Abstract
HIV-1 protease is an effective target for the design of drugs against AIDS. To help this process of drug design, three-dimensional structures have been determined of complexes between HIV-1 protease and a variety of transition-state analogue inhibitors. The true transition state, however, has not been structurally characterized. The crystal structure of the C95M/C1095A HIV-1 protease tethered dimer shows a distinctive feature in which the two flaps of the enzyme are in a ‘closed conformation’ even in the unliganded state. This unique feature has been utilized here to study the structure of HIV-1 protease complexed to an oligopeptide substrate of amino acid sequence His-Lys-Ala-Arg-Val-Leu*NPhe-Glu-Ala-Nle-Ser (where* denotes the cleavage site, and NPhe and Nle denote p-nitrophenylalanine and norleucine residues respectively). The X-ray structure of the complex refined against 2.03 Å (0.203 nm) resolution synchrotron data shows that the substrate is trapped as a tetrahedral reaction intermediate in the crystal. The hydrogen-bonding interactions between the reaction intermediate and the catalytic aspartates are different from those observed previously using transition-state analogues. The reaction intermediate did not dissociate to release the products, possibly due to the inflexibility introduced in the flaps when the enzyme is packed inside crystals.
Keywords: catalysis, HIV-1 protease, inhibitor, reaction intermediate, transition state, X-ray crystallography
Abbreviations: HAART, highly active antiretroviral therapy
INTRODUCTION
Although HIV/AIDS was discovered two decades ago, a cure is still not imminent [1]. The current strategy is to improve the quality of life of infected individuals through suppressing viral replication and maintaining the virus at low to undetectable levels using HAART (highly active antiretroviral therapy) [2–4]. Protease inhibitors are essential components of most HAART protocols, and are often the first line of treatment of infected patients. Currently there are eight protease inhibitors that have been approved by U.S. FDA (Food and Drug Administration) as drugs against AIDS, and all of these drugs have resulted from structure-based drug design efforts [5]. However, drug-resistant mutations in HIV-1 protease present a new challenge to future structure-based drug design efforts, and additional input, in terms of understanding the interactions between an active enzyme and a true polypeptide substrate, are required to overcome the problem of drug-resistant mutations. However, the basic difficulty is that a true substrate cannot be crystallized with an active enzyme, due to substrate conversion before crystals are grown and data collected [6]. One possible solution is to render the enzyme inactive by mutating the catalytic aspartate residues. The crystal structures of six complexes between peptide substrates and such a mutant inactive D25N HIV-1 protease have been reported [7]. However, the question arises as to whether the interactions observed with the substrate at the catalytic site of the inactive enzyme represent what actually occurs in the active enzyme.
To address this question, we have determined the crystal structure of a complex of an active HIV-1 protease with a cleavable peptide substrate. In this structure, refined to 2.03 Å resolution (where 1 Å=0.1 nm), we find that the substrate is not cleaved, but is trapped as a tetrahedral reaction intermediate. We also find that the interactions of the intermediate with the catalytic aspartates of the active enzyme are significantly different from those of the substrate with inactive enzyme. Interesting interactions at the catalytic centre observed here should help to improve our understanding of the reaction catalysed by HIV-1 protease. Tetrahedral reaction intermediates have been observed only recently in the crystal structures of serine proteases, human dipeptidyl peptidase IV [8,9] and porcine pancreatic elastase [10]. The present structure is the first report of a reaction intermediate in the case of HIV-1 protease.
MATERIALS AND METHODS
Protease assay
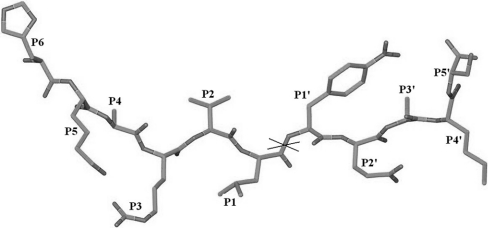
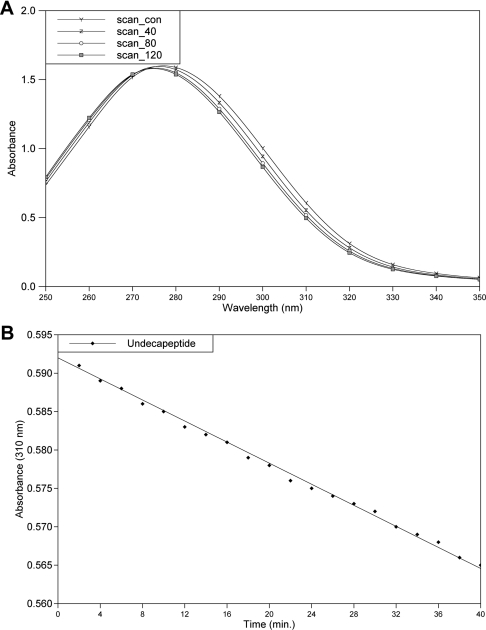
An 11-residue peptide of amino acid sequence His-Lys-Ala-Arg-Val-Leu*NPhe-Glu-Ala-Nle-Ser (where * denotes the cleavage site, and NPhe and Nle denote p-nitrophenylalanine and norleucine residues respectively) (Figure 1), which acts as a substrate for HIV-1 protease, was synthesized and purified at the National Institute for Research in Reproductive Health, Parel, Mumbai, India. The spectrophotometric assay [11] of this peptide was performed using a Shimadzu UV-160 spectrophotometer. A stock solution of the peptide substrate was prepared in distilled water, and the enzyme was present in 50 mM sodium acetate buffer (pH 4.5) containing 5 mM 2-mercaptoethanol. The reaction was carried out by incubating the substrate (190 μM) with the enzyme at room temperature. UV absorption spectra for the wavelength range 250–350 nm were recorded for the substrate and for the reaction mixture (substrate+enzyme) at 40, 80 and 120 min after the start of the reaction (Figure 2A). A decrease in absorbance at 310 nm resulting from hydrolysis of the scissile peptide bond was monitored for 40 min (Figure 2B). As a control, absorbance at 310 nm was measured for the substrate alone for the same period of time. The almost linear decrease in absorbance at 310 nm, and the shift in the absorption maximum to a lower wavelength, demonstrate that the undecapeptide used is being cleaved by the protease used here for structural studies.
Figure 1. The 11-residue peptide substrate of HIV-1 protease used in soaking experiments.
The scissile peptide bond is indicated by a cross (‘X’).
Figure 2. Spectrophotometric assay of the substrate peptide.
(A) Time course (120 min) of changes in the UV absorption spectra of the undecapeptide substrate. Scan_con spectrum is for the substrate alone. Scan_40, Scan_80 and Scan_120 spetra were recorded at 40, 80 and 120 min from the start of the reaction (addition of the protease). The absorption maximum is seen to be shifted from 280 to 275.2 nm after 120 min. (B) Plot of the decrease in absorbance (0–40 min) at 310 nm and the least-squares-fitted line.
Crystallization and soaking experiments
The cloning, site-directed mutagenesis, expression, purification and crystallization of the HIV-1 protease tethered dimer has been reported previously [12–14]. Briefly, the cloned gene was expressed into inclusion bodies, to yield a 29 kDa gene product, which corresponds to the 22 kDa HIV-1 protease tethered dimer with an extension of 54 amino acids on the N-terminal side. The N-terminal extension is autocleaved during renaturation. Crystals of a C95M/C1095A mutant HIV-1 protease tethered dimer were grown by the hanging-drop vapour diffusion method. The protein was present at a concentration of 5 mg/ml in 50 mM sodium acetate buffer, pH 4.5, containing 1 mM DTT. The reservoir solution was 5% saturated ammonium sulphate in 0.2 M sodium phosphate/0.1 M sodium citrate buffer, pH 6.2. Aliquots of 2 μl of protein solution and 2 μl of reservoir solution were mixed together and placed on a siliconized coverslip, which was then inverted and sealed with vacuum grease over a well containing 800 μl of the reservoir solution. Diffraction-quality crystals, which were obtained in 2–3 days, grew in the form of hexagonal rods of size 0.3 mm×0.05 mm×0.05 mm.
Lyophilized powder of the peptide was dissolved in 10% (v/v) acetic acid to give a 5 mM concentration of the peptide. For soaking experiments, 10 μl of this peptide solution was placed on a coverslip, and a protease crystal was transferred into this solution using a loop. The coverslip was inverted and sealed over the same reservoir well where the crystals had appeared. The crystal was incubated in the peptide solution for 1 week at room temperature before it was used for data collection.
X-ray data collection
The soaked crystal was equilibrated for 5 min in cryoprotectant solution (25% glycerol and 75% reservoir solution) before collection of diffraction data at 100 K on a beam line I711 instrument, at MAX-II Synchrotron, Lund University, Sweden. A total of 150 images were collected, each of 0.5° oscillation and 2 min of exposure. The crystals diffracted to 2.0 Å resolution.
The diffraction data were indexed and integrated using the program DENZO, and scaled using the program SCALEPACK of the HKL program suite [15]. The final statistics are given in Table 1.
Table 1. Reflection data statistics.
| Parameter | Value |
|---|---|
| Resolution | 2.03 Å |
| Space group | P61 |
| Unit cell parameters | a=b=63.06 Å; c=83.34 Å |
| Total number of reflections measured | 235151 |
| Number of unique reflections | 11960 |
| Completeness to highest resolution | 94.0% |
| Completeness in the highest-resolution shell | 80.5% (in 2.03–2.15 Å shell) |
| Mosaicity | 0.54 |
| Rmerge | 5.6% |
| 〈I/σ〉 (all reflections) | 21.43 |
| 〈I/σ〉 (highest-resolution shell) | 4.70 (in 2.03–2.15 Å shell) |
Refinement of the structure
The structure was refined in the software suite CNS (Crystallography and NMR System) using a maximum likelihood target function involving structure factor amplitudes and standard protocols [16]; 5% of the reflections were set aside for cross-validation. The motif refined was a 1:1 complex between the tethered protease dimer and the substrate oligopeptide molecule bound in the active-site cavity. The substrate was modelled in three different ways: (1) as a regular peptide uncleaved at the scissile bond, (2) as a peptide cleaved at the scissile bond with the two product peptides bound in the active site, and (3) as an uncleaved hydrated peptide with the scissile carbonyl converted into a gem-diol. Thus the three models differ from each other only at the scissile peptide linkage. Each of these three models for the complex between HIV-1 protease and the substrate was refined separately. The tetrahedral intermediate model refined to an R-factor of 21.51% with an Rfree of 29.02%. The number of reflections and the R-factor in different resolution shells are shown in Table 2. The final model in the asymmetric unit consists of the HIV-1 protease dimer, one substrate molecule and 207 water molecules. The root mean square deviations from the ideal values for bond length and bond angle were 0.016 Å and 1.91° respectively. The entire model building and structural superposition were carried out using the software O [17]. For structural comparisons with other substrate–inhibitor complexes, only the protein Cα atoms were used in the superposition.
Table 2. R-factors for the reaction intermediate model in different resolution shells.
| Bin | Resolution range | No. of reflections | R-factor |
|---|---|---|---|
| 1 | 20.00–4.43 | 1076 | 0.1931 |
| 2 | 4.43–3.53 | 1086 | 0.1846 |
| 3 | 3.53–3.08 | 1066 | 0.2132 |
| 4 | 3.08–2.80 | 1065 | 0.2182 |
| 5 | 2.80–2.60 | 1073 | 0.2290 |
| 6 | 2.60–2.45 | 1089 | 0.2282 |
| 7 | 2.45–2.32 | 979 | 0.2831 |
| 8 | 2.32–2.22 | 1011 | 0.2692 |
| 9 | 2.22–2.14 | 1036 | 0.2602 |
| 10 | 2.14–2.06 | 1008 | 0.2798 |
| 11 | 2.06–2.03 | 469 | 0.2591 |
| Overall | 20.00–2.03 | 10958 | 0.2151 |
RESULTS AND DISCUSSION
HIV-1 protease is a homodimer in which each monomer contains 99 amino acid residues; in the three-dimensional structure, the C-terminus of the first monomer and the N-terminus of the second monomer are separated by less than 5 Å. The tethered dimer of HIV-1 protease used in the present study is a single polypeptide chain where the C-terminus of the first monomer is linked to the N-terminus of the second monomer through a pentapeptide (-GGSSG-) linker. The enzyme activity of such a construct is comparable with that of the native homodimer [12]. In the tethered dimer, we have numbered the residues in the first monomer 1–99, and those in the second monomer 1001–1099. Crystal structures of a functional HIV-1 protease tethered dimer (PDB ID: 1LV1 and 1G6L), as reported previously [13,14,18], showed that the two flaps are in ‘closed conformation’ even when no ligands are bound in the active-site cavity. This observation presented us with a unique opportunity whereby any potential inhibitor could be soaked in these crystals efficiently without the need for their co-crystallization to elucidate the three-dimensional structure of the complex [19]. Here we have utilized this feature to study the complex between HIV-1 protease and a true substrate oligopeptide. An 11-residue peptide (Figure 1) was used for the soaking experiments. Since the soaked crystals were isomorphous with those of the unliganded enzyme (a=b=63.06 Å, c=83.42 Å), the co-ordinates of the unliganded enzyme (PDB ID: 1LV1) were used directly for refinement and Fourier map calculations.
Nature and modelling of the ‘substrate’ in the electron density map
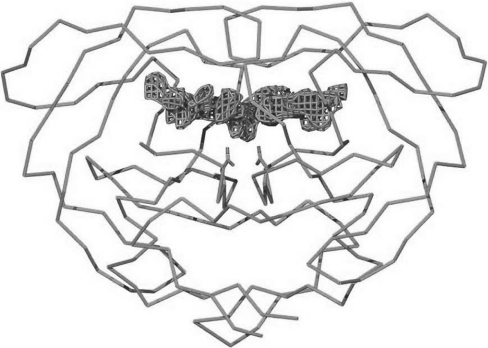
The Fo–Fc map showed good electron density in the active site (Figure 3), corresponding to the bound substrate. This difference electron density in the active site was almost symmetrical around the two-fold symmetry axis of the HIV-1 protease dimer, suggesting that the substrate peptide was bound in two orientations, related to each other by rotation around the two-fold symmetry axis of the protease dimer. As mentioned in the Materials and methods section, three different scenarios were considered for the atomic model of the substrate: (1) the substrate was bound uncleaved as a normal peptide, (2) the substrate was cleaved and the products were bound in the active site, and (3) the substrate was trapped as a tetrahedral reaction intermediate in the active-site cavity. When the substrate was modelled and refined as a regular peptide, there was good electron density for the P1 and P1′ residues of the substrate, and the Cα atoms of these residues could be placed unambiguously. However, the planar peptide between the P1 and P1′ residues did not fit properly, being at the edge of the observed electron density. Even after a series of careful model building and refinement exercises, the fit could not be improved (Figure 4a). We then realized that refinement brings the atoms outside the density to make the peptide group between the P1 and P1′ residues planar.
Figure 3. Difference electron density in the active site when the Fo–Fc map is calculated without using the substrate co-ordinates.
The map is contoured at 3σ. The Cα trace of the HIV-1 protease dimer and the side-chain atoms of active-site aspartates are also shown.
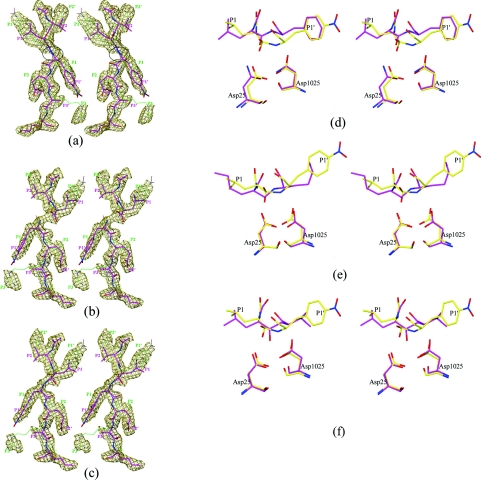
Figure 4. The tetrahedral peptidolysis intermediate.
(a)–(c) Stereo diagrams of 2Fo–Fc maps (contoured at 0.8σ). (a) The substrate is refined as a regular peptide. The region of the model between the P1 and P1′ residues, which does not fit properly in the density, is circled. (b) The substrate is refined as a cleaved peptide; (c) the substrate is refined as a reaction intermediate. Two orientations of the substrate are shown with magenta and green carbon atoms; sticks in the magenta model have been made thicker to allow easy tracing of the peptide chain. (d)–(f) Stereo diagrams of structural superposition of the reaction intermediate (yellow carbon) on to: (d) the regular peptide from the structure 1kjh (PDB code) (magenta carbon), (e) the reduced amide inhibitor MVT101 from the structure 4hvp (magenta carbon) and (f) the hydroxyethylene inhibitor U85548E from the structure 8hvp (magenta carbon). Only one orientation of the substrate is shown for the sake of clarity. Similar features are also present in the other orientation. Only protein Cα atoms were used for superposition.
Since the crystals of an inherently active enzyme were soaked in a substrate that was cleavable by the enzyme, it was thought that the substrate may have been cleaved in the crystal, and therefore the scenario of a cleaved peptide was considered next. We started afresh with the co-ordinates of the unliganded enzyme, to get rid of model bias, and tried to build a substrate model in which the peptide bond between the P1 and P1′ residues was cleaved. After a few cycles of refinement and model building, most of the residues of the two products fitted nicely in the electron density map. Good electron density for the side-chain atoms of the P1 and P1′ residues dictated the position of Cα atoms of these residues (Figure 4b). However, this cleaved-peptide model has the following drawbacks: (a) it doesn't account for the continuous electron density between the P1 and P1′ residues, and (b) the distance between the C-atom of the P1 residue and the N-atom of the P1′ residue was 2.48 Å, which is much shorter than the required van der Waals separation. These features suggested that the species present in the active site was not a cleaved peptide.
We then considered the possibility of a reaction intermediate being present in the crystal. In this tetrahedral reaction intermediate, two oxygen atoms are attached to the carbonyl carbon of the scissile peptide bond. For the purposes of crystallographic refinement, the residues at positions P1 and P1′ were grouped together as a single residue, named ‘PHL’, with two oxygen atoms attached to the central carbon. The force constants in the CNS (Crystallography and NMR System) parameter file for this residue were relaxed at the scissile peptide bond, so that the geometry of the reaction intermediate was guided primarily by the experimental electron density. After a few cycles of refinement, the reaction intermediate model fitted better in the electron density, as shown in Figure 4(c). The C–N distance of the scissile peptide bond converged to 1.65 Å. The crystallographic R-factors obtained for the three models are shown in Table 3. The water molecules included in these three refinement models were the same. Since the difference in the three models is limited to only a few atoms around the scissile bond, one cannot expect to see large differences in R-factors. Both Rwork and Rfree were slightly lower for the reaction intermediate model of the substrate. Thus the R-values also give an indication that the species present in the crystal is a reaction intermediate.
Table 3. Final R-factors for different models of the substrate used in the refinement.
| Substrate model | R-factor (%) | Rfree (%) |
|---|---|---|
| Regular peptide | 22.17 | 29.61 |
| Cleaved peptide | 22.08 | 29.37 |
| Tetrahedral intermediate | 21.51 | 29.02 |
Trapping of a reactive intermediate
HIV-1 protease catalyses the peptide hydrolysis reaction through nucleophilic attack by a water molecule on the carbonyl carbon of the scissile peptide bond [20]. This water molecule is believed to be the one that is hydrogen-bonded to both of the catalytic aspartates in the unliganded enzyme structure. When the substrate peptide moves into the active site of the enzyme and is oriented suitably, the water molecule attacks the carbon atom of the scissile peptide bond. The carbon atom then becomes tetravalent, with two oxygens attached to it. This leads to a highly strained structure, and observation of such a species outside the enzyme's pocket would, in general, not be possible. However, when the entire substrate is enveloped by the enzyme, as is the case here, such a strained substrate structure would be stabilized by many different interactions with the enzyme. Stabilization of such a reaction intermediate by the enzyme is the key to rate enhancement in a catalytic reaction [21]. The natural question that arises here is: if a reaction intermediate is formed in the crystal, why does it not dissociate to form the products? The answer possibly lies in the flap regions of the enzyme. The dynamics of the flaps are very important for the functioning of this enzyme [22]. However, the flap dynamics (flexibility) are lost in the present hexagonal crystal, where flaps from symmetry-related molecules are in van der Waals contact. The rigidity thus introduced is also reflected by well-ordered electron density for the flaps even in unliganded hexagonal crystal structures [13,18]. Uncleaved reaction intermediates bound in the active site have been observed in crystals of acyl–enzyme complexes of serine proteases [8–10].
Conformation of the reaction intermediate
In the crystal structures of complexes between substrate oligopeptides and the inactive mutant D25N HIV-1 protease, the substrate is retained as a regular peptide [7]. Because of the hydration of the substrate peptide in the present crystal, the conformation of the reaction intermediate was significantly different from that of a normal peptide, especially in the region P1–P1′ (Figure 4d). It may be seen that the major change is in the orientation of the scissile amide proton relative to the catalytic aspartates. While this proton is pointing away from the aspartates in the unproductive regular peptide complex [7], it is pointing towards the aspartates in the reaction intermediate model (Figure 4d). In fact, there is a strong hydrogen bond between the scissile nitrogen atom and the outer oxygen of one of the aspartates (O…N distance 2.80 and 2.90 Å). This change is caused largely by the decrease in the torsion angle about the scissile C–N bond, from a value of 180° in the regular peptide to 56° in the reaction intermediate model. Figures 4(e) and 4(f) show structural comparisons of the present tetrahedral reaction intermediate with reduced amide (MVT101) and hydroxyethylene (U85548E) transition-state analogue inhibitors. It may be seen that, even in the case of the reduced amide compound, the amide proton is not pointing towards the catalytic aspartates. In the case of the hydroxyethylene compound, the hydroxy group is located symmetrically between the two aspartates, unlike in the present structure. Thus there are significant differences both in the conformation and in the interactions with the catalytic aspartates of the reaction intermediate and the transition-state analogues.
Hydrogen-bonding interactions in the active site
It is well known that rate enhancement in enzyme-catalysed reactions is due mainly to a higher binding affinity of the enzyme to the transition-state analogue as compared with an unaltered substrate. We have therefore compared the hydrogen-bonding interactions observed in the active site of the present structure and those observed in D25N protease–substrate complexes [7]. The hydrogen bonds can be divided into two groups: group 1, involving residues P1–P1′, binding to catalytic aspartates (Figure 5), and group 2, involving substrate residues farther away from the scissile bond (Figure 6).
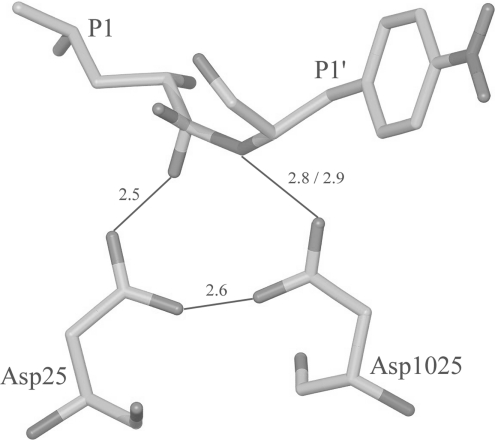
Figure 5. Hydrogen-bonding interactions at the catalytic site.
Marked distances are in Å.
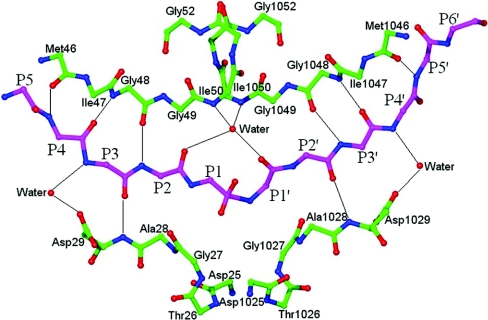
Figure 6. Hydrogen-bonding interactions between main-chain atoms of the substrate and the protease molecule.
Side-chain atoms are not shown for most of the residues for the sake of clarity.
In the present structure, there are eight group 2 hydrogen bonds between the main-chain atoms of the substrate residues and the main-chain atoms of the protein residues (Figure 6). Of these eight, four are on the unprimed side, involving protein atoms 46CO, 48NH, 29NH and 48CO from the first monomer, and four are on the primed side, involving the same protein atoms from the other monomer. One water molecule in the active site, popularly known as the flap water, links the reaction intermediate to the protein through four hydrogen bonds, two to the flap amide groups of Ile-50 and Ile-1050, and two to the CO groups of residues P2 and P1′ from the reaction intermediate. The location and geometry of these group 2 hydrogen bonds is very similar to what has been observed in the crystal structures of inactive D25N mutant HIV-1 protease complexed with oligopeptide substrates [7]. There are, however, some additional water-mediated group 2 hydrogen bonds that are observed only in the present structure. The NH groups of the residues at positions P3 and P4′ are hydrogen-bonded to water molecules, which in turn are hydrogen-bonded to the carboxy oxygen atom of side-chain Asp-29/1029 from the enzyme (Figure 6).
There are differences in the group 1 hydrogen bonds observed in the present structure and in the inactive protease–substrate complexes [7]. Based on the distance criteria alone, there are two strong hydrogen bonds involving catalytic aspartates: (a) between the O1 atom of the tetrahedral intermediate and the outer oxygen atom (OD1) of one of the catalytic aspartates (distance 2.5 Å), and (b) between the substrate nitrogen and the outer oxygen (OD1*) of the second catalytic aspartate (distance 2.8 Å). In contrast, in the inactive enzyme complex, there is only one hydrogen bond at the catalytic centre, and that is between the scissile carbonyl oxygen and the outer carboxyl oxygen of the catalytic aspartate. Further, the hydrogen bond to the catalytic aspartate in the present structure is much shorter and therefore is likely to be stronger. These differences in the number and strength of both group 1 and group 2 hydrogen bonds for altered and unaltered substrates might contribute to the higher affinity of the transition-state intermediates for the enzyme.
Substrate-induced changes in protease structure
To identify the changes in protease structure brought about by substrate binding, the structure of the protease was least-squares superposed on the unliganded structure using the software O [17]. The root mean square deviation for 198 Cα atom pairs was 0.29 Å. To identify significant changes, error-scaled difference distance matrix plots were also examined (results not shown). Only minor changes in the protease structure were observed on substrate binding, and these were located mainly around the P1/P1′ site of the substrate. The changes in the two monomers of the protease were very similar. To accommodate the P1 residue of the substrate, the main-chain atoms of residues 78–84 moved by an average of 0.46 Å. The Cα atom of Gly-48 from the flaps was shifted by 0.35 Å. The side-chain atoms of Arg-8 also shifted to accommodate the P1/P1′ residue of the substrate. The shift of the Cz atom of Arg-8 was 0.70 Å. Other changes seen in the protease structure were in flexible regions, which had relatively high temperature factors.
Of the six water molecules in the active site of the unliganded protease, four were replaced in the complex structure by the substrate. The water molecule that was hydrogen-bonded to both of the catalytic aspartates in unliganded structure [13,18], and believed to be involved in catalysis, is missing from the present structure. It is plausible that this water molecule has attacked the carbonyl carbon of the scissile peptide bond of the substrate to form the reaction intermediate.
Significance of the reaction intermediate structure
To understand the chemistry of an enzyme-catalysed reaction, it is important to know the atomic dispositions of the substrate in the transition state. The geometry of the transition-state intermediate carries information about how exactly the reactants could have acted to form this intermediate and, further, how exactly this intermediate will dissociate to form the products. However, the transition state is inherently transient in nature, and it is not always possible to obtain information about the atomic dispositions in this state. However, in an elegant experiment, a tetrahedral reaction intermediate has been observed in crystallographic studies on porcine pancreatic elastase, where the reaction in the crystals of the acyl–enzyme complex was triggered by an increase in pH [10]. More recently, a tetrahedral intermediate has been observed in medium-resolution crystallographic studies of human dipeptidyl peptidase IV by two groups [8,9]. In all of these structures, the enzyme is a serine protease and the observed tetrahedral intermediate is covalently bound to the enzyme. However, in the structure reported in the present study, the tetrahedral intermediate is non-covalently bound to the enzyme. The ‘closed-flap’ structure of unliganded HIV-1 protease seems to have provided us with an opportunity to trap this non-covalently bound tetrahedral intermediate in the crystals. It is generally accepted [20] that peptide bond cleavage by HIV-1 protease involves two steps: (1) formation of tetrahedral intermediate through nucleophilic attack on the scissile carbonyl carbon, and (2) protonation of the scissile amide nitrogen. However, what is not established is whether these two steps occur simultaneously or sequentially. The structure presented here provides direct experimental support for the formation of a tetrahedral intermediate and for the view that the two steps are sequential. The structure also identifies the water molecule that is involved in the nucleophilic attack.
In addition, as a substrate in the transition state can often possess 1000-fold higher affinity for the enzyme than a substrate in the ground state [21], most effective inhibitors of HIV-1 protease use the concept of transition-state mimics. However, the structure and interactions of a true transition-state substrate with the protease is not known. The present structure shows that the interactions with HIV-1 protease of a true reaction intermediate and of transition-state analogues are significantly different. The structure of the reaction intermediate reported here thus could also provide a valuable reference point for guiding the design of tighter-binding inhibitors.
Acknowledgments
We thank the National Facility for Macromolecular Crystallography, SSPD, BARC, for the X-ray diffraction and biochemistry equipment. We thank Dr Derek Logan and Yngve Cerenius for their help in collecting data on the I711 beamline on the MAX-II synchrotron in Lund, Sweden. We thank Dr R. Chidambaram, Dr K. K. Kannan and Dr Bindu Pillai for scientific discussions, and S. R. Jadhav for technical help. M.V.H. thanks Anders Liljas and the Wenner Gren Research Foundation for the award of a visiting fellowship.
References
- 1.Heagarty M. C. AIDS: The battle rages on. Issues in Science and Technology Online. 2003 Summer 2003, http://www.nap.edu/issues/19.4/heagarty.html#top.
- 2.Porter K., Babiker A., Bhaskaran K., Darbyshire J., Pezzotti P., Porter K., Walker A. S. (CASCADE Collaboration) Determinants of survival following HIV-1 seroconversion after the introduction of HAART. Lancet. 2003;362:1267–1274. doi: 10.1016/s0140-6736(03)14570-9. [DOI] [PubMed] [Google Scholar]
- 3.Carrieri P., Spire B., Duran S., Katlama C., Peyramond D., Francois C., Chene G., Lang J. M., Moatti J. P., Leport C. APROCO Study Group. Health-related quality of life after 1 year of highly active antiretroviral therapy. J. Acquired Immune Defic. Syndr. 2003;32:38–47. doi: 10.1097/00126334-200301010-00006. [DOI] [PubMed] [Google Scholar]
- 4.Lampe F. C., Johnson M. A., Lipman M., Loveday C., Youle M., Ransom D., Sabin C. A., Tyrer M., Phillips A. N. Viral breakthrough after suppression with highly active antiretroviral therapy: experience from 233 individuals with viral loads of less than 50 copies/ml followed for up to 4 years. AIDS. 2003;17:759–777. doi: 10.1097/01.aids.0000050876.71999.11. [DOI] [PubMed] [Google Scholar]
- 5.Wlodawer A., Vondrasek J. Inhibitors of HIV-1 protease: a major success of structure-assisted drug design. Annu. Rev. Biophys. Biomol. Struct. 1998;27:249–284. doi: 10.1146/annurev.biophys.27.1.249. [DOI] [PubMed] [Google Scholar]
- 6.Rose R. B., Craik C. S., Douglas N. L., Stroud R. M. Three-dimensional structures of HIV-1 and SIV protease product complexes. Biochemistry. 1996;35:12933–12944. doi: 10.1021/bi9612733. [DOI] [PubMed] [Google Scholar]
- 7.Jeyabalan M. P., Nalivaika E., Schiffer C. A. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure. 2002;10:369–381. doi: 10.1016/s0969-2126(02)00720-7. [DOI] [PubMed] [Google Scholar]
- 8.Aertgeerts K., Ye S., Tennant M. G., Kraus M. L., Rogers J., Sang B. C., Skene R. J., Webb D. R., Prasad G. S. Crystal structure of human dipeptidyl peptidase IV in complex with a decapeptide reveals details on substrate specificity and tetrahedral intermediate formation. Protein Sci. 2004;13:412–421. doi: 10.1110/ps.03460604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thoma R., Loffler B., Stihle M., Huber W., Ruf A., Hennig M. Structural basis of proline-specific exopeptidase activity as observed in human dipeptidyl peptidase-IV. Structure. 2003;11:947–959. doi: 10.1016/s0969-2126(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 10.Wilmouth R. C., Edman K., Neutze R., Wright P. A., Clifton I. J., Schneider T. R., Schofield C. J., Hajdu J. X-ray snapshots of serine protease catalysis reveal a tetrahedral intermediate. Nat. Struct. Biol. 2001;8:689–694. doi: 10.1038/90401. [DOI] [PubMed] [Google Scholar]
- 11.Tomaszek T. A., Jr, Magaard V. W., Bryan H. G., Moore M. L., Meek T. D. Chromophoric peptide substrates for the spectrophotometric assay of HIV-1 protease. Biochim. Biophy. Res. Commun. 1990;168:274–280. doi: 10.1016/0006-291x(90)91704-v. [DOI] [PubMed] [Google Scholar]
- 12.Cheng Y.-S. E., Yin F. H., Foundling S., Blomstrom D., Kettner C. A. Stability and activity of human immunodeficiency virus protease: comparison of a natural dimer with a homologous single chain tethered dimer. Proc. Natl. Acad. Sci. U.S.A. 1990;87:9660–9664. doi: 10.1073/pnas.87.24.9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar M., Kannan K. K., Hosur M. V., Bhavesh N. S., Chatterjee A., Mittal R., Hosur R. V. Effects of remote mutation on the autolysis of HIV-1 PR: X-ray and NMR investigations. Biochem. Biophys. Res. Commun. 2002;294:395–401. doi: 10.1016/S0006-291X(02)00482-5. [DOI] [PubMed] [Google Scholar]
- 14.Kumar M., Hosur M. V. Adaptability and flexibility of HIV-1 protease. Eur. J. Biochem. 2003;270:1231–1239. doi: 10.1046/j.1432-1033.2003.03483.x. [DOI] [PubMed] [Google Scholar]
- 15.Otwinowski Z. Oscillation data reduction program. In: Swyer L., Isaacs N., Baily S., editors. Proceedings of the CCP4 Study Weekend: Data Collection and Processing. Warrington, U.K.: SERC Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 16.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J.-S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography and NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 17.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 18.Pillai B., Kannan K. K., Hosur M. V. 1.9 Å X-ray study shows closed flap conformation in crystals of tethered HIV-1 PR. Proteins. 2001;43:57–64. doi: 10.1002/1097-0134(20010401)43:1<57::aid-prot1017>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 19.Pillai B., Bhat S. V., Kannan K. K., Hosur M. V. Rapid screening for HIV-1 protease inhibitor leads through X-ray diffraction. Acta Crystallogr. D Biol. Crystallogr. 2004;60:594–596. doi: 10.1107/S0907444903029676. [DOI] [PubMed] [Google Scholar]
- 20.Dunn B. M. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem. Rev. 2002;102:4431–4458. doi: 10.1021/cr010167q. [DOI] [PubMed] [Google Scholar]
- 21.Wolfenden R. Conformational aspects of inhibitor design: enzyme-substrate interactions in the transition state. Bioorg. Med. Chem. 1999;7:647–652. doi: 10.1016/s0968-0896(98)00247-8. [DOI] [PubMed] [Google Scholar]
- 22.Nicholson L. K., Yamazaki T., Torchia D. A., Grzesiek S., Bax A., Stahl S. J., Kaufman J. D., Wingfield P. T., Lam P. Y., Jadhav P. K., et al. Flexibility and function in HIV-1 protease. Nat. Struct. Biol. 1995;2:274–280. doi: 10.1038/nsb0495-274. [DOI] [PubMed] [Google Scholar]