acid synthetase and lipoate protein ligase B, known in eukarya and bacteria to act in the biosynthesis and attachment (to the E2 subunit) of lipoic acid cofactors (47).
Glycolysis/Gluconeogenesis.
As with the other completed archaeal genomes, several of the genes involved in glycolysis/gluconeogenesis have been identified; however, no ORFs with sequence similarity to phosphofructokinase or fructose bisphosphate aldolase are found. On the basis of a 25-aa N-terminal sequence from a recently characterized ATP-dependent 6-phosphofructokinase from the hyperthermophilic archaeon Desulfurococcus amylolyticus (48), we have annotated a P. aerophilum ORF (PAE0835) as a possible phosphofructokinase (Pfk). This candidate Pfk has a high-scoring blocks (49) match to the interpro (50) PfkB family of carbohydrate kinases (Entry IPR002173) and is of similar molecular mass (32 kDa) to the D. amylolyticus Pfk (33 kDa). PAE0835 has a reasonable match at the N-terminal to the D. amylolyticus N-terminal fragment (7 of 25 residues are identical with no gaps); however, a strong homolog of PAE0835 (E = 10−27), in the genome of the crenarchaeon A. pernix (APE0012), has a more conclusive match to the D. amylolyticus N-terminal (11 identities of 25 with no gaps). Homologs of APE0012 and PAE0835, generally annotated as sugar kinases, are found in all of the completed archaeal genomes as well as in many bacteria and eukaryotes.
Respiratory Proteins.
Although the cultivated thermophilic crenarchaeota are almost exclusively anaerobic respirers, the three crenarchaeons with genome sequences available, P. aerophilum, A. pernix (51), and S. solfataricus (http://www-archbac.u-psud.fr/projects/sulfolobus/) are able to grow aerobically. Numerous respiratory chain proteins are found in the P. aerophilum genome, including a cluster containing cytochrome C oxidase subunits and assembly factor, cytochrome b/b6, senC, nosD, and two iron sulfur proteins. Three putative Rieske iron–sulfur proteins were identified in the genome, one of which, designated ParR, has been expressed and partially characterized (52). P. aerophilum can also grow anaerobically, by nitrate reduction, using the denitrification pathway (refs. 1 and 53). Five putative nitrate reductase subunits were found in the genome, with the major (α and β) subunits sharing 50–55% amino acid identity to their Bacillus subtilis homologs. Three putative nitrite reductase subunits were found as well as a nitric oxide reductase subunit with 28% amino acid identity to the NorZ of the β proteobacteria Ralstonia eutropha. Surprisingly, a set of nitrate reductase genes similar to the P. aerophilum genes (60% amino acid identity for the major subunits) were found in the A. pernix genome, although A. pernix has been reported to be strictly aerobic (54).
Sulfur Metabolism.
The presence of elemental sulfur is lethal to cultures of P. aerophilum (1). This feature distinguishes P. aerophilum from its close phylogenetic neighbors, including two other cultured species of Pyrobaculum (islandicum and organotrophum). P. aerophilum does have a set of three sulfur metabolism genes that, when intact, could be used for either sulfate reduction or sulfur oxidation. However, both of the adenylylsulfate reductase subunits, for the middle step, are disrupted. The α subunit has a premature stop codon three quarters into the gene, and the β subunit has a frameshift near the middle of the gene. The proteins involved in this potentially bidirectional pathway (sulfate adenylyltransferase, adenylylsulfate reductase, and sulfite reductase) are well conserved across phylogenetically distant prokaryotes. It is possible that the loss of function of the adenylylsulfate reductase is responsible for the organism's sensitivity to elemental sulfur, and that introducing a functional replacement enzyme could restore resistance and/or add a new definable metabolic capacity. In this case, these genes could provide a plasmid-borne selectable marker that would be a significant step toward the development of a genetic system. The closely related P. islandicum (Fig. 1) is a likely source of the genes, as sequences of the three subunits of the neighboring sulfite reductase have been reported (55) (66, 69, and 78% nucleotide identity to P. aerophilum).
These sulfur metabolism genes are all found within a 100-kb region of the P. aerophilum genome. Putative enzymes thiosulfate sulfurtransferase and phosphoadenosine phosphosulfate reductase (PAPS reductase) were also found in this region.
PaRep2 Family.
P. aerophilum does not have any large families of exceptionally well conserved (98–100% nucleotide identity) repeats with ORFs, reminiscent of active transposons or insertion elements. However, the genome does have a very large and unusual family, comprising 3.3% of the genome, whose members are remarkably variable in both length and level of conservation. The repeat generally carries three ORFs (paRep2a2, 2a1, and 2b) divergently oriented from a central point between 2a1 and 2b. There are 22 copies that contain both 2a and 2b regions, 9 with only 2a regions, and 14 with only 2b regions. The highest conservation of each family member with another ranges from 100% nucleotide identity over 876 bases (and 96% identity over 3,770 bases) to less than 30% amino acid identity. They are extremely variable in size, ranging from 98 to 5,532 bases, primarily because of deletions from the outer 3′ ends of the paRep2as and the central 5′ ends of the paRep2bs. Multiple alignments (Fig. 6, which is published as supporting information on the PNAS web site) show numerous frameshifts and internal stop codons, perhaps indicating that most family members are no longer functional and are undergoing degradation. Nucleotide alignment of the three largest and most conserved copies (Fig. 6c) reveals several interesting features of this family. The unequal pattern of conservation across the length of the alignment (ranging from 55% in some regions to 100% in others) suggests that partial recombination has occurred between at least two of these sequences. All of the eight small insertion/deletions between the three sequences, found distributed over the 4,700-bp shared region, are in multiples of 3 (thus not causing frameshifts), suggesting that a selective pressure has maintained the ORFs because of the duplication/recombination events that created them. Surprisingly, however, all three of the sequences share an identical frameshift within the 4,050-bp paRep2b ORF (at approximately position 1020). A graphical outline of the 45 paRep2 regions (Fig. 7, which is published as supporting information on the PNAS web site) shows further the surprising patterns and variation among this family.
T/A Box Families.
A few distantly related families of P. aerophilum specific proteins have runs of 8–11 As or Ts in the 24 bases preceding their start codons (Table 3 and Fig. 3). Remarkably, whether the run is of As or of Ts (on the strand coding for the gene) seems to depend on its location in the genome. There are 10 upstream A runs found starting at genome positions 839489 until the last at position 1864462, covering roughly half of the genome, whereas the 10 upstream runs in the rest of the circular genome are all of Ts. The pattern does not depend on genome strand, as the genes fall on both strands within each region (Table 3). Many of the associated genes share similar characteristics and are part of a gene pair in which the downstream genes also share similar characteristics with each other (Table 4, which is published as supporting information on the PNAS web site). The first gene of each of the pairs codes for a large protein of between 77 and 90 kDa and usually has a single transmembrane helix predicted at the N terminus. The second gene of each of the pairs codes for a small protein of between 16 and 23 kDa and always has from four to six predicted transmembrane helices. There are other members of these gene families that do not have T or A runs upstream. No gene pair families with similar molecular weights and numbers of transmembrane helices were evident in any of the published microbial genomes. The reason for the striking dichotomous distribution of the T/A boxes is unclear. Further characterization of these interesting families could possibly lead to insights about genome organization.
Table 3.
T/A box families
| Genome position | Run | Genome strand | No. bases from start codon | Gene identifications |
|---|---|---|---|---|
| 1864462 | 8Ts | − | −24 | PAE3129,3125 |
| 1895538 | 8Ts | + | −7 | PAE3182 |
| 1957470 | 8Ts | − | −7 | PAE3261,3260 |
| 159693 | 9Ts | − | −5 | PAE0282,0280 |
| 163987 | 8T/As | +/− | −20/−5 | PAE0293/0292 |
| 577313 | 8Ts | − | −3 | PAE0985,0984 |
| 624765 | 8Ts | + | −9 | PAE1067,1068 |
| 633675 | 8Ts | + | −6 | PAE1079,1081 |
| 639492 | 8Ts | + | −6 | PAE1089,1092 |
| 806449 | 8Ts | + | −4 | PAE1358 |
| 839489 | 9As | + | −22 | PAE1407 |
| 1007991 | 8As | + | −21 | PAE1720 |
| 1013101 | 8As | − | −21 | PAE1727 |
| 1028282 | 11As | − | −20 | PAE1756,1755 |
| 1049571 | 8As | − | −17 | PAE1783 |
| 1059643 | 9As | + | −15 | PAE1802,1803 |
| 1071224 | 8As | + | −21 | PAE1819 |
| 1181593 | 9As | + | −22 | PAE2005,2006 |
| 1293892 | 9As | + | −20 | PAE2186 |
| 1736810 | 8As | − | −24 | PAE2924 |
Intergenic runs of eight or more As or Ts and their associated gene families are listed in the order in which they appear on the circular genome, starting at the switch between As and Ts. The eight runs that are not listed either fell within genes or were 73 bases or more away from a start codon. The run at 163987 is upstream of two start codons, one on each strand. Listed pairs of genes have the run upstream of the first gene and are always immediate neighbors (and in the same orientation), even if the gene identifiers are not contiguous numbers.
Figure 3.
Upstream regions of T/A box family genes. T/A boxes are underlined. Start codons for the associated genes are in bold. The second possible associated start codon, on the reverse strand, is shown underlined for PAE0293.
A genome sequence is important, because it provides both an invaluable tool for ongoing biological research and a generator of new hypotheses for further research. The P. aerophilum genome not only provides valuable basic data in an understudied branch of life but also poses many questions warranting several new lines of research.
Supplementary Material
Acknowledgments
We thank Mark Borodovsky and John Besemer for generous help with gene prediction, including development of the genemarks program to take advantage of P. aerophilum's unusually strong upstream signals for start site prediction. We thank Terry Gaasterland for help with implementation, maintenance, and use of the magpie system and for useful discussions. We thank Todd Lowe for manual tRNA and sRNA predictions and discussions. We thank Fredrick Blattner and DNAstar (Madison, WI) for use of the genvision software (see the supporting information on the PNAS web site, www.pnas.org). J.H.M. was supported by grants from the U.S. Office of Naval Research (ONR) and the National Institutes of Health (NIH) (GM57917). K.O.S. was supported by grants from the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie. S.T.F. received support from the National Aeronautics and Space Administration through the Astrobiology Institute and from grants to J.H.M. from the ONR and NIH (GM57917). The bulk of the raw sequence was obtained at the California Institute of Technology sequencing facility and was funded by grants to M.S. from the Genome Project of the U.S. Department of Energy.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AE009441).
References
- 1.Völkl P, Huber R, Drobner E, Rachel R, Burggraf S, Trincone A, Stetter K O. Appl Environ Microbiol. 1993;59:2918–2926. doi: 10.1128/aem.59.9.2918-2926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitz-Gibbon S, Choi A, Miller J H, Stetter K O, Simon M, Swanson R, Kim U-J. Extremophiles. 1997;1:36–51. doi: 10.1007/s007920050013. [DOI] [PubMed] [Google Scholar]
- 3.Takai K, Horikoshi K. Appl Environ Microbiol. 1999;65:5586–5589. doi: 10.1128/aem.65.12.5586-5589.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaasterland T, Sensen C W. Biochimie. 1996;78:302–310. doi: 10.1016/0300-9084(96)84761-4. [DOI] [PubMed] [Google Scholar]
- 5.Borodovsky M, McIninch J. Comput Chem. 1993;17:123–133. [Google Scholar]
- 6.Besemer J, Lomsadze A, Borodovsky M. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bateman A, Birney E, Durbin R, Eddy S R, Finn R D, Sonnhammer E L. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowe T M, Eddy S R. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Omer A D, Lowe T M, Russell A G, Ebhardt H, Eddy S R, Dennis P P. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 12.Parsons J D. Comput Appl Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- 13.Sonnhammer E L, von Heijne G, Krogh A. Intel Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 14.Modrich P. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 15.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 16.Kolodner R. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharyya N P, Skandalis A, Ganesh A, Groden J, Meuth M. Proc Natl Acad Sci USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fishel R, Lescoe M K, Rao M R, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 19.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomäki P, Sistonen P, Aaltonen L A, Nyström-Lahti M, et al. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 20.Papadopoulos N, Nicolaides N C, Wei Y F, Ruben S M, Carter K C, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, et al. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 21.Cupples C G, Cabrera M, Cruz C, Miller J H. Genetics. 1990;125:275–280. doi: 10.1093/genetics/125.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Funchain P, Yeung A, Stewart J L, Lin R, Slupska M M, Miller J H. Genetics. 2000;154:959–970. doi: 10.1093/genetics/154.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strand M, Prolla T A, Liskay R M, Petes T D. Nature (London) 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 24.Parkhill J, Wren B W, Mungall K, Ketley J M, Churcher C, Basham D, Chillingworth T, Davies R M, Feltwell T, Holroyd S, et al. Nature (London) 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 25.Mao E F, Lane L, Lee J, Miller J H. J Bacteriol. 1997;179:417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LeClerc J E, Li B, Payne W L, Cebula T A. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 27.Sniegowski P D, Gerrish P J, Lenski R E. Nature (London) 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 28.Taddei F, Matic I, Godelle B, Radman M. Trends Microbiol. 1997;5:427–428. doi: 10.1016/S0966-842X(97)01157-8. [DOI] [PubMed] [Google Scholar]
- 29.Eisen J A, Hanawalt P C. Mutat Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller J H, Suthar A, Tai J, Yeung A, Truong C, Stewart J L. J Bacteriol. 1999;181:1576–1584. doi: 10.1128/jb.181.5.1576-1584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karunakaran P, Davies J. J Bacteriol. 2000;182:3331–3335. doi: 10.1128/jb.182.12.3331-3335.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sartori A A, Schar P, Fitz-Gibbon S, Miller J H, Jiricny J. J Biol Chem. 2001;276:29979–29986. doi: 10.1074/jbc.M102985200. [DOI] [PubMed] [Google Scholar]
- 33.Sandigursky M, Franklin W A. J Biol Chem. 2000;275:19146–19149. doi: 10.1074/jbc.M001995200. [DOI] [PubMed] [Google Scholar]
- 34.Yang H, Fitz-Gibbon S, Marcotte E M, Tai J H, Hyman E C, Miller J H. J Bacteriol. 2000;182:1272–1279. doi: 10.1128/jb.182.5.1272-1279.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H J, Phan I T, Fitz-Gibbon S, Shivji M K K, Wood R D, Clendenin W M, Hyman E C, Miller J H. Nucleic Acids Res. 2001;29:604–613. doi: 10.1093/nar/29.3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cann I K, Komori K, Toh H, Kanai S, Ishino Y. Proc Natl Acad Sci USA. 1998;95:14250–14255. doi: 10.1073/pnas.95.24.14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kahler M, Antranikian G. J Bacteriol. 2000;182:655–663. doi: 10.1128/jb.182.3.655-663.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgell D R, Klenk H P, Doolittle W F. J Bacteriol. 1997;179:2632–2640. doi: 10.1128/jb.179.8.2632-2640.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poplawski A, Gullbrand B, Bernander R. Gene. 2000;242:357–367. doi: 10.1016/s0378-1119(99)00517-x. [DOI] [PubMed] [Google Scholar]
- 40.Horn C, Paulmann B, Kerlen G, Junker N, Huber H. J Bacteriol. 1999;181:5114–5118. doi: 10.1128/jb.181.16.5114-5118.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernander R. Trends Microbiol. 2000;8:278–283. doi: 10.1016/s0966-842x(00)01760-1. [DOI] [PubMed] [Google Scholar]
- 42.Slupska M M, King A G, Fitz-Gibbon S, Besemer J, Borodovsky M, Miller J H. J Mol Biol. 2001;309:347–360. doi: 10.1006/jmbi.2001.4669. [DOI] [PubMed] [Google Scholar]
- 43.Curnow A W, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin T M, Soll D. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burggraf S, Larsen N, Woese C R, Stetter K O. Proc Natl Acad Sci USA. 1993;90:2547–2550. doi: 10.1073/pnas.90.6.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serrano J A, Bonete M J. Biochim Biophys Acta. 2001;1520:154–162. doi: 10.1016/s0167-4781(01)00263-9. [DOI] [PubMed] [Google Scholar]
- 46.She Q, Singh R K, Confalonieri F, Zivanovic Y, Allard G, Awayez M J, Chan-Weiher C C, Clausen I G, Curtis B A, De Moors A, et al. Proc Natl Acad Sci USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. . (First Published June 26, 2001; 10.1073/pnas.141222098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jolley K A, Maddocks D G, Gyles S L, Mullan Z, Tang S L, Dyall-Smith M L, Hough D W, Danson M J. Microbiology. 2000;146:1061–1069. doi: 10.1099/00221287-146-5-1061. [DOI] [PubMed] [Google Scholar]
- 48.Hansen T, Schönheit P. Arch Microbiol. 2000;173:103–109. doi: 10.1007/s002039900114. [DOI] [PubMed] [Google Scholar]
- 49.Henikoff S, Henikoff J G. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- 50.Apweiler R, Attwood T K, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti T, Corpet F, Croning M D R, et al. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, et al. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. , 145–152. [DOI] [PubMed] [Google Scholar]
- 52.Henninger T, Anemüller S, Fitz-Gibbon S, Miller J H, Schäfer G, Schmidt C L. J Bioenerg Biomembr. 1999;31:119–128. doi: 10.1023/a:1005447710894. [DOI] [PubMed] [Google Scholar]
- 53.Afshar S, Kim C, Monbouquette H G, Schroder I. Appl Environ Microbiol. 1998;64:3004–3008. doi: 10.1128/aem.64.8.3004-3008.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sako Y, Nomura N, Uchida A, Ishida Y, Morii H, Koga Y, Hoaki T, Maruyama T. Int J Syst Bacteriol. 1996;46:1070–1077. doi: 10.1099/00207713-46-4-1070. [DOI] [PubMed] [Google Scholar]
- 55.Molitor M, Dahl C, Molitor I, Schäfer U, Speich N, Huber R, Deutzmann R, Trüper H G. Microbiology. 1998;144:529–541. doi: 10.1099/00221287-144-2-529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.