Abstract
The peptidoglycan-recognition protein LCa (PGRP-LCa) is a transmembrane receptor required for activation of the Drosophila immune deficiency pathway by monomeric Gram-negative peptidoglycan. We have determined the crystal structure of the ectodomain of PGRP-LCa at 2.5-Å resolution and found two unique helical insertions in the LCa ectodomain that disrupt an otherwise L-shaped peptidoglycan-docking groove present in all other known PGRP structures. The deficient binding of PGRP-LCa to monomeric peptidoglycan was confirmed by biochemical pull-down assays. Recognition of monomeric peptidoglycan involves both PGRP-LCa and -LCx. We showed that association of the LCa and LCx ectodomains in vitro depends on monomeric peptidoglycan. The presence of a defective peptidoglycan-docking groove, while preserving a unique role in mediating monomeric peptidoglycan induction of immune response, suggests that PGRP-LCa recognizes the exposed structural features of a monomeric muropeptide when the latter is bound to and presented by the ectodomain of PGRP-LCx. Such features include N-acetyl glucosamine and the anhydro bond in the glycan of the muropeptide, which have been demonstrated to be critical for immune stimulatory activity.
Keywords: innate immunity, peptidoglycan
Bacterial pathogens initiate host defense responses through pattern-recognition receptors that directly recognize unique structures of conserved bacterial components and activate innate immune signaling cascades. In mammals, a diversity of conserved bacterial structures is recognized by Toll-like receptors. The wide variety of such pathogen-associated molecular patterns exploited by Toll-like receptors for bacterial detection includes peptidoglycan, lipopolysaccharide, lipoteichoic acids, lipoproteins, bacterial CpG DNA, and flagellin (1).
Insects such as Drosophila seem to focus on peptidoglycan as the main target for innate immune recognition of bacterial infection. Peptidoglycan is an essential and unique cell-wall polymer present in both Gram-positive and -negative bacteria. It is composed of long glycan chains made of two alternating sugars and crosslinked by short peptides. The polymeric peptidoglycan consists of repeating subunits known as muropeptides, which are composed of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) attached with a stem peptide chain consisting of d- and l- (or meso-) amino acids, with the third amino acid being more frequently lysine in Gram-positive bacteria and diaminopimelic acid (DAP) in Gram-negative bacteria. Free monomeric muropeptides are present in bacterial cells, because they are constantly synthesized de novo or hydrolyzed from peptidoglycan and recycled during cell division (2, 3).
In Drosophila, the pattern-recognition receptors responsible for detection of peptidoglycan belong to a family of peptidoglycan-recognition proteins (PGRPs) (4, 5). PGRP members are expressed as either secreted, cytosolic, or transmembrane forms, which share a conserved PGRP domain with a structural fold similar to bacteriophage T7 lysozyme (6, 7). Proteins with PGRP domains are widespread in organisms ranging from insects to humans. To date, crystal structures of Drosophila PGRP-LB and -SA, and human PGRP-IαC and -S have been reported (8–11). Notably, all these structures adopt a conserved surface groove that has been demonstrated to be the peptidoglycan-docking groove by mutational and structural analysis (9, 12).
The critical role of several PGRPs in the innate immune signaling pathways in Drosophila has been uncovered by genetics studies. PGRP-SA, expressed as a soluble protein in the hemo-lymph (the insect blood), is required for activating the Toll pathway by recognizing Gram-positive lysine-type peptidoglycan (13, 14). PGRP-SD, another humoral protein, also functions as a pattern-recognition receptor for Toll activation by detecting certain Gram-positive bacteria (15). Activation of the Toll pathway triggers the nuclear localization of the NF-κB-like factors Dif and Dorsal, which induce the transcription of several antimicrobial peptide genes such as Drosomycin (16, 17).
The transmembrane receptor PGRP-LC is required for activating the Drosophila immune deficiency (Imd) pathway by Gram-negative bacteria (18–21). Activation of the PGRP-LC/Imd pathway leads to the proteolytic processing of another NF-κB-related protein, Relish, which activates antibacterial peptide genes such as Diptericin (22, 23). Through alternative splicing, the PGRP-LC gene encodes three isoforms, PGRP-LCa, -LCx, and -LCy, which share identical cytoplasmic and transmembrane domains but contain unique PGRP-like ectodomains with <40% sequence identity (6). Both polymeric and monomeric forms of Gram-negative DAP-type peptidoglycan serve as potent ligands of the Imd pathway (24, 25). Recognition of polymeric peptidoglycan requires only PGRP-LCx; however, monomeric peptidoglycan recognition requires both PGRP-LCa and -LCx (24). The function of PGRP-LCy is unknown; suppressing -LCy expression by RNA interference does not affect the Imd signaling upon challenge with peptidoglycan (24, 26). Given that all PGRP structures known to date contain a similar peptidoglycan-docking groove, it was unclear how the ectodomains of PGRP-LCa and -LCx can mediate distinctive recognition toward monomeric and polymeric forms of peptidoglycan.
We determined the crystal structure of the ectodomain of PGRP-LCa to understand the structural basis of the unique role of PGRP-LCa in recognition of monomeric peptidoglycan. The structure shows that the LCa ectodomain does not possess a typical peptidoglycan-docking groove found in other PGRP domains, including PGRP-LCx. Rather, its groove is obstructed by the presence of two unique helical insertions found only in PGRP-LCa but not in other PGRPs. Our structural data highlight the significant influence of the amino acid insertions on the structure of the peptidoglycan-docking groove in PGRPs and explain the functional difference between PGRP-LCa and -LCx in recognizing monomeric peptidoglycan, which is further confirmed by biochemical pull-down assays. Based on these data, we suggest a model of ligand recognition mediated by the two receptors for the activation of the Imd pathway.
Materials and Methods
Protein Expression, Purification, and Crystallization. cDNA fragments corresponding to the ectodomains of PGRP-LCa (from Phe-355 to Ser-520) and PGRP-LCx (from Val-335 to Ile-500) were subcloned into the pFastbac HTa transfer vector (GIBCO/BRL) premodified by insertion of a gene encoding the signal sequence from honey bee mellitin upstream to the 6xHis codons. Recombinant baculoviral DNA was then obtained by using the Bac-to-bac system (Invitrogen). Insect Hi-5 cells (Invitrogen) grown in serum-free conditions were infected with the recombinant baculovirus. After further growth for 72 h, cell culture supernatant was concentrated to a volume of 400 ml and dialyzed overnight against 6 liters of PBS buffer, pH 7.4 (prepared from PBS tablets from Sigma) supplemented with 200 mM NaCl. The supernatant was mixed with 4 ml of Talon metal affinity resins (Clontech) for 1 h at 4°C. The resins were washed with PBS buffer and the protein eluted at 250 mM imidazole. The protein was then subjected to Tev protease cleavage to remove the 6xHis tag followed by buffer exchange and loaded onto the Talon resins. The flowthrough fractions containing either the untagged LCa or LCx ectodomains were further purified by Mono Q and Superdex 75 columns (Amersham Pharmacia); both columns were also used for purifying His-tagged LCa used for binding assays. The identity and purity of the purified recombinant proteins were analyzed by N-terminal sequencing and mass spectrometry. The LCa ectodomain was concentrated and mixed with 2 mM GlcNAc-MurNAc(anhydro) GlcNAc-MurNAc(anhydro)-l-Ala-γ-d-Glu-meso-DAP-d-Ala [GM(anh)-tetraDAP], which was isolated as described (25), at a total final concentration of 15 mg/ml. Crystallization was carried out at 25°C by the hanging-drop vapor diffusion technique. Small cubic crystals were grown over a reservoir containing 1.7 M ammonium sulfate and 100 mM phosphate citrate, pH 4.2. The same condition could not support crystal formation when concentrated protein sample was prepared without the presence of 2 mM of GM(anh)-tetraDAP. Crystals were cryoprotected in reservoir solution supplemented with 20% glycerol before data collection.
Structure Determination. A complete 2.5-Å data set was collected by using synchrotron radiation source at the PF-BL5 beamline at the Photon Factory in Tsukuba (Japan). The diffraction images were processed and scaled with the hkl2000 package (27). The structure of the LCa ectodomain was determined by molecular replacement with the program phaser (28) by using the PGRP-IαC structure as the search model (PDB ID code 1TWQ). The initial solution contains one molecule per asymmetric unit and gave an R factor of 49% at a resolution range of 50–2.5 Å. The molecular replacement model was iteratively rebuilt manually by using the program o (29) and refined with refmac5 of ccp4 (30). The final model consists of 166 protein residues, one N-acetylglucosamine residue, two sulfate ions from the crystallization solution, and nine water molecules. Data collection and refinement statistics are listed in Table 1, which is published as supporting information on the PNAS web site. Pictures of surface representation in the article were generated by using pymol 0.97 (www.pymol.org). Pictures of ribbon models were prepared with bobscript (31), gl_render (E. Esser, personal communication), and pov-ray (Persistence of Vision Raytracer, Ver. 3.1g, Pov-Ray, Williamstown, Australia).
Affinity Pull-Down Assay. The assay was performed at 4°C by incubating 15 μg of His-tagged LCa ectodomain or His-tagged LCx ectodomain with 5-fold molar excess of GM(anh)-tetraDAP and 10 μl of Talon metal affinity resins (CLONTECH), in 300 μl of binding buffer containing PBS, pH 7.4, supplemented with 300 mM NaCl for 60 min. The resins were isolated by spinning at 600 × g for 5 min and washed with 1.0 ml of binding buffer twice. The ligand was eluted with 10 μl of 0.2% acetic acid. The bound GM(anh)-tetraDAP was detected by using MALDI-TOF mass spectrometry and identified by comparing the mass spectra obtained from the purified protein and the GM(anh)-tetraDAP samples alone. To detect interaction between the LCa and LCx ectodomains, 15 μg of His-tagged LCa ectodomain was incubated for 60 min at 4°C with 10 μl of the Talon resins and 2-fold molar excess of untagged LCx ectodomain with or without 5-fold molar excess of GM(anh)-tetraDAP. The resins were spun down at 600 × g for 5 min and washed with 1.0 ml of the binding buffer twice. The bound proteins retained in the resins were dissolved in 10 μl of SDS buffer for SDS/PAGE analysis and visualized by Coomassie blue staining.
Results
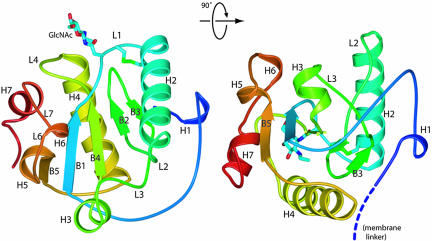
Overall Structure of the LCa Ectodomain. PGRP-LCa is a type II membrane protein of 520 amino acid residues, with N-terminal cytoplasmic and C-terminal ectodomains (24). The structure of the LCa ectodomain (residues 355–520) is composed of a central five-stranded (B1–5) mixed β-sheet sandwiched between two major helices (H2 and H4); this structural core is decorated with five short helices (H1, H3, H5, H6, and H7) (Fig. 1). Residues Cys-390 and -396 form a buried disulfide bond that tethers together the L1 loop and the H2 helix; an equivalent disulfide bridge is present in almost all known PGRPs except PGRP-LE. In the crystal, one glycosylation site was identified from the electron-density map showing an ordered GlcNAc. The GlcNAc bound to the side chain of Asn-389, which precedes the disulfide-forming Cys-390, is located between the L1 and L4 loops (Fig. 1). The side chain of Asn-515, another potential glycosylation site, is disordered in the crystal. Thus, PGRP-LCa is a glycoprotein with at least one N-linked glycosylation site in the ectodomain, and the surface containing the L1 and L4 loop may be covered with oligosaccharides. However, neither of the glycosylation sites is conserved in other PGRP-LC isoforms (see Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 1.
Overall structure of the ectodomain of PGRP-LCa in ribbon model. Ribbon diagram showing the front face (Left) and a side view (Right) of the LCa ectodomain. The ribbon is colored from N to C terminus in a progression from blue to red. The disulfide bridge and the N-linked sugar are shown as stick models. The linker connecting the N terminus of the structure to the transmembrane segment, which is not included in the protein construct, is indicated by the dashed coil.
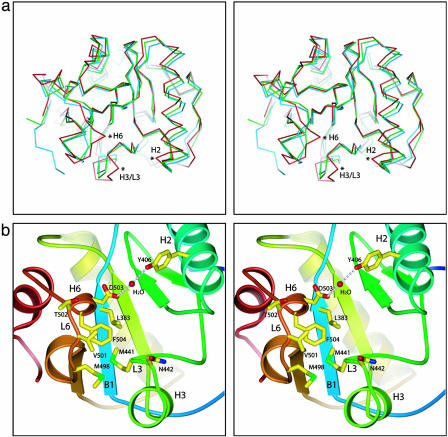
The disulfide-bond-linked L1 loop and H2 helix, together with the L2, L3, and L6 loops and the H6 helix, define the front face of the ectodomain. Mutational analysis of PGRP-SA demonstrated that a central groove in the front face demarcated by the H2 helix on one side and by the L3, L6, and L7 loops on the other side is the peptidoglycan-docking groove (9). Removing only the side chain of Ser-158 in the L7 loop was sufficient to eliminate peptidoglycan-binding activity, suggesting that the L6/L7 wall of the groove may contribute the bulk of the binding energy (9). Recently, the crystal structure of human PGRP-IαC with a muramyl tripeptide MTP bound to this central groove has been reported (12). Although the overall backbone conformation of the LCa ectodomain is similar to other PGRP structures, substantial structural differences in these groove-lining regions distinguish the LCa ectodomain from all other PGRPs (Fig. 2a). The major differences are in the C-terminal part of the H2 helix, the segment consisting of the L6-H6-L7 regions and the H3-L3 regions of the LCa ectodomain. Because of the changes in the positions of these structural segments, the overall structure of the LCa ectodomain adopts a more closed conformation than other PGRPs.
Fig. 2.
Structure analysis of the LCa ectodomain. (a) Stereoview of the LCa ectodomain (red) superimposed with PGRP-SA (blue) and -IαC (green) in Cα traces by using the central β-sheet as a reference. Regions with significant conformational changes are marked with asterisks. (b) Closeup stereoview of the front face of the LCa ectodomain. Side chains of the selected residues described are in stick representation.
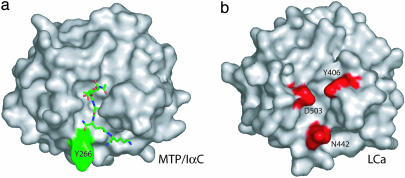
Two Unique Insertions in the LCa Ectodomain Disrupt an Otherwise Continuous Central Groove. All previously reported PGRP structures contain a conserved L-shaped peptidoglycan-docking groove (8–11, 32). This is not the case for the LCa ectodomain, which contains two insertions not present in other PGRPs, including the two other PGRP-LC isoforms (Fig. 6). The first insertion consists of Ile-444 and Asn-445 in the short α helix H3, whose presence results in a substantial conformational change in the immediately preceding H3/L3 loop region, which houses Asn-442 and Met-441 (Fig. 2b). In all other known PGRP structures, the helix H3 is absent, and the residue corresponding to Asn-442 of -LCa is exposed and located at the corner of the L-shaped groove, whereas its preceding residues are buried. This corner residue is a proline in PGRP-LB and a tyrosine in both PGRP-SA and -IαC (Fig. 3a). In the LCa ectodomain, the Cα of Asn-442 is moved by 3.0 Å from the corner wall of the groove toward the opposite wall (compared with the Cα of Tyr-266 in PGRP-IαC), such that its side chain occupies the center space of the corner of the L-shaped groove and disrupts the groove (Fig. 3b). Asn-442 thus would prevent docking interaction, because in the MTP-docked PGRP-IαC structure, the central d-isoglutamine residue of the stem peptide of the MTP compound is located in the same position where the residue lies (see Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 3.
Molecular surfaces of PGRP-IαC and the LCa ectodomain. (a) Surface representation of PGRP-IαC complexed to MTP (see text), which is shown as a stick model. Residue Tyr-266 is highlighted. (b) Surface representation of the front face of the LCa ectodomain. Residues Asn-442, Asp-503, and Tyr-406 are highlighted.
The second insertion in PGRP-LCa appears to provide an additional barricade for ligand binding. It consists of residues Asp-503–Phe-504 in the 310 helix H6 (residues 502–504). The side chain of Phe-504, located near the corner of the L-shaped groove, makes hydrophobic contacts with Leu-383 from the B1 strand, Met-441 from the L3 loop, and Met-498 and Val-501 from the L6 loop (Fig. 2b). The H6 region in the LCa ectodomain adopts a completely different conformation from the corresponding L4 loop in other PGRPs (Fig. 2a). In PGRP-SA, the L4 loop houses Ser-158, which is located within the upper end of the docking groove and is critical for peptidoglycan interaction and for L,Dcarboxypeptidase activity (9). By contrast, Thr-502, the corresponding residue in the LCa ectodomain, is completely solvent-exposed (Fig. 2b). The formation of the 310 helix positions Asp-503 at the helical corner facing the other wall of the groove and orients the side chain toward Tyr-406 in the H2 helix (Fig. 3b); the two residues are linked by hydrogen bonding through a water molecule (Fig. 2b). Based on the MTP-bound PGRP-IαC structure, the side chain of Asp-503 in the LCa ectodomain would be in steric clash with the MurNAc moiety of MTP (Fig. 7).
PGRP-LCx, the other -LC isoform required for sensing both polymeric and monomeric peptidoglycan, does not contain any insertion and is similar in sequence to both PGRP-SA and -IαC. Although the LCa ectodomain (residues 355–520) has only 28% sequence identity to both PGRP-SA and -IαC, the LCx ectodomain has 40% and 45% sequence identity to PGRP-SA and -IαC, respectively. Therefore, PGRP-LCx is likely to possess a peptidoglycan-docking groove structure and is expected to bind either polymeric or monomeric DAP-type peptidoglycans. By contrast, the unique structural features in LCa ectodomain would disallow the MTP–PGRP-IαC-like interactions with both the MurNAc and the stem peptide of the muropeptide. Although the LCa crystals were grown in the presence of 2 mM of GM(anh)-tetraDAP (2-fold molar excess), no electron density for the compound was found in the map. The two barricade regions were clearly visible in the election density map (data not shown). Because these regions are involved in intimate hydrophobic interactions with each other as well as with the protein core, it is unlikely that a substantial backbone conformational change could occur to accommodate the ligand.
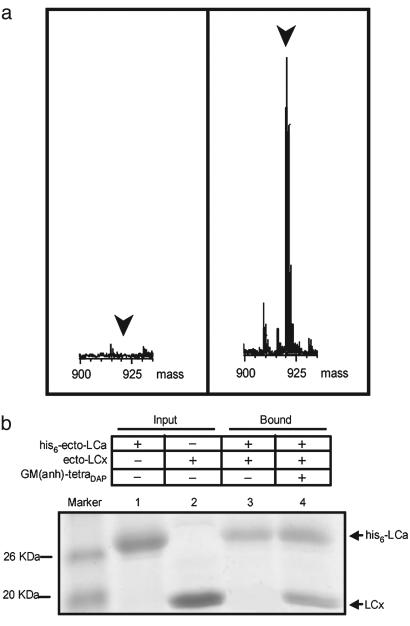
In Vitro Interactions of the LCa and LCx Ectodomains with Monomeric Peptidoglycan. To further confirm that the LCa, but not the LCx, ectodomain is deficient in binding to monomeric peptidoglycan, affinity pull-down assays were performed by using His-tagged LCa or LCx ectodomains with 5-fold molar excess of GM(anh)-tetraDAP. The ligand bound to the His-tagged ectodomains was detected by mass spectrometry. Indeed, the LCa ectodomain failed to bind GM(anh)-tetraDAP, whereas the LCx ectodomain displays ligand-binding activity (Fig. 4a).
Fig. 4.
Interaction of GM(anh)-tetraDAP with the ectodomains of PGRP-LCx and -LCa. (a) Mass spectra of the pull-down fractions of His-tagged LCa (Left) and LCx ectodomains (Right) incubated with Talon metal-affinity resins and GM(anh)-tetraDAP. Only the mass range from 900 to 935 Daltons is shown. The position of the GM(anh)-tetraDAP peak is indicated. (b) SDS/PAGE analysis of the pull-down fractions of His-tagged LCa ectodomain incubated with 2-fold molar excess of the untagged LCx ectodomain with or without GM(anh)-tetraDAP present. Gel was stained with Coomassie blue.
Because both the structural and biochemical results show that the PGRP-LCa cannot bind to GM(anh)-tetraDAP, PGRP-LCa is likely to be involved in ligand recognition by interaction with GM(anh)-tetraDAP-bound PGRP-LCx. We tested this hypothesis by using the affinity pull-down assay. When the His-tagged LCa ectodomain was mixed with 2-fold molar excess of untagged LCx ectodomain in the absence of GM(anh)-tetraDAP, only very weak association between the LCa and LCx ectodomains was observed (Fig. 4b, lane 3). However, after incubating with 2-fold molar excess of untagged LCx ectodomain in the presence of 5-fold excess of GM(anh)-tetraDAP, the His-tagged LCa ectodomain was able to pull down an ≈1:1 stoichiometric amount of the LCx ectodomain, suggesting the possible formation of a ternary LCa/GM(anh)-tetraDAP/LCx complex (Fig. 4b, lane 4).
Discussion
Recently, two groups have independently demonstrated that not only does polymeric DAP-typed peptidoglycan activate the PGRP/Imd pathway, but also monomeric DAP-typed muropeptide acts as a potent inducer of the pathway (24, 25) (see Fig. 8, which is published as supporting information on the PNAS web site, for the chemical structure of peptidogylcan). Both publications showed that a natural Escherichia coli monomeric disaccharide tetrapeptide with an internal 1,6-anhydro bond in the MurNAc residue, GlcNAc-MurNAc (anhydro) GlcNAc-MurNAc(anhydro)-l-Ala-γ-d-Glu-meso-DAP-d-Ala [GM(anh)-tetraDAP], strongly activates the PGRP-LC/Imd pathway in cells and flies. They also found that both the disaccharide moiety and the DAP residue at the third position of the stem peptide are critical for the immune induction effect of GM(anh)-tetraDAP. Stenbak et al. (25) further showed that the presence of both MurNAc anhydro bond and GlcNAc in GM(anh)-tetraDAP is essential for optimal stimulatory activity. The minimal active muropeptide was found to be a GM(anh)-triDAP, a homologue of GM(anh)-tetraDAP lacking the fourth Ala residue (25). Moreover, Kaneko et al. (24) showed that recognition of GM(anh)-tetraDAP requires both PGRP-LCa and -LCx receptors, whereas recognition of polymeric DAP-type peptidoglycan requires only PGRP-LCx. Based on these results, they proposed that PGRP-LC acts as a dimeric receptor such that the PGRP-LCx/LCa heterodimer recognizes GM(anh)-tetraDAP. Such receptor dimerization has been suggested to be required for PGRP-LC activation (21).
In this study, we show that the ectodomain of PGRP-LCa does not possess a typical peptidoglycan-docking groove found in all other PGRP structures and cannot interact directly with a muropeptide. Furthermore, pull-down assay results suggest that the LCa ectodomain forms a complex with the LCx ectodomain when the latter is bound to a monomeric muropeptide. Although Kaneko et al. (24) showed that the two receptors homo/heterodimerize in the absence of the muropeptide, full-length LCa and LCx were used; therefore, such homo/heterodimerization may be due to interaction via their identical cytoplasmic domains, which were shown to self-dimerize in a recent study (21). The binding of monomeric peptidoglycan may induce conformational change in the LCx ectodomain to allow interaction with the LCa ectodomain; however, no conformational change was observed in the PGRP-IαC structure when MTP was bound (12). Hence, for PGRP-LCa to participate in GM(anh)-triDAP recognition with PGRP-LCx without the need of a peptidoglycan-docking groove, the LCa ectodomain would most likely be involved in recognizing the muropeptide, when the latter is bound to the peptidoglycan-docking groove of -LCx. In other words, the LCa ectodomain is likely to recognize a GM(anh)-triDAP presented by the LCx ectodomain but not the muropeptide by itself (Fig. 5a). In this model, the GM(anh)-triDAP molecule would need to bind to the L-shaped docking groove on the LCx ectodomain first, in the same the way that MTP binds to PGRP-IαC and becomes half buried within the docking groove. This binary complex would then allow the LCa ectodomain to bind to the exposed part of the docked GM(anh)-triDAP, along with the surrounding LCx residues in the front face (Fig. 5b). As a consequence, monomeric peptidoglycan brings PGRP-LCa and -LCx receptors in close vicinity, allowing their cytoplasmic domains to interact for Imd activation (21). By contrast, polymeric DAP-type peptidoglycan allows PGRP-LC cytoplasmic domains to associate for signaling by bringing PGRP-LCx receptors, but not PGRP-LCa, in close proximity via its repeating muropeptide subunits (Fig. 5c). This model explains why PGRP-LCa is not required for polymeric peptidoglycan recognition but is needed for recognition of monomeric peptidoglycan, and why PGRP-LCx is required for recognition of both polymeric and monomeric peptidoglycan. This model also explains how the 1,6-anhydro bond of MurNAc is critically involved in recognition. In the crystal structure of the MTP–PGRP-IαC complex, the pyranose ring of MurNAc is in the chair conformation, with the C1-O5-C5-C6 side of the chair oriented toward the solvent space and accessible, while the other side is buried within the groove (Fig. 7). Therefore, upon binding to the LCx ectodomain, the exposed 1,6-anhydro bond of MurNAc(anhydro) would be available for interacting with the LCa ectodomain. In the MTP–PGRP-IαC structure, the GlcNAc was not included in the compound. However, because the hydroxyl group at C4 of the MurNAc is exposed in the MTP–PGRP-IαC structure, the GlcNAc would attach to C4 and also be accessible for interaction with the LCa ectodomain. In the proposed model, the important roles of the glycan residues are highlighted, because they represent the parts of a muropeptide that would be recognized by the ectodomains of both PGRP-LCa and -LCx. In contrast to other residues in the stem peptide of MTP, the side chain of the lysine residue at the third position is half packed against an open wall of the docking groove and half exposed in the MTP–PGRP-IαC structure (Fig. 7). This lysine would correspond to the DAP residue in GM(anh)-triDAP. Although the side chain of DAP is expected to be involved in specific interaction with the docking groove on PGRP-LCx, it may also be involved in interacting with the LCa ectodomain. In the future, confirming the proposed model by identifying the regions on the front face of the LCa ectodomain that are responsible for interacting with the muropeptide/-LCx complex by mutational analysis and determining the structure of the LCa-GM(anh)-triDAP-LCx ectodomain complex are required for a better understanding of muropeptide recognition by the PGRP-LCa and -LCx receptors.
Fig. 5.
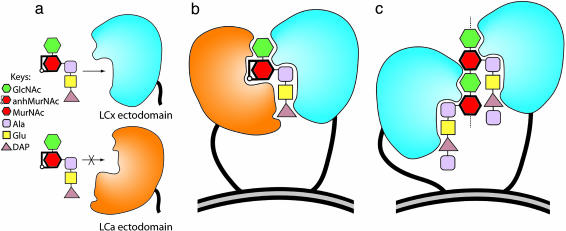
Proposed model of peptidoglycan recognition mediated by PGRP-LCa and -LCx receptors. Schematic illustration showing how the recognitions of monomeric DAP-type muropeptide GM(anh)-triDAP and polymeric DAP-type peptidoglycan are achieved by PGRP-LCa and -LCx while bringing two receptor molecules in close proximity. (a) The LCx ectodomain (sky blue) is able to engage a typical muropeptide-docking interaction as observed in the MTP–PGRP-IαC structure with a GM(anh)-triDAP molecule in a way such that the anhydro bond of MurNAc is exposed after docking to the LCx ectodomain. Such interaction cannot occur with the LCa ectodomain (orange), because the latter does not contain a canonical docking groove. In b, however, the surface structure of the LCa ectodomain would allow recognition of the exposed atoms of glycan, and perhaps the DAP, of the muropeptide when such ligand is docked to and presented by the LCx ectodomain. In the model, GM(anh)-triDAP brings PGRP-LCa and -LCx receptors in close vicinity, allowing their cytoplasmic domains (not shown) to dimerize for receptor activation (21). In c, two LCx ectodomains are engaged in direct docking interactions with two muropeptide subunits of polymeric peptidoglycan, which brings two receptors in close proximity. Only the ectodomain surface features for recognition are illustrated for PGRP-LCa and -LCx. Note the 1,6-anhydro bond of MurNAc present in the GM(anh)-triDAP molecule does not occur in the polymeric form of peptidoglycan except at the terminal MurNAc residue of the glycan chain, which is not shown.
Supplementary Material
Acknowledgments
We thank Maya Palnitkar for preparing the baculoviral transfer vector, Kirsten Fischer Lindahl for critical reading of the manuscript, and Bruno Lemaitre (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France) for providing the cDNA plasmid of PGRP-LCa and for critical comments on the manuscript. The laboratory of S.W. at the Photon Factory was supported by Special Coordination Funds for Promoting Science and Technology and Protein 3000 Project of the Ministry of Education, Culture, Sports, Science, and Technology of Japan. The laboratory of D.M.-L. was supported by the Centre National de la Recherche Scientifique. J.D. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: C.-I.C. and J.D. designed research; C.-I.C., K.I., and Y.C. performed research; D.M.-L. contributed new reagents/analytic tools; C.-I.C., K.I., Y.C., and S.W. analyzed data; and C.-I.C. wrote the paper.
Abbreviations: DAP, diaminopimelic acid; GlcNAc, N-acetylglucosamine; MurNAc, N-acetylmuramic acid; GM(anh)-tetraDAP, GlcNAc-MurNAc(anhydro) GlcNAc-MurNAc(anhydro)-l-Ala-γ-d-Glu-meso-DAP-d-Ala; Imd, immune deficiency; PGRP, peptidoglycan-recognition protein; MTP, MurNAc-l-Ala-d-Gln-l-Lys.
Data deposition: The structure factors and atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1Z6I).
References
- 1.Medzhitov, R. (2001) Nat. Rev. Immunol. 1, 135-145. [DOI] [PubMed] [Google Scholar]
- 2.Goodell, E. W. (1985) J. Bacteriol. 163, 305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodell, E. W. & Schwarz, U. (1985) J. Bacteriol. 162, 391-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang, D., Liu, G., Lundstrom, A., Gelius, E. & Steiner, H. (1998) Proc. Natl. Acad. Sci. USA 95, 10078-10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida, H., Kinoshita, K. & Ashida, M. (1996) J. Biol. Chem. 271, 13854-13860. [DOI] [PubMed] [Google Scholar]
- 6.Werner, T., Liu, G., Kang, D., Ekengren, S., Steiner, H. & Hultmark, D. (2000) Proc. Natl. Acad. Sci. USA 97, 13772-13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu, C., Xu, Z., Gupta, D. & Dziarski, R. (2001) J. Biol. Chem. 276, 34686-34694. [DOI] [PubMed] [Google Scholar]
- 8.Guan, R., Malchiodi, E. L., Wang, Q., Schuck, P. & Mariuzza, R. A. (2004) J. Biol. Chem. 279, 31873-31882. [DOI] [PubMed] [Google Scholar]
- 9.Chang, C. I., Pili-Floury, S., Herve, M., Parquet, C., Chelliah, Y., Lemaitre, B., Mengin-Lecreulx, D. & Deisenhofer, J. (2004) PLoS Biol. 2, E277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, M. S., Byun, M. & Oh, B. H. (2003) Nat. Immunol. 4, 787-793. [DOI] [PubMed] [Google Scholar]
- 11.Guan, R., Wang, Q., Sundberg, E. J. & Mariuzza, R. A. (2005) J. Mol. Biol. 347, 683-691. [DOI] [PubMed] [Google Scholar]
- 12.Guan, R., Roychowdhury, A., Ember, B., Kumar, S., Boons, G. J. & Mariuzza, R. A. (2004) Proc. Natl. Acad. Sci. USA 101, 17168-17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michel, T., Reichhart, J. M., Hoffmann, J. A. & Royet, J. (2001) Nature 414, 756-759. [DOI] [PubMed] [Google Scholar]
- 14.Leulier, F., Parquet, C., Pili-Floury, S., Ryu, J. H., Caroff, M., Lee, W. J., Mengin-Lecreulx, D. & Lemaitre, B. (2003) Nat. Immunol. 4, 478-484. [DOI] [PubMed] [Google Scholar]
- 15.Bischoff, V., Vignal, C., Boneca, I. G., Michel, T., Hoffmann, J. A. & Royet, J. (2004) Nat. Immunol. 5, 1175-1180. [DOI] [PubMed] [Google Scholar]
- 16.Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M. & Hoffmann, J. A. (1996) Cell 86, 973-983. [DOI] [PubMed] [Google Scholar]
- 17.Meng, X., Khanuja, B. S. & Ip, Y. T. (1999) Genes Dev. 13, 792-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choe, K. M., Werner, T., Stoven, S., Hultmark, D. & Anderson, K. V. (2002) Science 296, 359-362. [DOI] [PubMed] [Google Scholar]
- 19.Gottar, M., Gobert, V., Michel, T., Belvin, M., Duyk, G., Hoffmann, J. A., Ferrandon, D. & Royet, J. (2002) Nature 416, 640-644. [DOI] [PubMed] [Google Scholar]
- 20.Ramet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B. & Ezekowitz, R. A. (2002) Nature 416, 644-648. [DOI] [PubMed] [Google Scholar]
- 21.Choe, K. M., Lee, H. & Anderson, K. V. (2005) Proc. Natl. Acad. Sci. USA 102, 1122-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemaitre, B., Kromer-Metzger, E., Michaut, L., Nicolas, E., Meister, M., Georgel, P., Reichhart, J. M. & Hoffmann, J. A. (1995) Proc. Natl. Acad. Sci. USA 92, 9465-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hedengren, M., Asling, B., Dushay, M. S., Ando, I., Ekengren, S., Wihlborg, M. & Hultmark, D. (1999) Mol. Cell 4, 827-837. [DOI] [PubMed] [Google Scholar]
- 24.Kaneko, T., Goldman, W. E., Mellroth, P., Steiner, H., Fukase, K., Kusumoto, S., Harley, W., Fox, A., Golenbock, D. & Silverman, N. (2004) Immunity 20, 637-649. [DOI] [PubMed] [Google Scholar]
- 25.Stenbak, C. R., Ryu, J. H., Leulier, F., Pili-Floury, S., Parquet, C., Herve, M., Chaput, C., Boneca, I. G., Lee, W. J., Lemaitre, B., et al. (2004) J. Immunol. 173, 7339-7348. [DOI] [PubMed] [Google Scholar]
- 26.Werner, T., Borge-Renberg, K., Mellroth, P., Steiner, H. & Hultmark, D. (2003) J. Biol. Chem. 278, 26319-26322. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 28.Storoni, L. C., McCoy, A. J. & Read, R. J. (2004) Acta Crystallogr D 60, 432-438. [DOI] [PubMed] [Google Scholar]
- 29.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 31.Esnouf, R. M. (1999) Acta Crystallogr. D 55 (Pt 4), 938-940. [DOI] [PubMed] [Google Scholar]
- 32.Reiser, J. B., Teyton, L. & Wilson, I. A. (2004) J. Mol. Biol. 340, 909-917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.