Abstract
Background
The largest open reading frame in the Saccharomyces genome encodes midasin (MDN1p, YLR106p), an AAA ATPase of 560 kDa that is essential for cell viability. Orthologs of midasin have been identified in the genome projects for Drosophila, Arabidopsis, and Schizosaccharomyces pombe.
Results
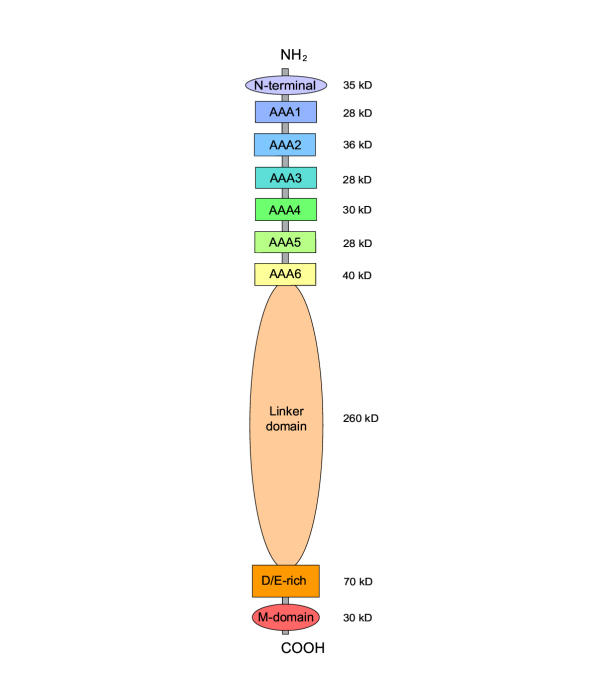
Midasin is present as a single-copy gene encoding a well-conserved protein of ~600 kDa in all eukaryotes for which data are available. In humans, the gene maps to 6q15 and encodes a predicted protein of 5596 residues (632 kDa). Sequence alignments of midasin from humans, yeast, Giardia and Encephalitozoon indicate that its domain structure comprises an N-terminal domain (35 kDa), followed by an AAA domain containing six tandem AAA protomers (~30 kDa each), a linker domain (260 kDa), an acidic domain (~70 kDa) containing 35–40% aspartate and glutamate, and a carboxy-terminal M-domain (30 kDa) that possesses MIDAS sequence motifs and is homologous to the I-domain of integrins. Expression of hemagglutamin-tagged midasin in yeast demonstrates a polypeptide of the anticipated size that is localized principally in the nucleus.
Conclusions
The highly conserved structure of midasin in eukaryotes, taken in conjunction with its nuclear localization in yeast, suggests that midasin may function as a nuclear chaperone and be involved in the assembly/disassembly of macromolecular complexes in the nucleus. The AAA domain of midasin is evolutionarily related to that of dynein, but it appears to lack a microtubule-binding site.
Background
Although the challenge represented by the largest open reading frame in the yeast genome (YLR106C) has existed for more than five years, there is surprisingly little known about the function of YLR106p, the protein that it encodes. The predicted amino acid sequence of YLR106p contains 4910 amino acids, with a molecular mass (MM) of 560 kDa. Systematic deletion studies of yeast proteins have indicated that YLR106p is required for viability [1]. YLR106p has recently been identified in a 60S pre-ribosomal particle involved in export of 60S ribosome subunits from the nucleus [2], but it is not yet clear whether this association is functional or adventitious. Different sets of polypeptides were found associated with YLR106p in a systematic study of the yeast proteome [3,4]. In a survey of the family of AAA ATPases, Neuwald and coworkers [5] noted that YLR106p is a member of the AAA ATPase family and is unusual in containing six tandem copies of the set of sequence motifs that characterize an AAA protomer.
The genome projects for Drosophila, Arabidopsis, and Schizosaccharomyces pombe have identified an ortholog of YLR106p in these organisms. The ortholog in these species, as well as the presence of an unassembled orthologous gene in humans, Caenorhabditis and Giardia, has been noted by Mocz and Gibbons [6], who discussed its evolutionary relationship to the heavy chain of dynein motor protein. The COOH-terminal region of the human ortholog of YLR106p has been obtained as clone kiaa0301 by Nagase and coworkers [7] as part of a comprehensive project to clone high molecular weight polypeptides in humans. These authors reported that the gene is expressed at a low level in most tissues, with higher levels present in testis and kidney [8].
In this study, we have used sequence alignments of YLR106p and its orthologs from widely diverse eukaryotes to identify and characterize five major functional domains in the protein. Expression of epitope-tagged YLR106p is shown to result in a polypeptide of the anticipated size that localizes principally in the nucleus.
Results and discussion
Preliminary screening of the genomic databases with the predicted amino acid sequence of the Saccharomyces protein YLR106p indicated that most eukaryotes, including both animals and plants, contain a single copy gene encoding a well conserved ortholog of size and structure comparable to YLR106p. One of the most characteristic features of this protein is the presence of a COOH-terminal domain possessing a full set of the sequence motifs that are indicative for the MIDAS (metal ion dependent adhesion site) conformation that occurs in the I-domain of vertebrate integrin α-chains [9,10]. In order to support a uniform terminology over the broad range of eukaryotes in which the ortholog of YLR106p occurs, we propose that it be given the name "midasin" and that the gene symbol in Saccharomyces be MDN1.
Amino acid sequence
In Saccharomyces, midasin (Mdn1p) is encoded as a predicted polypeptide of 4910 amino acids (accession S64942; MM 560 kDa). In Schizosaccharomyces pombe, it is encoded as a polypeptide of 4717 amino acids (accession CAB11610; MM 538 kDa). In Giardia intestinalis, it comprises a polypeptide of 4835 amino acids (accession AF494287; MM 540 kDa).
We have determined the sequence of the human midasin gene from PCR-amplified fragments of cDNA from testis. The coding region of the human gene for midasin (accession AF503925) comprises 102 exons spanning ~156,000 bp at map position 6q15 and encodes a predicted polypeptide of 5596 amino acids with a molecular mass of 631 kDa. All intron boundaries in the gene follow the gt-ag rule. Microheterogeneity of splice sites, including the omission of exons 71 and 86, was observed in some amplified fragments of cDNA.
Midasin has also been identified as an ortholog of YLR106p in the genome projects for Drosophila (gene CG13185; MM 605 kDa) and Arabidopsis (accession AAD10657; MM 583 kDa). In Caenorhabditis, there appears to be an unassembled midasin gene located on chromosome VIII (data not shown). However, the midasin sequences in these organisms have not yet been verified by experimental confirmation of the computer-predicted exon-intron boundaries and they are not used in this analysis.
Domain structure
Dot matrix plots comparing the amino acid sequence of midasin from different organisms reveal the presence of five major domains through an abrupt change in the level of sequence conservation at domain boundaries (Figs. 1, 2). A weakly conserved N-terminal domain is followed by a highly conserved AAA domain that contains six tandem AAA protomers. This AAA domain is connected by a large linker domain to a D/E-rich domain that has a conserved amino acid composition rich in aspartate and glutamate residues, but only moderately conserved sequence. The D/E-rich domain is followed by the COOH-terminal M-domain that is highly conserved and contains a set of MIDAS sequence motifs [9].
Figure 1.
Diagram of domain structure in midasin The values of molecular mass indicated are for midasin in humans. In other organisms, the principal difference is the length of the linker domain. In Giardia and Encephalitozoon, the N-terminal domain is truncated to ~25 residues. The mass of short regions of polypeptide joining the six protomers in the AAA domain (~10 kDa each) is not included in the values shown.
Figure 2.
Dot-matrix plots showing level of sequence conservation as a function of residue position in midasin The amino acid sequence of midasin in yeast, Saccharomyces cerevisiae (vertical) is compared with those in fission yeast, Schizosaccharomyces pombe (horizontal, left) and in humans (horizontal, right). Closely spaced dots forming a diagonal line indicate well-conserved regions of sequence. Horizontal dashed lines have been added to indicate the abrupt changes in conservation level that correspond to putative domain boundaries. The off-diagonal lines in the AAA domain correspond to sequence similarity between the six AAA protomers comprising this domain. The rectangular area containing numerous off-diagonal dots in the D/E-rich domain derives from the conserved high percentage of aspartate and glutamate residues in this domain, although the actual sequences are only moderately conserved. Note the short region of relatively well-conserved sequence near the middle of the otherwise less conserved linker domain. The matrices were generated with a stringency of 26% in a sliding window of 40 residues. Abbreviations: Sacce, Saccharomyces cerevisiae; pombe, Schizosaccharomyces pombe.
N-terminal domain
In most organisms, the region between the NH2-terminus of midasin and the beginning of the AAA domain comprises a weakly conserved domain of approximately 300 residues. However, in the compacted genome of Giardia [11,12]., the N-terminal domain is greatly truncated and consists of only about 25 residues. The N-terminal domain is the least conserved region of the midasin molecule (Table 1).
Table 1.
Amino Acid Conservation in Midasin Domains
| Domain | Yeast vs Human | Giardia vs Human | Encephalitozoon vs Human |
|---|---|---|---|
| (percent) | (percent) | (percent) | |
| N-terminal | 19–38 (14) | NA | NA |
| AAA (overall) | 41–57 (9) | 31–47 (10) | 27–42 (23) |
| AAA1 | 30–45 (10) | 29–49 (19) | 26–48 (29) |
| AAA2 | 43–61 (5) | 33–50 (10) | 28–50 (18) |
| AAA3 | 65–78 (0) | 50–68 (4) | 50–67 (4) |
| AAA4 | 53–67 (23) | 41–62 (12) | 47–65 (13) |
| AAA5 | 58–71 (1) | 49–67 (2) | 34–58 (8) |
| AAA6 | 33–51 (6) | 29–47 (12) | 27–45 (36) |
| Linker | 18–34 (30) | 17–33 (31) | 28–61 (57) |
| D/E-rich | 25–45 (11) | 18–39 (5) | 28–50 (58) |
| M-domain | 45–65 (3) | 32–53 (5) | 32–53 (25) |
| Overall | 29–46 (18) | 26–44 (14) | 18–29 (47) |
The first and second numbers in each cell give the percentages of identical and similar amino acids, respectively; the number in parenthesis gives the percentage of gaps in the alignment. All percentages were determined in pairwise alignments of the indicated midasin domain, as generated by the "Blast 2 Sequences" server at http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html with default parameters. In cases where the domains differed in length, identical and similar are calculated as percentages of the smaller domain; gap is calculated as a percentage of the larger domain. When necessary, dummy anchor sequences were added to NH2-termini and COOH-termini in order to force the server to align full length domains.
In many organisms, the N-terminal domain contains a cluster of basic residues; these include the sequences RWIKDSKKK (residues 75–83) in Saccharomyces, and RYGRRRMKLR (residues 137–146) in humans. However, the absence of such a basic cluster in the N-terminal domains of Giardia (residues 1–25) and Schizosaccharomyces pombe (data not shown) renders its significance questionable.
AAA domain
As first noted for the yeast sequence by Neuwald and coworkers [5], midasin is a member of the AAA ATPase family. The proteins of this family are unified by their sharing of a common structural organization that is based upon a conserved ATPase domain of ~225 residues referred to as an AAA protomer. The structure of an AAA protomer includes an ATP-binding site located in the cleft between a large α/β N-subdomain and a smaller all-α C-subdomain. In contrast to their shared structure, the AAA proteins participate in diverse cellular activities, including proteolysis, protein folding and unfolding, membrane trafficking, DNA replication, metal ion metabolism and intracellular motility [13-15]. Recent structural studies have revealed that the protomers of an AAA protein usually oligomerize into ring-shaped hexameric structures that constitute molecular platforms essential to their mode of action. In many cases, conformational changes occurring in the individual AAA protomers during an ATPase cycle function cooperatively to change the shape of the overall hexameric ring in a manner that exerts mechanical force on their protein or nucleic acid substrates. In most AAA proteins, the ring structure is formed by six identical polypeptide subunits, with each contributing a single AAA protomer [14]. However, the AAA motor dynein differs in having six distinct AAA protomers disposed in tandem on a single polypeptide subunit of unusually high molecular weight, and it is believed to form a unimolecular pseudo-hexameric AAA ring [5,6,16].
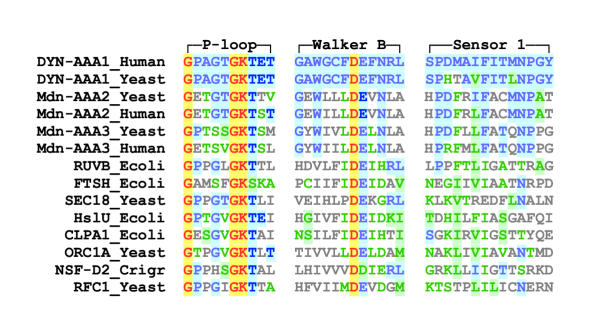
The AAA domain of midasin, like that of dynein, contains six tandem copies of the amino acid sequence motifs that characterize AAA protomers (Fig. 3; see also reference [5]). The Walker A motif GxxxxGK [T,S] and Walker B motif hhhhDExx (where h is any hydrophobic residue and x is any residue), that contain residues essential for ATP binding and hydrolysis in all P-type ATPases [17], are present in their canonical forms in the AAA2, AAA3, AAA4, and AAA5 protomers of midasin for all the organisms in Figure 3. The Walker A motif is present also in AAA1 and AAA6, although the Walker B motif in these protomers is deviant. The sensor 1 and sensor 2 motifs that are specific for members of the AAA ATPase family are present and highly conserved among organisms in AAA2, AAA3, AAA4 and AAA5, but are less conserved or absent in AAA1 and AAA6. It is notable that the functionally important Asn in sensor 1 and Arg in sensor 2 [15] are present and invariant among organisms in all protomers of midasin, except AAA1 and AAA6. These critical residues lie close to the γ-phosphate site in other AAA proteins with known structures and they are believed to trigger a change in the angle between the N- and C-subdomains of the protomer upon binding of ATP or dissociation of the γ-phosphate [18]. Indirect evidence suggests that such a conformational change in one protomer can be communicated to the adjacent protomers and result in a cooperative alteration in the shape of the overall hexameric ring [19].
Figure 3.
Sequence alignment of the six AAA protomers in midasin from humans, yeast and Giardia The alignment shows two aspects of sequence conservation: (1) the level of conservation for each single AAA protomer compared to the same protomer in other organisms; (2) the level of conservation between the six AAA protomers (AAA1, AAA2, AAA3, AAA4, AAA5, AAA6) compared one against another. The Motif line above the alignment indicates the sequence motifs that characterize members of the AAA ATPase family [5]. All AAA motifs occur in regions of relatively well-conserved sequence, with the exception of Box II (not shown) which is poorly conserved in midasin. Critical residues interacting with bound nucleotide in other AAA proteins are indicated by: ▲, residues in the Walker A and B motifs required for binding and hydrolysis of ATP [17]; ●, Asn in sensor 1 and Arg in sensor 2 believed to sense the γ-phosphate status [5]; ■, Asp and Arg in Boxes VI and VII that are believed to sense the γ-phosphate status in adjacent protomers of the hexameric structure [6,15,18]. The Structure line below the alignment indicates the secondary structural elements corresponding to the sequence motifs in other AAA ATPases whose structures have been resolved at the atomic level [5]. Colors indicate level of consensus for each residue position. In this context, consensus is defined as sequence conservation of a single AAA protomer in the three organisms analyzed; it does not refer to comparisons between different AAA protomers. Color key: red/yellow, invariant; blue/blue, majority consensus of identical residues; green/green, majority consensus of highly conserved residues; green/white, weakly conserved residues; gray/white, non-conserved residues. Organisms : HUMAN, Homo sapiens; SACCE: Sacchararomyces cerevisiae; GIARD, Giardia intestinalis.
In both midasin and dynein, the evolutionary fusion of the six AAA protomers into a single polypeptide has permitted the individual protomers in the hexameric assembly to acquire substantial structural and functional specialization. In dynein, this has included the acquisition of two substantial accessory structures that protrude asymmetrically from the hexameric AAA ring. Concomitant with this development of asymmetrical structure, the AAA1 protomer of the dynein motor unit evolved a functional dominance, in which it alone retains the full ability for binding and hydrolysis of ATP, while AAA2, AAA3 and AAA4 have lost the capability for hydrolysis and the most degenerate protomers AAA5 and AAA6 show no significant binding of ATP [20-22]. In midasin, the specialization of AAA protomers appears to have taken a less drastic course than in dynein. In all the available organisms, the four central protomers of the midasin polypeptide, AAA2, AAA3, AAA4 and AAA5, possess canonical Walker A and B motifs, as well as the critical Asn in sensor 1 and Arg in sensor 2 that are required for proper function in other AAA proteins [15]. The presence of these critical residues, taken together with their higher level of average sequence conservation (Table 1), suggests that the four central protomers all retain an active enzymatic function. Only in the two outer protomers, AAA1 and AAA6, do the AAA sequence motifs depart sufficiently from their canonical forms to suggest that the protomers containing them play a less active role in midasin function.
Phylogenetic analysis of the sequence differences between the six protomers in the AAA-domain of midasin (Fig. 4) shows that the evolutionary distance between any two protomers in a single organism is substantially greater than that between any single protomer taken from different organisms, even for such highly disparate organisms as humans and Giardia. This analysis also indicates that the odd-numbered protomers, AAA3 and AAA5, and the even numbered protomers, AAA2, and AAA4, form two separate groups in which, regardless of the organism they come from, the members in any one group are more closely related to each other than they are to any members of the other group.
Figure 4.
Tree of sequence relatedness for the six AAA protomers in midasin from humans, yeast and Giardia Asterisks indicate nodes supported with a bootstrap probability of 95% or better, plus signs indicate nodes with probability of 75–95%. The tree was calculated from the multiple alignment in Fig. 3.
Although this odd/even grouping is less obvious in the most divergent protomers, AAA1 and AAA6, it remains visible in such features as the extended loop inserted between the Box IV and Box IV' motifs of all the even-numbered protomers (Fig. 3). Taken together, these data support a phylogenetic model for midasin in which the hexa-protomeric structure of its AAA domain evolved through a trimeric assembly of pre-existing di-protomeric AAA polypeptides that had evolved previously. However, such primordial di-protomeric AAA polypeptides would have to have been simpler than those present in NSF, ClpA and p97 and other AAA proteins of the current era, for the latter assemble into two-tiered hexameric assemblies, with the axis joining the two protomers in each polypeptide oriented perpendicular to the plane of the hexameric ring [14]. We cannot, at present, exclude an alternative model for midasin structure in which the hexameric AAA ring assembly is a dimeric two-layered structure formed by two midasin polypeptides arranged laterally with all six odd-numbered protomers in one layer and all six even-numbered protomers in the other.
Linker domain
The COOH-end of the sixth protomer in the AAA domain is joined to the upstream end of the D/E-rich domain by a lengthy linker domain, ranging between 1700 and 2300 residues in most organisms. The sequence of this linker domain is moderately well conserved among organisms, with 13–19% identity and 26–34% similarity in pairwise alignments of the yeast sequence with those of human and Giardia (Table 1). The structure-prediction program PhD [23] suggests that the linker domain folds into a compact globular conformation, containing approximately 65% α-helix and less than 10% β-sheet. Screening of the translated non-redundant nucleotide database at GenBank with the sequence of the yeast linker domain detected the midasins in other organisms, but revealed no significant homology of this domain to other proteins.
The roughly constant length of the linker domain in most organisms, together with its moderate level of sequence conservation, suggests that it primarily fulfils a structural role in midasin. However, the presence of a short region of relatively well conserved sequence near the middle of the linker (Fig. 2) suggests a more active role for this region of the domain, perhaps acting as a hinge or as a binding site for other proteins.
D/E-rich domain
The D/E-rich domain of midasin comprises 430–630 residues located between the COOH-end of the linker domain and the NH2-end of the M-domain. It is characterized by having a highly acidic amino acid composition, with 35–40% of the residues being either aspartate or glutamate (Fig. 5). This composition gives the domain a predicted isoelectric point 3.7–4.0. The NH2-boundary of the D/E-rich domain is marked by a well-conserved glycine-rich motif GxGxGxGxG. The interior of the domain contains additional small clusters of glycines, but few other hydrophobic residues. The absence of any consistent pattern of heptad hydrophobic repeats indicates that there is little or no α-helical coiled-coil structure in the domain. The downstream boundary of the domain is usually marked by the presence of a small cluster of proline residues. Structure prediction with the program PhD [23,24]. suggests that the D/E-rich domain has a highly extended conformation containing about 30% α-helix and 70% coil.
Figure 5.
Distribution of aspartate and glutamate residues as a function of residue position in midasin The distribution is shown as the mole fraction of (aspartate + glutamate) in a sliding window of 31 residues.
The D/E-rich domain appears to be only weakly conserved, with ~20% identity in pairwise alignments of the sequence from different organisms (Table 1), although the biased amino acid composition often makes correct alignment ambiguous. Screening of the translated non-redundant database at GenBank with the amino acid sequence of the yeast D/E-rich domain selected numerous other aspartate-glutamate rich proteins, including neurofilament proteins and caldesmon. However, scrutiny of the putative alignments indicated that most or all were based upon fortuitous fitting of the abundant acidic residues.
M-domain
The M-domain of midasin is a highly conserved domain of ~280 residues (30 kDa) located at the COOH-terminus of the protein. The domain contains a full set of MIDAS sequence motifs consisting of hhhhDxSxS, followed after ~70 residues by a conserved threonine, followed after a further ~30 residues by hhhh[S,T]DG, where h is any hydrophobic residue and x is any residue (Fig. 6). The best-studied examples of MIDAS-containing domains in other proteins are the I-domains of integrin α-chains-α1, α2, α10, and α11, and the A-domains of von Willebrand factor [9,25-27]. In proteins whose structure has been determined to atomic resolution, the MIDAS-containing domain has a classic Rossman-fold, with a central hydrophobic β sheet flanked on both sides by amphipathic helices. The residues in the MIDAS motifs lie on three closely apposed loops located on the upper edge of the β-sheet, where they form the metal-binding site, with oxygen atoms in the aspartate, serine and threonine residues coordinating the metal ion [9]. In integrin α2β1, which is a collagen receptor, the metal-binding site also binds the collagen ligand through the conserved glutamate in a GFOGER motif (O = hydroxyproline) completing the coordination sphere of the metal [28]. In vivo, the binding of collagen at this site appears to be regulated through a conformational shift in which the loops forming the MIDAS site change from a closed to an open conformation [29-31]. Other proteins containing a domain with all three MIDAS motifs include the D-subunit of magnesium chelatase [32], Ca-activated chloride-channel protein [33,34], nitrate reductase (accession AAC79447), and the D subunit of nitric oxide reductase (accession AAC45374). The ability of a MIDAS protein to bind its protein ligand is tightly linked with the presence of intact MIDAS motifs: mutations to the metal-coordinating residues in these motifs, weaken or eliminate ligand binding [35,36]. An additional set of MIDAS-like proteins, that includes the integrin β-chain [37,38] and the von Willebrand Factor A domain [27], lacks one of the three motifs and appears to bind ligands in a somewhat different manner [27,39,40]. In midasin, the presence of all three MIDAS motifs in the M-domain, with the putative metal ion-binding residues in the motifs invariant in the available organisms, indicates that midasin belongs to the family of MIDAS-containing proteins.
Figure 6.
Multiple sequence alignment of midasin M-domains with selected magnesium chelatases and integrin I-domains The conserved residues in the MIDAS sequence motifs (hhhhDxSxS, [S/T] and hhhh[S/T]DG) are indicated by ▲ under the alignment. In all midasins and chelatases, the MIDAS-containing domain is located at the COOH-terminus of the protein (asterisk). The Structure line below the alignment indicates the position of secondary structural elements in the I-domain of integrin α2 (pdb: 1AOX_A). The coloring indicates the level of consensus for each residue position in the sets of three midasins, three chelatases, and three integrins, with each set being considered separately. Color key: red/yellow, invariant; blue/blue, majority of identical residues; green/green, majority of highly conserved residues; green/white, weakly conserved residues; gray/white, non-conserved residues. Abbreviations: MIDA, midasin; YEAST, Saccharomyces cerevisiae; GIARD, Giardia intestinalis; BCHD_RHOCA, D-subunit of Mg chelatase from Rhodobacter capsulatus (accession P26157); BCHD_CHLFL, D-subunit of Mg chelatase from Chloroflexus aurantiacus (accession AAG15217); CHLD_ARATH, D-subunit of Mg chelatase from Arabidopsis thaliana (accession Q9SJE1). ITAL, I-domain of integrin αL (accession P20701). ITAM, I-domain of integrin αM (accession P11215); ITA2, I-domain of integrin α2 (accession P17301). All residue numbers indicate position in full-length subunits.
In addition to this conserved framework, the midasin M-domain possesses an extension of ~75 residues at the NH2-end of the hhhhDxSxS motif. The sequence in this NH2 extension is highly conserved, with 46% of the residues invariant among midasins from the available organisms. Since this region of well-conserved sequence continues unbroken between the NH2-extension and the hhhhDxSxS motif, the NH2-extension and the carboxyl region of the M-domain probably form two parts of a single structural domain. The structure prediction program PhD [23,24] suggests that the COOH region of the M-domain has an α/β conformation, similar to that expected from its sequence homology to the integrin I-domain, whereas the NH2-extension is predicted to be largely α-helical. Although the sequence of the midasin NH2-extension shows no statistically significant similarity to other proteins in the GenBank database, its high density of basic and hydrophobic residues generally resembles that in the equivalent region of the magnesium chelatases (Fig. 6), suggesting that the two proteins share a common fold in this region.
The highly conserved sequence in the midasin M-domain indicates that it plays a critical role in midasin function. One possibility is that the MIDAS site serves to attach protein ligands through a mechanism involving participation of a glutamate or aspartate residue on the ligand in the coordination of a Mg2+ ion at the MIDAS site of midasin, in a manner similar to that by which the MIDAS site in integrin I-domains appears to mediate attachment of collagen through the glutamate in the GFOGER binding motif [36]. Interestingly, the related RGE ligand-binding motif [41] occurs in a conserved region of the midasin AAA-domain (yeast, residues 1835–1838), as well as in the AAA-domain of Mg chelatases [32]. The presence of this consensus binding motif in the AAA domain raises the possibility of the midasin molecule folding back onto itself, with the M-domain becoming attached to one face of the AAA domain and perhaps regulating access to its central chamber, in a manner analogous to that in which the 19S proteasome regulator, also an AAA protein, controls access to the central proteolytic chamber of the proteasome [42].
A compacted midasin in Encephalitozoon caniculi
The parasitic microsporidean Encephalitozoon caniculi has been under severe and sustained evolutionary pressure to reduce the size of its genome, which is the smallest of any sequenced eukaryote, with an overall length of only 3 Mbp [43]. One result of this pressure has been to compact many of the essential genes that the organism could not afford to eliminate [44].
We have examined the sequence of midasin in Encephalitozoon as one approach to probing why midasin is so large and to identifying which regions of its domains are essential to their function. Midasin in Encephalitozoon is encoded by a gene of 8496 bp, corresponding to a protein of 2832 residues (accession CAD26493) with a predicted molecular mass of 324 kDa. This corresponds to an overall 42% reduction compared to that of yeast midasin. Sequence alignment of Encephalitozoon midasin with that from the other available organisms shows that the different domains of the protein have been affected to conspicuously different extents by this compaction (See Additional File 1: FullAlignment). The N-terminal domain is reduced by 90% to ~25 residues, indicating that much of this domain plays a non-essential role. However, the abbreviated domain retains a cluster of basic residues (KFKKHKK, residues 2–8), similar to those present in this domain of humans and yeast. The AAA domain has been affected relatively little: all six AAA protomers are retained and the overall length of the domain is reduced by only ~19%. Most of this reduction involves a shortening of the lengthy loops between the Walker A and Walker B motifs of even-numbered protomers in normal organisms (Fig. 3). On the other hand, the linker domain, which is believed to serve a structural function, is reduced by more than 50% to 537 residues, indicating that it has undergone major reorganization through compaction. The D/E-rich domain is reduced in size by ~50%, while retaining its acidic character with a high percentage of aspartate and glutamate residues. The M-domain is reduced by ~20%, mostly through loss of a 40 residue section of the less conserved region immediately upstream of the hhhh [S,T]DG motif. All the MIDAS sequence motifs in this domain are retained and most of the residues in the highly conserved NH2-extension of the M-domain are unaffected.
Although all domains in midasin have yielded something to the pressure for gene compaction during evolution of Encephalitozoon, it is the N-terminal, linker and D/E-rich domains, in which the sequence is relatively less conserved among organisms, that have yielded the most. The essentially complete retention of the M-domain and the hexameric assembly of protomers in the AAA domain emphasizes the critical importance of these domains to the proper functioning of midasin.
Relationship of midasin to other AAA proteins
The early evolutionary divergence of the different major branches of the AAA family makes it difficult to evaluate the phylogenetic relationships among them [45]. However, by restricting the analysis to residues in the more highly conserved regions of the AAA structure that interact directly with ATP, we have been able to improve the signal to noise ratio sufficiently to probe the phylogenetic relationship of midasin and dynein to representative members in other branches of the AAA family. Such analysis of the AAA protomers that are best conserved among organisms (AAA2, AAA3, AAA4 and AAA5 in midasin, and AAA1 in dynein) shows that the AAA protomers of midasin and dynein are substantially more closely related to each other than they are to those in the other branches of the AAA family examined (Figs 7, 8). This result supports the view that midasin and dynein evolved from a common AAA ancestral protein that had already developed a subunit structure of six AAA protomers in a single polypeptide. However, insufficient information exists to relate this last common ancestor to any particular other branch of the presently existing AAA family.
Figure 7.
Sequence alignment of the AAA protomers in midasin with those in other branches of AAA-ATPase family The alignment is limited to residues contained in the P-loop, Walker B, and sensor 1 motifs of AAA2 and AAA3 protomers of midasin from humans and yeast, together with the corresponding residues in the AAA1 protomer of cytoplasmic dynein [6], and in a variety of other AAA proteins. The positions and lengths of the P-loop, Walker B, and sensor 1 motifs are taken from the review of Neuwald et al [5]. The color used for each residue in the alignment indicates its similarity to that present at the corresponding position in yeast cytoplasmic dynein. Color key: Yellow/red, all AAA proteins shown have identical residue; blue/blue, identical to AAA1 of dynein; green/green, closely similar to AAA1 of dynein; green/white, moderately similar; gray/white, not similar.
Figure 8.
Tree of sequence relatedness of AAA protomers in midasin to those in other branches of AAA family The numbers beside the nodes in the tree represent the statistical significance of the node, assayed by the number of times the identical node appeared in 1000 bootstrap trials [57]. Nodes lacking a number are less than 90% significant. The tree is calculated for the alignment in Fig. 7.
Apart from the AAA domain, the other domains of midasin appear to show little or no relationship to dynein. The polypeptide joining the adjacent AAA protomers in midasin is approximately 100 residues long between all protomer pairs. There is no indication that any of these joining polypeptides contains a ATP-sensitive microtubule-binding site resembling that located between the AAA4 and AAA5 protomers of dynein [46]. The position of the N-terminal domain of midasin relative to the AAA domain is similar to that of the stem domain of dynein, and in several organisms the midasin N-terminal domain contains a cluster of basic residues that has the potential to bind microtubules in an ATP-insensitive manner. However, the major truncation of the midasin N-terminal domain in Giardia (Fig. 3) and Encephalitozoon, together with the absence of a cluster of basic residues in the N-terminal of Schizosaccharomyces pombe makes the domain of questionable functional significance.
Expression and localization of midasin in yeast
In order to verify the expression of the midasin gene as a single polypeptide and to make a preliminary study of its function, we modified the chromosomal MDN1 gene in a diploid strain of yeast by adding an oligonucleotide encoding a hemagglutamin (HA) epitope tag to the 3'-end of one copy of the gene. Subsequent sporulation and tetrad dissection yielded 4 viable spores from most tetrads, indicating that the HA-tagged midasin is able to function and maintain viability in a haploid strain. Preliminary characterization of the haploid cultures at temperatures ranging from 14°C to 37°C indicates that strains with the modified MDN1(HA) gene grow somewhat more slowly than those with the native MDN1(wt) gene, but appear otherwise normal.
Western blots obtained after electrophoresis of a crude homogenate of yeast containing the MDN1(HA) gene demonstrated the presence of midasin as a single immunostained band with a molecular weight of greater than 500 kDa (Fig. 9A); there was no comparable immunostained band in homogenates of yeast with the MDN1(wt) gene. Differential centrifugation of the homogenate showed that the midasin remained in the supernatant fraction upon centrifugation at 2,000 × g for 2 min, but mostly passed into the pellet fraction upon centrifugation at 18,000 × g for 20 min. Attempts to solubilize the midasin under relatively mild conditions by extracting the 18,000 × g pellet with 0.6 M NaCl, 0.1 M Na2CO3, or 1% Triton X100 were unsuccessful (data not shown). However, the midasin did pass into solution upon extraction of the 18,000 × g pellet with 8 M urea and it then remained in the supernatant fraction after centrifugation at 150,000 × g for 20 min (Fig. 9B).
Figure 9.
Electrophoresis gel showing expression of HA-tagged midasin in haploid yeast Preparations made from wild type yeast (wt) and from yeast with a HA epitope tag on the MDN1 gene (HA). Gel A: S0, supernatant fraction after homogenizing yeast with glass beads and centrifuging at 2000 × g for 2 min; S1, P1, supernatant and pellet fractions after centrifugation of S0 fraction at 18,000 × g for 20 min. Gel B: S2, P2, supernatant and pellet fractions obtained by resuspending P1 fraction in 8 M urea and centrifuging at 150,000 × g for 20 min. The midasin polypeptide (arrow) migrates somewhat more slowly than the dynein heavy chain (dyn) used as a molecular mass standard of ~525 kDa. The 45 kDa band in the 8 M urea supernatant is a cross-reacting protein present in wild type yeast. The 3–8% polyacrylamide gels were electrophoresed in the presence of 0.1% Na dodecyl SO4 and blotted onto Immobilon membrane. The blot was stained with monoclonal antibody against the HA epitope. The molecular mass standards were run in a noncontiguous lane of gel A.
The localization of midasin in yeast was examined by fluorescence microscopy of immunostained cells from exponentially growing cultures of MDN1(HA) yeast, with comparable immunostained cells from parallel cultures of MDN1(wt) yeast used as controls. The results demonstrated that the tagged midasin localizes predominantly to the nucleus (Fig. 10). The midasin signal covered a slightly larger area than the DAPI signal, and sometimes appeared granular or punctate. In most cells, this additional stained area appears nearly symmetrically disposed around the DNA (Fig. 10a). However, in cells that appeared to have recently divided, a "comet tail" of immunostained material was frequently visible trailing behind the separated nuclei (Fig. 10b,10c), This "tail" possibly corresponds to the finger of nuclear envelope and matrix that formerly enclosed the anaphase spindle [47,48].
Figure 10.
Light microscopy of immunostained yeast cellsA: cells in interphase and during division; B, C, selected cells in which the region staining for midasin is larger (arrows) than the region staining for DNA. "Comet tails" are conspicuous in several of these cells, with short tails remaining post-division into early interphase. All fields show haploid yeast containing the MDN1(HA) gene double-stained with HA-antibody and DAPI. D, differential interference images; M, localization of HA-tagged midasin; P, localization of DNA, as stained by DAPI. All antibody-stained and DAPI fields are matched levels taken from through-focal series with steps of 0.3 μm.
The localization of midasin to the nucleus in yeast, together with the difficulty in solubulizing it under moderately dissociating conditions, suggests that midasin occurs primarily in association with one of the cytoskeletal assemblies that constitute the nuclear matrix and the nuclear pore complex. Consistent with the lack of a consensus hydrophobic segment in its sequence, the solubility of midasin in 8 M urea indicates that it is not an intrinsic membrane protein.
When the amino acid sequence of yeast midasin is screened with the PredictNLS server at http://cubic.bioc.columbia.edu, it reveals two potential nuclear localization signals. One is the cluster of basic residues KKKKRR (residues 768–773), located in a relatively non-conserved region of the AAA domain between AAA3 and AAA4. The second is the highly conserved sequence RKDKIWLRRTKPSKRQ (residues 4687–4702) located in the NH2-extension of the M-domain, immediately upstream from the first MIDAS motif.
General discussion
Stripped to its fundamentals, the structure of midasin consists of a pseudo-hexameric assembly of AAA protomers associated with a MIDAS-containing M-domain. This basic domain organization shows a striking parallel to that of magnesium chelatase, a heterotrimeric enzyme containing BchD, BchI and BchH subunits, that performs ATP-dependent insertion of Mg2+ into the protoporphyrin IX ring in the course of chlorophyll biosynthesis [49]. In particular, the BchD subunit resembles midasin in possessing a single AAA protomer close to its NH2-terminus, together with a short aspartate-glutamate-rich region and a MIDAS-containing domain at its carboxy-terminus. The BchI subunit also possesses a single AAA protomer, but contains no MIDAS domain [32]. The third subunit BchH is able to bind the protoporphyrin ring in either the presence or absence of ATP. The initial step of the insertion can proceed in the presence of either Mg.ATP or a non-hydrolyzable ATP-analog and involves oligomerization of the BchD and BchI subunits to form an oligomeric ring of AAA protomers that resembles the ring structure of NSF and other AAA proteins [14,32]. The second step of the insertion involves an obligatory hydrolysis of ATP that is tightly coupled with the transfer of the chelated Mg2+ to the protoporphyrin ring by BchH [49]. Although the details differ from midasin, particularly with respect to the subunit composition of the AAA ring, this chelation reaction provides a structural model suggesting that the function of the midasin M-domain may be to regulate the ATPase activity of the AAA protomers in the pseudo-hexameric ring and thus couple ATP hydrolysis to the binding of a protein ligand at the MIDAS site.
Studies of the yeast proteome have the potential to clarify the function of midasin by identifying the protein partners with which it interacts in vivo. In a recent large-scale screen with 1739 yeast proteins used as bait [3], midasin (YLR106p) was identified in the polypeptide sets that copurified with four bait proteins, ESS1p, RPT1p, YML029p and HRT1p, that function principally in RNA metabolism and in the regulation of proteasomes (which are mostly intranuclear in yeast [50]). However, the interpretation of these proteomic data is clouded by the fact that no protein partners were identified in the complementary experiment in which midasin was itself used as bait. Substoichiometric amounts of midasin have also been detected in isolated preparations of 60S preribosomal particles suggesting that it may play a role in maturation of 60S ribosome subunits [2]. Such a role for midasin as a nuclear chaperone involved in the assembly/disassembly of macromolecular complexes in the nucleus would be consistent with our localization data and with the known functions of other AAA proteins. As part of this role, the function of the highly extended D/E-rich domain in midasin could be to interact with positively-charged nuclear protein substrates in a manner analogous to the acidic regions of the nuclear transport GTPase Ran [51], the nuclear transporter Tpr [52], and the chromatin remodeling proteins of the SWI/SNF family [53]. However, there are alternative possibilities that need to be considered.
At least part of the present dearth of direct information linking midasin to a specific cell function seems likely to be a consequence of its unusually high molecular weight. Many libraries used in genetic complementation screens do not contain inserts as large as 15 kb and so would be unable to detect midasin. Adequate migration of the 560 kDa midasin polypeptide on electrophoresis gels requires use of lower percentage gels than standard, especially if blotting is involved. Without such special handling, midasin may have been undetected in some cell fractionation experiments. However, the recent increased availability of high sensitivity mass-spectrometry for peptide identification greatly lessens the difficulty of detecting midasin in semi-purified cell fractions. It is to be expected that more detailed information about its function will be available shortly.
Conclusions
The highly conserved structure of midasin in eukaryotes, taken in conjunction with its nuclear localization in yeast, suggests that midasin may function as a nuclear chaperone and be involved in the assembly/disassembly of macromolecular complexes in the nucleus. However, other possibilities remain to be evaluated. The AAA domain of midasin is evolutionarily related to that of dynein, but it appears to lack a microtubule-binding site.
Materials and methods
Sequence determination
The sequence of the human midasin gene has been determined from PCR-amplified fragments of cDNA from human testis (Clontech). We used regions of amino acid sequence conserved between the midasin genes in yeast and Schizosaccharomyces pombe to make a rough map of the gene onto the August 2001 freeze of the assembled human genome with the public domain Blat server at http://genome.ucsd.edu/index.html [54,55]. The resultant partial nucleotide sequence identified about four-fifths of the exons and was used to design the requisite PCR primers to verify all exon-intron boundaries in the gene by physical sequencing of appropriate PCR-amplified cDNA fragments.
The midasin gene in Giardia intestinalis (accession AF494287) was cloned in silico from sets 1–11 of the unassembled genomic nucleotide sequences kindly made available by the Giardia Genome Project [11]. Regions of conserved amino acid sequence in the human and yeast genes for midasin were used as input to the Tblastn server at Genbank [56] to identify clones that encoded homologous regions in Giardia. These starting clones were extended by repeated cycles of searching to obtain neighboring, overlapping clones until the complete coding sequence of midasin was included. The resultant sequence was supported by double stranded raw data with a depth of four-fold or greater over approximately 85% of the gene. The sequence of the remaining regions was verified by physical sequencing of PCR amplified fragments of genomic DNA.
Sequence alignment
The overall alignment of the six AAA protomers in midasin from human, yeast and Giardia was created by first making separate alignment of the three organisms for each of the six AAA protomers. The resultant six partial alignments were then combined by using with the profile alignment procedure of ClustalX [57] with the Blosum100 scoring matrix. The overall alignment of the midasin M-domain from humans, yeast and Giardia with other MIDAS-containing proteins (three Mg-chelatases and the I-domains of three integrins) was created by first aligning the midasins, Mg-chelatases and integrins separately with the program T-Coffee [58]. The resultant three partial alignments were then combined by the profile alignment procedure of ClustalX, as above.
Tagging of the midasin gene in yeast
Diploid strain DDY1102 of Saccharomyces cerevisiae with genotype Mat a/α his3Δ200/his3Δ200 ura3-52/ura3-52 leu2-3,112/leu2-3,113 ade2-1/ADE2 LYS2/lys2-80 MDN1/MDN1 [59] was used. The 3'-end of the coding region of the midasin gene was fused to a tag encoding 3× hemagglutamin (HA) by using the PCR-mediated cassette procedure of Longtine and colleagues [60]. The PCR primers used to generate the cassette were 5'-ACTGATTTTG CGTCAATACT TTACAGACCT GGCATCCAGC CGGATCCCCG GGTTAATTAA-3' and 5'-TCGTGTAGTA AACCTCCTCT TCTTGGTTTT CACGATATAC GAATTCGAGC TCGTTTAAAC-3'. The site of subsequent integration by homologous recombination was verified by PCR amplification with one primer located in the transformation cassette and the second primer located in the midasin gene. The resultant MDN1(HA)/MDN1 diploid strain was sporulated and the tetrads dissected. Haploid MDN1(HA) clones for subsequent work were derived from tetrads that had yielded 4 viable spores with the anticipated mixture of genotypes.
In order to confirm the yeast database entry indicating that the MDN1 (YLR106C) gene is essential for viability in yeast, we performed tetrad analysis of the heterozygous midasin knock-out strain CMEY072(HE) (Mat a/α ura3-52/ura3-52 his3D1/his3D1 leu2-3_112/leu2-3_112 trp1-289/trp1-289; MDN1(4,14727)::kanMX4/MDN1) obtained from EuroScarf. PCR was used to verify that one copy of the midasin gene had been deleted and replaced with the selectable marker. After sporulation and tetrad dissection, all 15 of the 15 tetrads dissected yielded 2 viable spores. In a parallel analysis of the wild type strain (CEN.PK2), dissection yielded 9 tetrads with 4 viable spores, 1 tetrad with 3 viable spores and 1 tetrad with 2 viable spores: average germination 93%. These data confirm the database entry that the MDN1 gene is required for viability in Saccharomyces.
Western Blots
Cultures of the haploid yeast containing MDN1(HA) or MDN1 were harvested in log phase. The cells were washed once with buffer containing 50 mM Tris-HCl, pH7.5, 100 mM NaCl, 1 mM EGTA, and 4 mM MgSO4. Cells were then resuspended in 0.6 ml of the same buffer plus 1% β-mercaptoethanol, 0.5 mM phenyl methyl sulfonyl fluoride, 50 μg/ml leupeptin, 4 μg/ml aprotinin, and 15 μl/ml of protease inhibitor cocktail P8215 (Sigma). After the cells had been homogenized by vortexing with glass beads for 2.5 min at 4°C, they were centrifuged for 2 min at 1000 × g to remove the glass beads and large cell debris. The resultant supernatant was then centrifuged for 20 min at 18,000 × g.
Protein samples were subjected to electrophoresis on 3–8% gradient polyacrylamide gels (Novex) in Tris-acetate buffer containing 0.1% Na dodecyl SO4, according to the manufacturer's directions. The electrophoresed samples were transferred onto ImmobilonP membranes (Millipore) by electroblotting for 1 hr in buffer containing 25 mM Bicine, 25 mM Bis-Tris, 1 mM EDTA, 10% methanol and 0.0025% Na dodecyl SO4. Prestained molecular mass markers (161–0372, BioRad) were supplemented with axonemal dynein from cilia of Tetrahymena (MM 528 kDa) in order to obtain a standard of sufficiently high molecular mass.
Blots were probed with monoclonal antibody against the HA tag (H9658, Sigma) diluted 1:11,000 in PBSB (0.14 M NaCl, 1% bovine serum albumin, 0.05% Tween-20, 10 mM phosphate buffer, pH 7.4), for 1–2 hrs at room temperature. Blots were then incubated with anti-mouse peroxidase-linked secondary antibody (170–6516, BioRad) for 1 hr at a dilution of 1:4,000 in PBSB.
Immunocytochemistry
Haploid yeast was grown to log phase and fixed with 4% formaldehyde for 1 hr. Cells were harvested by centrifugation, and washed twice with buffer containing 40 mM potassium phosphate, pH 7.5, 0.5 mM MgCl2, and 1.2 M sorbitol. Cell walls were digested for 20–30 min at 29°C with 50 μg/ml Zymolyase 100 T in the same buffer with the addition of 2 μl/ml β-mercaptoethanol. Cells were attached to poly-lysine coated slides and dehydrated with methanol at -20°C for 6 min, followed by acetone for approximately 30 sec at the same temperature. After blocking in PBSB, the cells were incubated with anti-HA antibody (H9658, Sigma) at 1:900 in PBSB for 1 hr, washed 3–4 times in PBSB buffer, then incubated with anti-mouse antibody linked to Texas Red (Molecular Probes) at 1:600 in PBSB for 1 hr. The cells were then washed 3–4 times in PBSB and mounted in Gel/Mount (biomeda) containing 4',6 diamidino-2-phenylindole (DAPI) at 1:10,000, under cover slips. Cells were visualized on a Zeiss Axiovert S100 microscope with a 63X objective. Data were analyzed using Openlab software (Improvision).
In all preparations, control samples of haploid yeast containing the MDN1(wt) gene were processed identically in order to confirm the specificity of antibody staining. In some cases, samples of MDN(HA) yeast were processed with the primary HA antibody omitted as an additional control.
Authors' contributions
JEG performed the experimental work on yeast and performed or supervised the nucleotide sequencing. IRG performed the sequence alignment and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
FullAlignment
Contributor Information
Joan E Garbarino, Email: joangarb@uclink4.berkeley.edu.
I R Gibbons, Email: igibbons@socrates.berkeley.edu.
Acknowledgements
We thank Amy Corsa for assistance with the nucleotide sequencing, Keith Kozminski, Ching Shang, Saturo Uzawa, Meredith Johnson and Avital Rodal for materials and much help with the yeast work, and Scott Dawson for Giardia DNA. We are grateful to Zac Cande, J. Richard McIntosh and Karsten Weis for helpful discussion. IRG is grateful to Beth Burnside for accommodating him within her laboratory space. This work has been supported by research grant GM30401 from the National Institute of General Medical Sciences.
References
- Winzeler EA. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;3:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Bassler J. et al. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell. 2001;3:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- Gavin AC. et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;3:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Ho Y. et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;3:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;3:27–43. [PubMed] [Google Scholar]
- Mocz G, Gibbons IR. Model for the motor component of dynein heavy chain based on homology to the AAA family of oligomeric ATPases. Structure (Camb) 2001;3:93–103. doi: 10.1016/S0969-2126(00)00557-8. [DOI] [PubMed] [Google Scholar]
- Nagase T. et al. Prediction of the coding sequences of unidentified human genes. VII. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1997;3:141–150. doi: 10.1093/dnares/4.2.141. [DOI] [PubMed] [Google Scholar]
- Kikuno R, Nagase T, Waki M, Ohara O. HUGE: a database for human large proteins identified in the Kazusa cDNA sequencing project. Nucleic Acids Res. 2002;3:166–168. doi: 10.1093/nar/30.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Rieu P, Arnaout MA, Liddington R. Crystal structure of the A domain from the α subunit of integrin CR3 (CD11b/CD18). Cell. 1995;3:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Evolution of the integrin alpha and beta protein families. J Mol Evol. 2001;3:63–72. doi: 10.1007/s002390010134. [DOI] [PubMed] [Google Scholar]
- McArthur AG. et al. The Giardia genome project database. FEMS Microbiol Lett. 2000;3:271–273. doi: 10.1016/S0378-1097(00)00299-8. [DOI] [PubMed] [Google Scholar]
- Iwabe N, Miyata T. Overlapping genes in parasitic protist Giardia lamblia. Gene. 2001;3:163–167. doi: 10.1016/S0378-1119(01)00767-3. [DOI] [PubMed] [Google Scholar]
- Patel S, Latterich M. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 1998;3:65–71. doi: 10.1016/S0962-8924(97)01212-9. [DOI] [PubMed] [Google Scholar]
- Vale RD. AAA proteins. Lords of the ring. J. 2000;3:F13–F20. doi: 10.1083/jcb.150.1.F13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure – diverse function. Genes Cells. 2001;3:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- King SM. AAA domains and organization of the dynein motor unit. J. 2000;3:2521–2526. doi: 10.1242/jcs.113.14.2521. [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;3:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva EA, May AP, He P, Whiteheart SW. Uncoupling the ATPase activity of the N-ethylmaleimide sensitive factor (NSF) from 20S complex disassembly. Biochemistry. 2002;3:530–536. doi: 10.1021/bi015632s. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, Chung CH. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure (Camb) 2001;3:1107–1116. doi: 10.1016/S0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- Gibbons IR, Lee-Eiford A, Mocz G, Phillipson CA, Tang WJ, Gibbons BH. Photosensitized cleavage of dynein heavy chains. Cleavage at the "V1 site" by irradiation at 365 nm in the presence of ATP and vanadate. J Biol Chem. 1987;3:2780–2786. [PubMed] [Google Scholar]
- Mocz G, Gibbons IR. Phase partition analysis of nucleotide binding to axonemal dynein. Biochemistry. 1996;3:9204–9211. doi: 10.1021/bi960662u. [DOI] [PubMed] [Google Scholar]
- Mocz G, Helms MK, Jameson DM, Gibbons IR. Probing the nucleotide binding sites of axonemal dynein with the fluorescent nucleotide analogue 2'(3')-O-(-N-Methylanthraniloyl)-adenosine 5'-triphosphate. Biochemistry. 1998;3:9862–9869. doi: 10.1021/bi9730184. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;3:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Conservation and prediction of solvent accessibility in protein families. Proteins. 1994;3:216–226. doi: 10.1002/prot.340200303. [DOI] [PubMed] [Google Scholar]
- Leitinger B, Hogg N. Integrin I domains and their function. Biochem Soc Trans. 1999;3:826–832. doi: 10.1042/bst0270826. [DOI] [PubMed] [Google Scholar]
- Celikel R, Varughese KI, Madhusudan, Yoshioka A, Ware J, Ruggeri ZM. Crystal structure of the von Willebrand factor A1 domain in complex with the function blocking NMC-4 Fab. Nat Struct Biol. 1998;3:189–194. doi: 10.1038/nsb0398-189. [DOI] [PubMed] [Google Scholar]
- Bienkowska J, Cruz M, Atiemo A, Handin R, Liddington R. The von Willebrand factor A3 domain does not contain a metal ion-dependent adhesion site motif. J Biol Chem. 1997;3:25162–25167. doi: 10.1074/jbc.272.40.25162. [DOI] [PubMed] [Google Scholar]
- Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Structural basis of collagen recognition by integrin α2β1. Cell. 2000;3:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- Lee JO, Bankston LA, Arnaout MA, Liddington RC. Two conformations of the integrin A-domain (I-domain): a pathway for activation? Structure. 1995;3:1333–1340. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- Lupher ML Jr, Harris EA, Beals CR, Sui LM, Liddington RC, Staunton DE. Cellular activation of leukocyte function-associated antigen-1 and its affinity are regulated at the I domain allosteric site. J Immunol. 2001;3:1431–1439. doi: 10.4049/jimmunol.167.3.1431. [DOI] [PubMed] [Google Scholar]
- Xiong JP, Li R, Essafi M, Stehle T, Arnaout MA. An isoleucine-based allosteric switch controls affinity and shape shifting in integrin CD11b A-domain. J Biol Chem. 2000;3:38762–38767. doi: 10.1074/jbc.C000563200. [DOI] [PubMed] [Google Scholar]
- Fodje MN, Hansson A, Hansson M, Olsen JG, Gough S, Willows RD, Al-Karadaghi S. Interplay between an AAA module and an integrin I domain may regulate the function of magnesium chelatase. J Mol Biol. 2001;3:111–122. doi: 10.1006/jmbi.2001.4834. [DOI] [PubMed] [Google Scholar]
- Gandhi R, Elble RC, Gruber AD, Schreur KD, Ji HL, Fuller CM, Pauli BU. Molecular and functional characterization of a calcium-sensitive chloride channel from mouse lung. J Biol Chem. 1998;3:32096–32101. doi: 10.1074/jbc.273.48.32096. [DOI] [PubMed] [Google Scholar]
- Fuller CM, Ji HL, Tousson AM, Elble RC, Pauli BU, Benos DJ. Ca(2+)-activated Cl(-) channels: a newly emerging anion transport family. Pflugers Arch. 2001;3:S107–S110. doi: 10.1007/s004240100655. [DOI] [PubMed] [Google Scholar]
- Kamata T, Liddington RC, Takada Y. Interaction between collagen and the α(2) I-domain of integrin α(2)α(1). Critical role of conserved residues in the metal ion-dependent adhesion site (MIDAS) region. J Biol Chem. 1999;3:32108–32111. doi: 10.1074/jbc.274.45.32108. [DOI] [PubMed] [Google Scholar]
- Smith C, Estavillo D, Emsley J, Bankston LA, Liddington RC, Cruz MA. Mapping the collagen-binding site in the I domain of the glycoprotein Ia/IIa (integrin α(2)β(1)). J Biol Chem. 2000;3:4205–4209. doi: 10.1074/jbc.275.6.4205. [DOI] [PubMed] [Google Scholar]
- Xiong YM, Zhang L. Structure-function of the putative I-domain within the integrin β2 subunit. J Biol Chem. 2001;3:19340–19349. doi: 10.1074/jbc.M008903200. [DOI] [PubMed] [Google Scholar]
- Xiong JP. et al. Crystal structure of the extracellular segment of integrin αVβ3. Science. 2001;3:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EC, Ratnikov BI, Tsai PM, Gonzalez ER, McDonald S, Pelletier AJ, Smith JW. Evidence that the integrin β3 and β5 subunits contain a metal ion-dependent adhesion site-like motif but lack an I domain. J Biol Chem. 1997;3:14236–14243. doi: 10.1074/jbc.272.22.14236. [DOI] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science. 2002;3:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;3:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- Gorbea C, Taillandier D, Rechsteiner M. Assembly of the regulatory complex of the 26S proteasome. Mol Biol Rep. 1999;3:15–19. doi: 10.1023/A:1006957802028. [DOI] [PubMed] [Google Scholar]
- Biderre C, Pages M, Metenier G, Canning EU, Vivares CP. Evidence for the smallest nuclear genome (2.9 Mb) in the microsporidium Encephalitozoon cuniculi. Mol Biochem Parasitol. 1995;3:229–231. doi: 10.1016/0166-6851(95)02495-6. [DOI] [PubMed] [Google Scholar]
- Katinka MD. et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;3:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Frohlich KU. An AAA family tree. J Cell Sci. 2001;3:1601–1602. doi: 10.1242/jcs.114.9.1601. [DOI] [PubMed] [Google Scholar]
- Koonce MP, Tikhonenko I. Functional elements within the dynein microtubule-binding domain. Mol. Biol. Cell. 2000;3:523–529. doi: 10.1091/mbc.11.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;3:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;3:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Gibson LC, Hunter CN. ATPase activity associated with the magnesium-protoporphyrin IX chelatase enzyme of Synechocystis PCC6803: evidence for ATP hydrolysis during Mg2+ insertion, and the MgATP-dependent interaction of the ChlI and ChlD subunits. Biochem J. 1999;3(Pt 1):127–134. doi: 10.1042/0264-6021:3390127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A, Janek K, Braun B, Kloetzel PM, Enenkel C. 20 S proteasomes are imported as precursor complexes into the nucleus of yeast. J Mol Biol. 2002;3:401–13. doi: 10.1006/jmbi.2002.5443. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Arndt A, Kutay U, Gorlich D, Wittinghofer A. Structural view of the Ran-Importin β interaction at 2.3 A resolution. Cell. 1999;3:635–646a. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- Zimowska G, Paddy MR. Structures and dynamics of Drosophila tpr inconsistent with a static, filamentous structure. Exp Cell Res. 2002;3:223–232. doi: 10.1006/excr.2002.5525. [DOI] [PubMed] [Google Scholar]
- Ruhf ML, Braun A, Papoulas O, Tamkun JW, Randsholt N, Meister M. The domino gene of Drosophila encodes novel members of the SWI2/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development. 2001;3:1429–1441. doi: 10.1242/dev.128.8.1429. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Haussler D. Assembly of the working draft of the human genome with GigAssembler. Genome Res. 2001;3:1541–1548. doi: 10.1101/gr.183201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES. et al. Initial sequencing and analysis of the human genome. Nature. 2001;3:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;3:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;3:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;3:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Kozminski KG, Chen AJ, Rodal AA, Drubin DG. Functions and functional domains of the GTPase Cdc42p. Mol Biol Cell. 2000;3:339–354. doi: 10.1091/mbc.11.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;3:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FullAlignment