Abstract
In the course of evolution, mutations do not affect both strands of genomic DNA equally. This imbalance mainly results from asymmetric DNA mutation and repair processes associated with replication and transcription. In prokaryotes, prevalence of G over C and T over A is frequently observed in the leading strand. The sign of the resulting TA and GC skews changes abruptly when crossing replication-origin and termination sites, producing characteristic step-like transitions. In mammals, transcription-coupled skews have been detected, but so far, no bias has been associated with replication. Here, analysis of intergenic and transcribed regions flanking experimentally identified human replication origins and the corresponding mouse and dog homologous regions demonstrates the existence of compositional strand asymmetries associated with replication. Multiscale analysis of human genome skew profiles reveals numerous transitions that allow us to identify a set of 1,000 putative replication initiation zones. Around these putative origins, the skew profile displays a characteristic jagged pattern also observed in mouse and dog genomes. We therefore propose that in mammalian cells, replication termination sites are randomly distributed between adjacent origins. Taken together, these analyses constitute a step toward genome-wide studies of replication mechanisms.
Keywords: replication termination, wavelet transform, compositional bias, skewness
Comprehensive knowledge of genome evolution relies on understanding mutational processes that shape DNA sequences. Nucleotide substitutions do not occur at similar rates, and, in particular, owing to strand asymmetries of the DNA mutation and repair processes, they can affect each of the two DNA strands differently. Asymmetries of substitution rates coupled to transcription have been observed in prokaryotes (1–3) and in eukaryotes (4–6). Strand asymmetries (i.e., G ≠ C and T ≠ A) associated with the polarity of replication have been found in bacterial, mitochondrial, and viral genomes, where they have been used to detect replication origins (7–9). In most cases, the leading replicating strand presents an excess of G over C and of T over A. Along one DNA strand, the sign of this bias changes abruptly at the replication origin and at the terminus. In eukaryotes, the situation is unclear. Several studies failed to show compositional biases related to replication, and analyses of nucleotide substitutions in the region of the β-globin replication origin did not support the existence of mutational bias between the leading and the lagging strands (8, 10, 11). In contrast, strand asymmetries associated with replication were observed in the subtelomeric regions of Saccharomyces cerevisiae chromosomes, supporting the existence of replication-coupled asymmetric mutational pressure in this organism (12). Here, we present analyses of strand asymmetries flanking experimentally determined human replication origins, as well as the corresponding mouse and dog homologous regions. Our results demonstrate the existence of replication-coupled strand asymmetries in mammalian genomes. Multiscale analysis of skew profiles of the human genome using the wavelet transform methodology reveals the existence of numerous putative replication origins associated with randomly distributed termination sites.
Data and Methods
Human Replication Origins. Nine replication origins were examined: namely, those situated near the genes MCM4 (13), HSPA4 (14), TOP1 (15), MYC (16), SCA-7 (17), AR (17), DNMT1 (18), LaminB2 (19), and β-globin (20).
Sequences. Sequence and annotation data were retrieved from the Genome Browser of the University of California, Santa Cruz, for the human (May 2004), mouse (May 2004), and dog (July 2004) genomes. To delineate the most reliable intergenic regions, transcribed regions were retrieved from “all_mrna,” one of the largest sets of annotated transcripts. To obtain intronic sequences, we used the KnownGene annotation (containing only protein-coding transcripts); when several transcripts presented common exonic regions, only common intronic sequences were retained. For the dog genome, only preliminary gene annotations were available, precluding the analysis of intergenic and intronic sequences. To avoid biases intrinsic to repeated elements, all sequences were masked with repeatmasker, leading to 40–50% sequence reduction.
Strand Asymmetries. The TA and GC skews were calculated as STA = (T – A)/(T + A) and SGC = (G – C)/(G + C), and the total skew was calculated as S = STA + SGC in nonoverlapping, 1-kbp windows. (All values are given in percent.) The cumulated skew profiles ΣTA and ΣGC were obtained by cumulative addition of the values of the skews along the sequences. To calculate the skews in transcribed regions, only central regions of introns were considered (after removal of 530 nt from each extremity) to avoid the skews associated with splicing signals (6). To calculate the skews in intergenic regions, only windows that did not contain any transcribed region were retained. To eliminate the skews associated with promoter signals and with transcription downstream of poly(A) sites, transcribed sequences were extended by 0.5 kbp and 2 kbp at 5′ and 3′ extremities, respectively (6).
Sequence Alignments. Mouse and dog regions homologous to the six human regions shown in Fig. 1 were retrieved from University of California, Santa Cruz (Human Synteny). Mouse intergenic sequences were individually aligned by using pipmaker (21), leading to a total of 150 conserved segments >100 bp in size (>70% identity) and corresponding to a total of 26 kbp (5.3% of intergenic sequences).
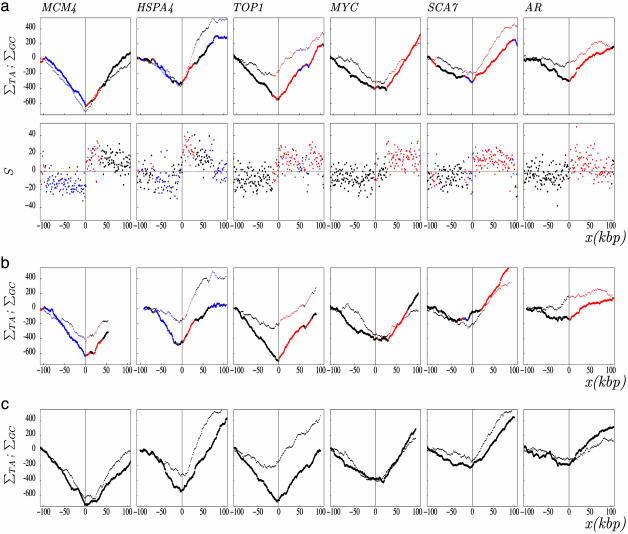
Fig. 1.
TA and GC skew profiles around experimentally determined human replication origins. (a) The skew profiles were determined in 1-kbp windows in regions surrounding (±100 kbp without repeats) experimentally determined human replication origins (see Data and Methods). (Upper) TA and GC cumulated skew profiles ΣTA (thick line) and ΣGC (thin line). (Lower) Skew S calculated in the same regions. The ΔS amplitude associated with these origins, calculated as the difference of the skews measured in 20-kbp windows on both sides of the origins, are: MCM4 (31%), HSPA4 (29%), TOP1 (18%), MYC (14%), SCA7 (38%), and AR (14%). (b) Cumulated skew profiles calculated in the six regions of the mouse genome homologous to the human regions analyzed in a.(c) Cumulated skew profiles in the six regions of the dog genome homologous to human regions analyzed in a. The abscissa (x) represents the distance (in kilobase pairs) of a sequence window to the corresponding origin; the ordinate represents the values of S given in percent. Red, (+) genes (coding strand identical to the Watson strand); blue, (–) genes (coding strand opposite to the Watson strand); black, intergenic regions. In c, genes are not represented.
Wavelet-Based Analysis of the Human Genome. The wavelet transform methodology is a multiscale discontinuities tracking technique (22, 23) (for details, see the supporting information, which is published on the PNAS web site). The main steps involved in detection of jumps were the following. We selected the extrema of the first derivative S′ of the skew profile S smoothened at large scale (i.e., computed in large windows). The scale 200 kbp was chosen as being just large enough to reduce the contribution of discontinuities associated with transcription [i.e., larger than most human genes (24)] yet as small as possible so as to capture most of the contributions associated with replication. To delineate the position corresponding to the jumps in the skew S at smaller scale, we then progressively decreased the size of the analyzing window and followed the positions of the extrema of S′ across the whole range of scales down to the shortest scale analyzed (the precision was limited by the noisy background fluctuations in the skew profile). As expected, the set of extrema detected by this methodology corresponded to similar numbers of upward and downward jumps. The putative replication origins were then selected among the set of upward jumps on the basis of their ΔS amplitude.
Results and Discussion
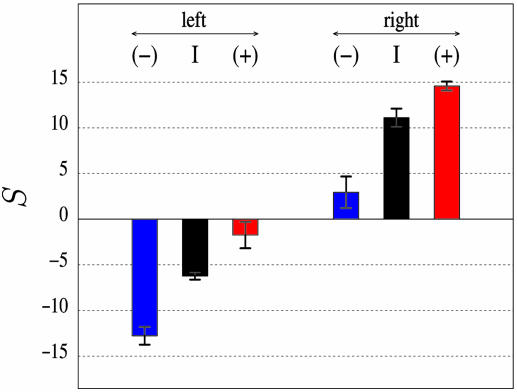
Strand Asymmetries Associated with Replication. We examined the nucleotide strand asymmetries around nine replication origins experimentally determined in the human genome (see Data and Methods). For most of them, the S skew measured in the regions situated 5′ to the origins on the Watson strand (lagging strand) presented negative values that shifted abruptly (over few kilobase pairs) to positive values in regions situated 3′ to the origins (leading strand), displaying sharp upward transitions with large ΔS amplitudes as observed in bacterial genomes (7–9) (Fig. 1a). This pattern was particularly clear with the cumulated TA and GC skews that presented decreasing (or increasing) profiles in regions situated 5′ (or 3′) to the origins, displaying characteristic V shapes pointing to the initiation zones. These profiles could, at least in part, result from transcription, as shown in previous work (6). To measure compositional asymmetries that would result only from replication, we calculated the skews in intergenic regions on both sides of the origins. The mean intergenic skews shifted from negative to positive values when crossing the origins (Fig. 2). This result strongly suggested the existence of mutational pressure associated with replication, leading to the mean compositional biases STA = 4.0 ± 0.4% and SGC = 3.0 ± 0.5% (note that the value of the skew could vary from one origin to another, possibly reflecting different initiation efficiencies) (Table 1). In transcribed regions, the S bias presented large values when transcription was cooriented with replication fork progression [(+) genes on the right and (–) genes on the left] and close to zero values in the opposite situation (Fig. 2). In these regions, the biases associated with transcription and replication added to each other when transcription was cooriented with replication fork progression, giving the skew Slead; they subtracted from each other in the opposite situation, giving the skew Slag (Table 1). We could estimate the mean skews associated with transcription by subtracting intergenic skews from Slead values, giving STA = 3.6 ± 0.7% and SGC = 3.8 ± 0.9%. These estimations were consistent with those obtained with a large set of human introns STA = 4.49 ± 0.01% and SGC = 3.29 ± 0.01% in ref. 6, further supporting the existence of replication-coupled strand asymmetries.
Fig. 2.
Skew S in regions situated on both sides of human replication origins. The mean values of S were calculated in intergenic regions and in intronic regions situated 5′ (Left) and 3′ (Right) of the six origins analyzed in Fig. 1a. Colors are as in Fig. 1; mean values are in percent ± SEM.
Table 1. Strand asymmetries associated with human replication origins.
| STA | SGC | S | I | G+C, % | |
|---|---|---|---|---|---|
| Intergenic (H.s.) all | 3.9 ± 0.4 | 3.0 ± 0.4 | 6.9 ± 0.4 | 487 | 42 |
| Intergenic (H.s.) ncr. | 4.0 ± 0.4 | 3.0 ± 0.5 | 7.0 ± 0.5 | 461 | 42 |
| Intergenic (M.m.) ncr. | 3.6 ± 0.4 | 2.2 ± 0.5 | 5.8 ± 0.5 | 441 | 42 |
| Slead (H.s. introns) | 7.5 ± 0.3 | 6.8 ± 0.4 | 14.3 ± 0.4 | 358 | 40 |
| Slag (H.s. introns) | -1.9 ± 1.0 | -0.3 ± 1.4 | -2.2 ± 1.3 | 49 | 44 |
The skews were calculated in the regions flanking the six human replication origins (Fig. 1a) and in the corresponding homologous regions of the mouse genome. Intergenic sequences were always considered in the direction of replication fork progression (leading strand); they were considered in totality (all) or after elimination of conserved regions (ncr.) between human (Homo sapiens, H.s.) and mouse (Mus musculus, M.m.) (see Data and Methods). To calculate the mean skew in introns, the sequences were considered on the nontranscribed strand. For Slead, the orientation of transcription was the same as the replication fork progression; for Slag, the situation was the opposite. The mean values of the skews STA, SGC, and S are given in percent (±SEM). l, total sequence length in kilobase pairs.
Could the biases observed in intergenic regions result from the presence of as yet undetected genes? Two pieces of evidence argued against this possibility. First, we retained as transcribed regions one of the largest sets of transcripts available, resulting in a stringent definition of intergenic regions. Second, several studies have demonstrated the existence of hitherto unknown transcripts in regions where no protein coding genes had been previously identified (25–28). Taking advantage of the set of non-protein-coding RNAs identified in the “H-Inv” database (29), we checked that none of them was present in the intergenic regions studied here. Another possibility was that the skews observed in intergenic regions result from conserved DNA segments. Indeed, comparative analyses have shown the presence of nongenic sequences conserved in humans and mice (30), which could present biased sequences, possibly contributing to the observed intergenic skews. We examined the mouse genome regions homologous to the six human replication zones (Fig. 1b). Alignment of corresponding intergenic regions revealed the presence of homologous segments, but these segments accounted for only 5.3% of all intergenic sequences. Removal of these segments did not change significantly the skew in intergenic regions; therefore, the possibility that intergenic skews are due to conserved sequence elements was eliminated (Table 1).
Conservation of Replication-Coupled Strand Asymmetries in Mammalian Genomes. We analyzed the skew profiles in DNA regions of mammalian genomes homologous to the six human origins (Fig. 1). The human, mouse, and dog profiles were strikingly similar to each other, suggesting that in mice and dogs, these regions also corresponded to replication initiation zones (indeed, they were very similar in primate genomes). Examination of mouse intergenic regions showed, as for humans, significant skew S values with opposite signs on each side of these putative origins, suggesting the existence of a compositional bias associated with replication S = 5.8 ± 0.5% (Table 1). Human and mouse intergenic sequences situated at these homologous loci presented significant skews, even though they presented almost no conserved sequence elements. This presence of strand asymmetry in regions that strongly diverged from each other during evolution further supported the existence of compositional bias associated with replication in both organisms: In the absence of such a process, intergenic sequences would have lost a significant fraction of their strand asymmetry.
Together, these results establish, in mammals, the existence of strand asymmetries associated with replication in germ-line cells. They determine that most replication origins experimentally detected in somatic cells coincide with sharp upward transitions of the skew profiles. The results also imply that for the majority of experimentally determined origins, the positions of initiation zones are conserved in mammalian genomes [a recent study confirmed the presence of a replication origin in the mouse MYC locus (31)]. Among nine human origins examined, three do not present typical V-type cumulated profiles. For the first one (DNMT1), the central part of the V profile is replaced by a large horizontal plateau (several tens of kilobase pairs), possibly reflecting the presence of several origins dispersed over the whole plateau. Dispersed origins have been observed, for example, in the hamster DHFR initiation zone (32). By contrast, the skew profiles of the LaminB2 and β-globin origins present no upward transition, suggesting that they might be inactive in germ-line cells or less active than neighboring origins (data not shown).
Detection of Putative Replication Origins. Human experimentally determined replication origins coincided with large-amplitude upward transitions of skew profiles. The corresponding ΔS ranged between 14% and 38%, owing to possible different replication initiation efficiencies and/or different contributions of transcriptional biases (Fig. 1a). Are such discontinuities frequent in human sequences, and can they be considered diagnostic of replication initiation zones? In particular, can they be distinguished from the transitions associated with transcription only? Indeed, strand asymmetries associated with transcription can generate sharp transitions in the skew profile at both gene extremities. These jumps are of the same amplitude and of opposite signs: e.g., upward (or downward) jumps at 5′ (or 3′) extremities of (+) genes (6). Upward jumps resulting from transcription only might thus be confused with upward jumps associated with replication origins.
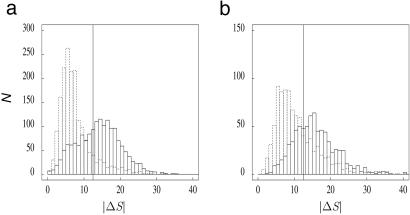
To address these questions, systematic detection of discontinuities in the S profile was performed with the wavelet transform methodology, leading to a set of 2,415 upward jumps and, as expected, to a similar number of downward jumps (see Data and Methods). The distributions of the |ΔS| amplitude of these jumps were then examined, and they showed strong differences between upward and downward jumps. For large |ΔS| values, the number of upward jumps exceeded by far the number of downward jumps (Fig. 3). This excess likely resulted from the fact that, contrasting with prokaryotes, where downward jumps result from precisely positioned replication termination, in eukaryotes, termination appears not to occur at specific positions but to be randomly distributed (this point will be detailed below) (33, 34). Accordingly, the small number of downward jumps with large |ΔS| resulted from transcription, not from replication. These jumps were due to highly biased genes that also generated a small number of large-amplitude upward jumps, giving rise to false-positive candidate replication origins. The number of large downward jumps was thus taken as an estimation of the number of false positives. In a first step, we retained as acceptable a proportion of 33% of false positives. This value resulted from the selection of upward and downward jumps presenting an amplitude |ΔS| ≥ 12.5%, corresponding to a ratio of downward jumps over upward jumps r = 0.33. The values of this ratio r were highly variable along the chromosomes (Fig. 3). In G+C-poor regions (G + C < 37%), we observed the smallest r values (r = 0.15). In regions with 37% ≤ G + C ≤ 42%, we obtained r = 0.24, contrasting with r = 0.53 in regions with G + C > 42%. In these latter regions (accounting for ≈40% of the genome) with high gene density and small gene length (24), the skew profiles oscillated rapidly with large upward and downward amplitudes (Fig. 5d), resulting in a too large number of false positives (53%). In a final step, we retained as putative origins upward jumps (with |ΔS| ≥ 12.5%) detected in regions with G + C ≤ 42%. This selection led to a set of 1,012 candidates among which we could estimate the proportion of true replication origins to 79% (r = 0.21; Fig. 3a).
Fig. 3.
Histograms of the |ΔS| amplitudes of the jumps in the S profile. Using the wavelet transform, a set of 5,101 discontinuities was detected (2,415 upward jumps and 2,686 downward jumps; see Data and Methods). The |ΔS| amplitude was calculated as in Fig. 1a. (a) |ΔS| distributions of the jumps presenting G + C < 42%, corresponding to 1,647 upward jumps and 1,755 downward jumps; the threshold |ΔS| ≥ 12.5% (vertical line) corresponded to 1,012 upward jumps that were retained as putative replication origins and to 211 downward jumps (r = 0.21). (b) |ΔS| distributions of the jumps presenting G + C > 42%, with |ΔS| ≥ 12.5% corresponding to 528 upward jumps and 280 downward jumps (r = 0.53). The G+C content was measured in the 100-kbp window surrounding the jump position. Upward jumps are shown in black, and downward jumps are shown with dots. The abscissa represents the values of the |ΔS| amplitudes calculated in percent.
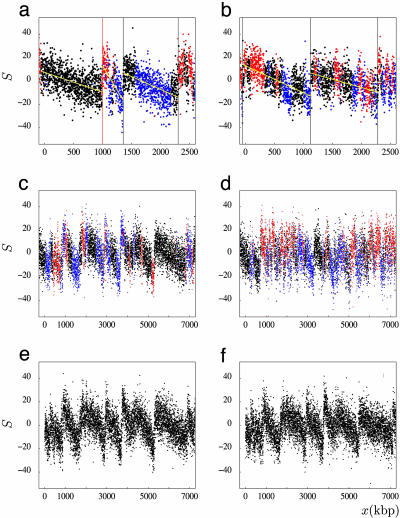
Fig. 5.
S profiles along mammalian genome fragments. (a) Fragment of chromosome 20 including the TOP1 origin (red vertical line). (b and c) Chromosome 4 and chromosome 9 fragments, respectively, with low G+C content (36%). (d) Chromosome 22 fragment with larger G+C content (48%). In a and b, vertical lines correspond to selected putative origins; yellow lines are linear fits of the S values between successive putative origins. Black, intergenic regions; red, (+) genes; blue, (–) genes. Note the fully intergenic regions upstream of TOP1 in a and from positions 5,290–6,850 kbp in c.(e) Fragment of mouse chromosome 4 homologous to the human fragment shown in c.(f) Fragment of dog chromosome 5 syntenic to the human fragment shown in c. In e and f, genes are not represented.
The mean amplitude of the jumps associated with the 1,012 putative origins was 18%, consistent with the range of values observed for the six origins in Fig. 1. Note that these origins were all found in the detection process. In close vicinity of the 1,012 putative origins (±20 kbp), most DNA sequences (55% of the analyzing windows) are transcribed in the same direction as the progression of the replication fork. By contrast, only 7% of sequences are transcribed in the opposite direction (38% are intergenic). These results show that the |ΔS| amplitude at putative origins mostly results from superposition of biases (i) associated with replication and (ii) with transcription of the gene proximal to the origin. Determining whether transcription is cooriented with replication at larger distances will require further studies.
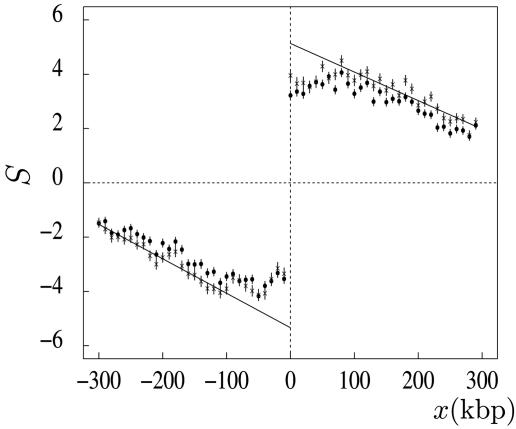
We then determined the skews of intergenic regions on both sides of these putative origins. As shown in Fig. 4, the mean skew profiles calculated in intergenic windows shift abruptly from negative to positive values when crossing the jump positions. To avoid the skews that could result from incompletely annotated gene extremities (e.g., 5′ and 3′ UTRs), 10-kbp sequences were removed at both ends of all annotated transcripts. The removal of these intergenic sequences did not significantly modify the skew profiles, indicating that the observed values do not result from transcription. On both sides of the jump, we observed a steady decrease of the bias, with some flattening of the profile close to the transition point. Note that, due to (i) the potential presence of signals implicated in replication initiation and (ii) the possible existence of dispersed origins (32), one might question the meaningfulness of this flattening that leads to a significant underestimate of the jump amplitude. As shown in Fig. 4, extrapolating the linear behavior observed at a distance >100 kb from the jump would lead to a skew of 5.3%, a value consistent with the skew measured in intergenic regions around the six origins (7.0 ± 0.5%; Table 1). Overall, the detection of upward jumps with characteristics similar to those of experimentally determined replication origins and with no downward counterpart further support the existence, in human chromosomes, of replication-coupled strand asymmetries, leading to the identification of numerous putative replication origins active in germline cells.
Fig. 4.
Mean skew profile of intergenic regions around putative replication origins. The skew S was calculated in 1-kbp windows (Watson strand) around the position (±300 kbp without repeats) of the 1,012 upward jumps (Fig. 3); 5′ and 3′ transcript extremities were extended by 0.5 and 2 kbp, respectively (filled circles), or by 10 kbp at both ends (stars) (see Data and Methods). The abscissa represents the distance (in kilobase pairs) to the corresponding origin; the ordinate represents the skews calculated for the windows situated in intergenic regions (mean values for all discontinuities and for 10 consecutive 1-kbp window positions). The skews are given in percent (vertical bars, SEM). The lines correspond to linear fits of the values of the skew (stars) for x <–100 kbp and x > 100 kbp.
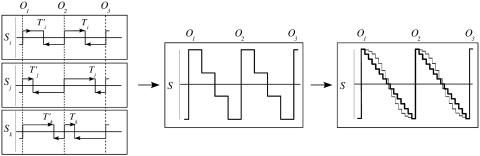
Random Replication Termination in Mammalian Cells. In bacterial genomes, the skew profiles present upward and downward jumps at origin and termination positions, respectively, separated by constant S values (7–9). Contrasting with this step-like shape, the S profiles of intergenic regions surrounding putative origins did not present downward transitions but decreased progressively in the 5′ to 3′ direction on both sides of the upward jump (Fig. 4). This pattern was typically found along S profiles of large genome regions showing sharp upward jumps connected to each other by segments of steadily decreasing skew (Fig. 5 a–c). The succession of these segments, presenting variable lengths, displayed a jagged motif reminiscent of the shape of “factory roofs” that was observed around the experimentally determined human origins (Fig. 5a and data not shown) as well as around a number of putative origins (Fig. 5 b and c). Some of these segments were entirely intergenic (Fig. 5 a and c), clearly illustrating the particular profile of a strand bias resulting solely from replication. In most other cases, we observed the superposition of this replication profile and of the transcription profile of (+) and (–) genes, appearing as upward and downward blocks standing out from the replication pattern (Fig. 5c). Overall, this jagged pattern could not be explained by transcription only but was perfectly explained by termination sites more or less homogeneously distributed between successive origins. Although some replication terminations have been found at specific sites in Schizosaccharomyces pombe (35), they occur randomly between active origins in S. cerevisiae and in Xenopus egg extracts (33, 34). Our results indicate that this property can be extended to replication in human germ-line cells. According to our results, we propose a scenario of replication termination relying on the existence of numerous termination sites distributed along the sequence (Fig. 6). For each termination site (used in a small proportion of cell cycles), strand asymmetries associated with replication will generate a skew profile with a downward jump at the position of termination and upward jumps at the positions of the adjacent origins, separated by constant values (as in bacteria). Various termination positions will correspond to elementary skew profiles (Fig. 6 Left). Addition of these profiles will generate the intermediate profile (Fig. 6 Center), and further addition of many elementary skews will generate the final profile (Fig. 6 Right). In a simple picture, we can suppose that termination occurs with constant probability at any position on the sequence. This behavior can result from the binding of some termination factor at any position between successive origins, leading to a homogeneous distribution of termination sites during successive cell cycles. The final skew profile is then a linear segment decreasing between successive origins (Fig. 6 Right, black line). In a more elaborate scenario, termination would take place when two replication forks collide, which would also lead to various termination sites, but the probability of termination would then be maximum at the middle of the segment separating neighboring origins and decrease toward extremities. Considering that firing of replication origins occurs during time intervals of the S phase (36) could result in some flattening of the skew profile at the origins, as sketched in Fig. 6 Right (gray curve). In the present state, our results clearly support the hypothesis of random replication termination in mammalian cells, but further analyses will be necessary to determine what scenario is precisely at work.
Fig. 6.
Model of replication termination. Schematic representation of the skew profiles associated with three replication origins O1, O2, and O3; we suppose that these replication origins are adjacent, bidirectional origins with similar replication efficiency. The abscissae represent the sequence positions; the ordinates represent the S values (arbitrary units). Upward (or downward) steps correspond to origin (or termination) positions. For convenience, the termination sites are symmetric relative to O2.(Left) Three different termination positions Ti, Tj, and Tk, leading to elementary skew profiles Si, Sj, and Sk.(Center) Superposition of these three profiles. (Right) Superposition of a large number of elementary profiles leading to the final factory-roof pattern. In the simple model, termination occurs with equal probability on both sides of the origins, leading to the linear profile (thick line). In the alternative model, replication termination is more likely to occur at lower rates close to the origins, leading to a flattening of the profile (gray line).
Importantly, the factory-roof pattern was not specific to human sequences; it was also observed in numerous regions of the mouse and dog genomes (e.g., Fig. 5 e and f), indicating that random replication termination is a common feature of mammalian germline cells. Moreover, this pattern was displayed by a set of 1,000 upward transitions, each flanked on each side by DNA segments of ≈300 kbp (without repeats), which can be roughly estimated to correspond to 20–30% of the human genome. In these regions, which are characterized by low and medium G+C contents, the skew profiles revealed a portrait of germ-line replication, consisting of putative origins separated by long DNA segments ≈1–2 Mbp long. Although such segments are much larger than could be expected from the classical view of ≈50- to 300-kbp-long replicons (37), they are not incompatible with estimations showing that replicon size can reach up to 1 Mbp (38, 39) and that replicating units in meiotic chromosomes are much longer than those engaged in somatic cells (40). Finally, it is not unlikely that in G+C-rich (gene-rich) regions, replication origins would be closer to each other than in other regions, further explaining the greater difficulty in detecting origins in these regions.
In conclusion, analyses of strand asymmetries demonstrate the existence of mutational pressure acting asymmetrically on the leading and lagging strands during successive replicative cycles of mammalian germ-line cells. Analyses of the sequences of human replication origins show that most of these origins, determined experimentally in somatic cells, are likely to be active also in germ-line cells. In addition, the results reveal that the positions of these origins are conserved in mammalian genomes. Finally, multiscale studies of skew profiles allow us to identify a large number (1,012) of putative replication initiation zones and provide a genome-wide picture of replication initiation and termination in germ-line cells.
Supplementary Material
Acknowledgments
We thank O. Hyrien for very helpful discussions. This work was supported by the Action Concertée Incitative Informatique, Mathématiques, Physique en Biologie Moléculaire 2004, the Centre National de la Recherche Scientifique, the French Ministère de l'Education et de la Recherche, and the Programmes d'Actions Intégrées Tournesol.
Author contributions: A.A. and C.T. designed research; M.T., S.N., B.A., E.-B.B.o.B., and Y.d.-C. performed research; M.T., S.N., B.A., E.-B.B.o.B., Y.d.-C., A.A., and C.T. analyzed data; and C.T. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Freeman, J. M., Plasterer, T. N., Smith, T. F. & Mohr, S. C. (1998) Science 279, 1827–1830. [Google Scholar]
- 2.Beletskii, A., Grigoriev, A., Joyce, S. & Bhagwat, A. S. (2000) J. Mol. Biol. 300, 1057–1065. [DOI] [PubMed] [Google Scholar]
- 3.Francino, M. P. & Ochman, H. (2001) Mol. Biol. Evol. 18, 1147–1150. [DOI] [PubMed] [Google Scholar]
- 4.Green, P., Ewing, B., Miller, W., Thomas, P. J. & Green, E. D. (2003) Nat. Genet. 33, 514–517. [DOI] [PubMed] [Google Scholar]
- 5.Touchon, M., Nicolay, S., Arneodo, A., d'Aubenton-Carafa, Y. & Thermes, C. (2003) FEBS Lett. 555, 579–582. [DOI] [PubMed] [Google Scholar]
- 6.Touchon, M., Arneodo, A., d'Aubenton-Carafa, Y. & Thermes, C. (2004) Nucleic Acids Res. 32, 4969–4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lobry, J. R. (1996) Mol. Biol. Evol. 13, 660–665. [DOI] [PubMed] [Google Scholar]
- 8.Mrazek, J. & Karlin, S. (1998) Proc. Natl. Acad. Sci. USA 95, 3720–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tillier, E. R. & Collins, R. A. (2000) J. Mol. Evol. 50, 249–257. [DOI] [PubMed] [Google Scholar]
- 10.Bulmer, M. (1991) J. Mol. Evol. 33, 305–310. [DOI] [PubMed] [Google Scholar]
- 11.Francino, M. P. & Ochman, H. (2000) Mol. Biol. Evol. 17, 416–422. [DOI] [PubMed] [Google Scholar]
- 12.Gierlik, A., Kowalczuk, M., Mackiewicz, P., Dudek, M. R. & Cebrat, S. (2000) J. Theor. Biol. 202, 305–314. [DOI] [PubMed] [Google Scholar]
- 13.Ladenburger, E. M., Keller, C. & Knippers, R. (2002) Mol. Cell. Biol. 22, 1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taira, T., Iguchi-Ariga, S. M. & Ariga, H. (1994) Mol. Cell. Biol. 14, 6386–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller, C., Ladenburger, E. M., Kremer, M. & Knippers, R. (2002) J. Biol. Chem. 277, 31430–31440. [DOI] [PubMed] [Google Scholar]
- 16.Vassilev, L. & Johnson, E. M. (1990) Mol. Cell. Biol. 10, 4899–4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nenguke, T., Aladjem, M. I., Gusella, J. F., Wexler, N. S. & Arnheim, N. (2003) Hum. Mol. Genet. 12, 1021–1028. [DOI] [PubMed] [Google Scholar]
- 18.Araujo, F. D., Knox, J. D., Ramchandani, S., Pelletier, R., Bigey, P., Price, G., Szyf, M. & Zannis-Hadjopoulos, M. (1999) J. Biol. Chem. 274, 9335–9341. [DOI] [PubMed] [Google Scholar]
- 19.Giacca, M., Zentilin, L., Norio, P., Diviacco, S., Dimitrova, D., Contreas, G., Biamonti, G., Perini, G., Weighardt, F., Riva, S., et al. (1994) Proc. Natl. Acad. Sci. USA 91, 7119–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitsberg, D., Selig, S., Keshet, I. & Cedar, H. (1993) Nature 366, 588–590. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz, S., Zhang, Z., Frazer, K. A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R. & Miller, W. (2000) Genome Res. 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arneodo, A., Audit, B., Decoster, N., Muzy, J. F. & Vaillant, C. (2002) in The Science of Disaster (Springer, Berlin), pp. 27–102.
- 23.Nicolay, S., Brodie of Brodie, E.-B., Touchon, M., d'Aubenton-Carafa, Y., Thermes, C. & Arneodo, A. (2004) Physica A 342, 270–280. [Google Scholar]
- 24.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 25.Kapranov, P., Cawley, S. E., Drenkow, J., Bekiranov, S., Strausberg, R. L., Fodor, S. P. & Gingeras, T. R. (2002) Science 296, 916–919. [DOI] [PubMed] [Google Scholar]
- 26.Chen, J., Sun, M., Lee, S., Zhou, G., Rowley, J. D. & Wang, S. M. (2002) Proc. Natl. Acad. Sci. USA 99, 12257–12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rinn, J. L., Euskirchen, G., Bertone, P., Martone, R., Luscombe, N. M., Hartman, S., Harrison, P. M., Nelson, F. K., Miller, P., Gerstein, M., et al. (2003) Genes Dev. 17, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kampa, D., Cheng, J., Kapranov, P., Yamanaka, M., Brubaker, S., Cawley, S., Drenkow, J., Piccolboni, A., Bekiranov, S., Helt, G., et al. (2004) Genome Res. 14, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imanishi, T., Itoh, T., Suzuki, Y., O'Donovan, C., Fukuchi, S., Koyanagi, K. O., Barrero, R. A., Tamura, T., Yamaguchi-Kabata, Y., Tanino, M., et al. (2004) PLoS Biol. 2, e162.15103394 [Google Scholar]
- 30.Dermitzakis, E. T., Reymond, A., Lyle, R., Scamuffa, N., Ucla, C., Deutsch, S., Stevenson, B. J., Flegel, V., Bucher, P., Jongeneel, C. V., et al. (2002) Nature 420, 578–582. [DOI] [PubMed] [Google Scholar]
- 31.Girard-Reydet, C., Gregoire, D., Vassetzky, Y. & Mechali, M. (2004) Gene 332, 129–138. [DOI] [PubMed] [Google Scholar]
- 32.Vassilev, L. T., Burhans, W. C. & DePamphilis, M. L. (1990) Mol. Cell. Biol. 10, 4685–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santamaria, D., Viguera, E., Martinez-Robles, M. L., Hyrien, O., Hernandez, P., Krimer, D. B. & Schvartzman, J. B. (2000) Nucleic Acids Res. 28, 2099–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Little, R. D., Platt, T. H. & Schildkraut, C. L. (1993) Mol. Cell. Biol. 13, 6600–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Codlin, S. & Dalgaard, J. Z. (2003) EMBO J. 22, 3431–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White, E. J., Emanuelsson, O., Scalzo, D., Royce, T., Kosak, S., Oakeley, E. J., Weissman, S., Gerstein, M., Groudine, M., Snyder, M., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 17771–17776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huberman, J. A. & Riggs, A. D. (1968) J. Mol. Biol. 32, 327–341. [DOI] [PubMed] [Google Scholar]
- 38.Yurov, Y. B. & Liapunova, N. A. (1977) Chromosoma 60, 253–267. [DOI] [PubMed] [Google Scholar]
- 39.Berezney, R., Dubey, D. D. & Huberman, J. A. (2000) Chromosoma 108, 471–484. [DOI] [PubMed] [Google Scholar]
- 40.Callan, H. G. (1972) Proc. R. Soc. London 181, 19–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.