Abstract
The minimal requirements for a eukaryotic origin of replication are an initiator binding site and a region of helically unstable DNA [DNA unwinding element (DUE)]. Budding yeast origins consist of modular elements, and one of these elements, B2, has been proposed to act as a DUE. To test this hypothesis, we screened for sequences that function at the B2 element of ARS1. We found that the B2 element required A-rich sequences, but that the function of these identified sequences did not correlate with helical instability. Instead, the sequences that substituted fully for B2 function showed similarity to the ARS consensus sequence (ACS). The ACS is the binding site for the initiator origin recognition complex (ORC), but the selected sequences are not strong ORC binding sites in vitro. Nonfunctional B2 sequences show a corresponding loss in Mcm2-7p origin association. The function of these mutant sequences is rescued by Cdc6p overexpression. We propose that the B2 element requires specific sequences to bind a component of the pre-RC.
Origins of replication provide a site for DNA unwinding and the loading of the DNA replication machinery. The DNA sequences required for an origin of replication are referred to as a replicator and contain a binding site for an initiator protein combined with a structural DNA unwinding element (DUE) (1). The eukaryotic initiator is the origin recognition complex (ORC), which has been shown to bind origins in several species (2–4). DUEs have been identified at origins in many organisms (5–7) and are defined as helically unstable DNA sequences that can be replaced by any easily unwound DNA. Although an initiator binding site and a DUE appear to be present at all origins, other sequence elements are likely to be necessary for origin function.
Saccharomyces cerevisiae replicator sequences were identified based on their ability to confer autonomous replication on episomes and have been extensively characterized. Mutagenesis of these sequences revealed that they are composed of multiple 10- to 15-bp elements within a 100- to 150-bp region. These elements include the essential A element, containing the 11-bp autonomously replicating sequence (ARS) consensus sequence (ACS), ORC's binding site. ORC is bound to the origin throughout the cell cycle and is required to recruit the remaining DNA replication machinery to the origin. The A element is insufficient for origin function, and a variable number of B elements also contribute to function (8–11). B elements were identified by analysis of multiple origins and named based on their ability to functionally substitute for each other. The B1 element cooperates with the ACS to form ORC's bipartite binding site (54, 55). The function of the B2 element is unknown, but may include either binding a specific protein or acting as a site for initial unwinding of the origin DNA.
Whether the B2 element functions structurally or in a sequence-specific manner, significant evidence supports a role for this sequence in pre-replicative complex (pre-RC) formation. The pre-RC protects the A, B1, and B2 region of ARS1 in vivo during G1 (12) and contains ORC, Cdc6p, and the mini-chromosome maintenance (MCM) proteins (13–16). A linker scan mutation in the B2 element of ARS1 reduces MCM loading in vivo but has no effect on ORC binding (17, 18). The MCM proteins are loaded onto the origin by Cdc6p (14–16) in an ATP-dependent manner (19). In Schizosaccharomyces pombe, Xenopus laevis, and Drosophila melanogaster, Cdt1p also associates with Cdc6p and is required for MCM loading (20–22).
The B2 element has been proposed to act structurally as a DUE. Consistent with this model, the region of ARS305 that can functionally replace the B2 elements at ARS1 and ARS307 is over 50 bp long, and has no short sequence elements that can be identified by linker scan mutations. The ARS305 B2 region can be replaced with a random easily unwound sequence (11, 23). The much shorter 11-bp ARS1 B2 element also has some characteristics of a DUE, because single point mutations have no effect (9) and in vitro replication protein A-dependent unwinding of the ARS1 origin depends on the B2 element (24). Similarly, the shorter 20-bp ARS307 B2 element has been proposed to function as a DUE based on a correlation between the helical stability and replication defects of several B2 mutations (25).
Other evidence suggests that the B2 element has a function other than a DUE. At ARS307, a specific sequence requirement is suggested by a 3-bp mutation that has little effect on helical stability but a large effect on origin function (9, 25). In addition, the ARS1 B2 element cannot be inverted or replaced with a sequence of equal helical stability (25). Together, these data suggest that the B2 element has additional sequence requirements and binds one or more replication factors, such as components of the pre-RC.
Here, we describe a genetic screen to identify the sequence requirements of the ARS1 B2 element. Sequences that functioned as B2 elements were found to each include a partial inverted match to the ACS, and not simply helically unstable DNA. Despite their similarity to the ACS, these sequences were not specifically bound by ORC in vitro. Origin function of the mutant B2 sequences correlated with Mcm2-7p loading, and function of defective mutants was significantly rescued by overexpressing Cdc6p. We propose a model whereby all yeast origins require large DUEs with a sequence related to the ACS embedded within them to function as a binding site for a pre-RC component.
Materials and Methods
Plasmids, Strains, and Library Construction.
Strains (W303 background) and plasmids were prepared by using standard laboratory methods (26). Plasmids pARS1/785-92ADE2 and pARS1/802-810ADE2 were created by cloning the SalI/NcoI fragment of pASZ10 (27) containing the ADE2 gene into the SalI and NcoI sites of pARS1/785-92 and pARS1/802-810 (8), replacing part of the URA3 gene. The plasmid pARS1/785-92ADE2SIR3 was created by cloning the XhoI to PstI fragment of SIR3 into the XhoI to PstI sites of pARS1/785-92ADE2 to facilitate cloning of the library. The ARS1 B2 mutant library was created by primer extension of the primers B2MUT1 (TATTACCTCGAGGTATTTNNNNNNNNNNNGTTTGTGCACTTGCCTGCAGGCCTTTT) and B2MUT2 (AAAAGGCCTGCAGGCAAGTGC), synthesized by Integrated DNA Technologies (Coralville, IA), and cloned into the XhoI and PstI sites of pARS1/785-92ADE2SIR3. Strains used in chromatin immunoprecipitation (ChIP) were created as described (18). Cdc6p overexpression strains were created as described (28).
B2+ Selection and B2− Screen.
B2− plasmids were identified by streaking yeast colonies from a transformation with the library onto minimal complete media, growing for 2 days at 30°C, and then incubating for a week at 4°C to allow the red color indicative of an ade2 mutation to develop. Under these conditions, streaks from pARS1/785-92ADE2 transformants were white, streaks from pARS1/802-810ADE2 transformants were pink, and streaks from pARS1/785-92ADE2SIR3 transformants were red. Streaks were replica plated to plates lacking adenine, and plasmids were recovered from replicas of streaks that turned red on minimal complete media. B2+ plasmids were selected by growing pools of ≈1,000 yeast library transformants together in liquid media including supplemental adenine. The cultures were maintained in log phase for 10 days by dilution, and then plated onto media lacking adenine.
Plasmid Loss Assays.
Plasmid loss assays for origin function were performed as described (8), with the exception that selection was for the ADE2 gene, not URA3. Media were supplemented with 0.1 mg/ml adenine. Plasmid loss rates with adenine selection were consistently lower than those with uracil selection, but the relative measurements were the same (data not shown).
Calculations.
Helical stability calculations were done with the program webthermodyn (http://wings.buffalo.edu/gsa/dna/dk/DWEBTHERMODYN/; ref. 29), by using the data sets of Santalucia et al. (30). Calculations included the 11 bases of the B2 element plus one base on either side. Statistical analyses, random sequence generations, and calculations of matches to the ACS were done by the program excel (Microsoft). The means of different samples were compared with two-tailed Student t tests, assuming equal variance.
ChIP.
ChIP was performed as described (28), except PCR was performed for 28 cycles on 1/50th of the immunoprecipitation or 1/10,000th of the input. Quantitation was done with a CHEMIIMAGER 500 (Alpha Innotech, San Leandro, CA) and accompanying software.
DNase I Protection Assays.
DNase I protection assays were performed as described (31), except that all reactions contained 1.4 mM ATP. Baculovirus ORC, and ORC1-5p were purified as described previously (32).
Results
A Screen for Sequences That Affect Origin Function.
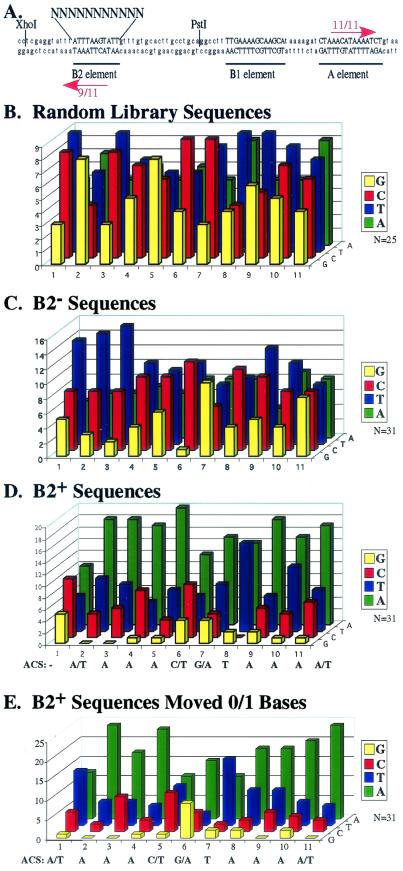
To determine the sequence requirements at the B2 element of S. cerevisiae origins, we took advantage of the well-defined B2 element of ARS1. Previous linker scan mutagenesis of ARS1 had identified B2 as a short 11-bp sequence (8) that can functionally substitute for the longer B2 elements at ARS307 and ARS305 (9, 25). To identify additional sequences that can function at this site, we constructed an ARS1 plasmid library with randomized sequences at the 11-bp B2 element (Fig. 1A). This library was amplified in Escherichia coli, and found to have a complexity of ≈40,000 clones. Because this represents a small fraction of all possible 11-bp sequences, we sequenced 25 clones to determine whether the library was enriched for any particular base. A slight enrichment of Ts and depletion of Gs on the bottom strand as depicted in Fig. 1A was observed (we will refer to the sequence of this strand throughout; Fig. 1B). However, the diversity of the library was sufficient to represent both functional and nonfunctional origins (see below and Fig. 1 C and D).
Figure 1.
ARS1 B2 mutant sequences. (A) The B2 region of ARS1. A XhoI to PstI fragment of ARS1 with randomized bases at the 11-bp B2 element was cloned into the XhoI to PstI region of pARS1/785-92. Note that the strand referred to throughout this paper is the Lower strand in this figure, the A-rich strand of the B2 element, read from right to left 5′ to 3′. The A and B2 element ACS matches are shown as red arrows. (B–E) The frequency of each nucleotide on the y axis is plotted against each of the 11 randomized positions on the x axis. (B) The library represents all 4 bases at all 11 positions. Twenty-five clones from the E. coli library were sequenced. (C) The B2− sequences are representative of the library as a whole. (D) The B2+ sequences are A-rich and weakly match up to the ACS starting at the second nucleotide. The ACS is aligned underneath the graph for comparison. (E) The B2+ sequences form a consensus that strongly matches the ACS if two-thirds of the sequences are shifted 1 base to the right. The B2+ sequences from D were examined to find the best match to the ACS within 1 base of the randomized 11 bases. The ACS is provided below for comparison.
Plasmid loss rates were used as a measure of function of the B2 mutant origins. Plasmids containing B2 element sequences that functioned poorly were identified by using a colorimetric assay for loss of the ADE2 gene (see Materials and Methods). Thirty-one plasmids were recovered with loss rates higher than the pARS1/802–810 B2 linker scan mutant (7.8 ± 1.0%/generation, Fig. 1C; and Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). B2 elements from these plasmids are termed B2− sequences. B2 sequences that functioned at wild-type levels were found in plasmids that were lost at a low rate and therefore retained for 10 days (see Materials and Methods). Thirty-one plasmids were recovered with plasmid loss rates within one standard deviation of wild-type ARS1 (3.0 ± 1.1%/generation, Fig. 1D; and Table 3, which is published as supporting information on the PNAS web site). The stable maintenance of these plasmids was confirmed by measuring their plasmid loss rates at least three times for each plasmid, and elements isolated in this selection are termed B2+ sequences.
B2+ Sequences Are A:T Rich and Contain a Sequence Related to the ACS.
To characterize the B2+ and B2− sequences, each group was compared with randomly sequenced clones from the library. A two-tailed Student t test was applied to the data in each case to calculate the probability that the differences between the selected and random sequences were due to chance alone (P value). A P value of less than 0.05 was considered significant.
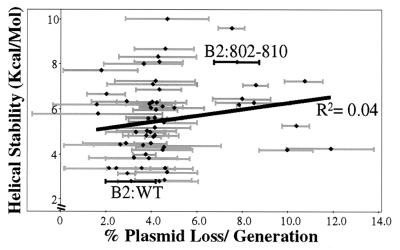
The B2+ sequences had a significant bias for A:T rich DNA. They contained an average of 5.6 ± 1.4 As on the bottom strand, which was much higher than the composition of the library as a whole (P = 2.9 × 10−11, Table 1). In contrast, there was no selection for Ts on the bottom strand (Table 1). Although these sequences had calculated helical stabilities lower than those of random library sequences, most of this helical instability could be accounted for by the bias for A-rich DNA (Table 1). Furthermore, there was no correlation between the rate of plasmid loss per generation and calculated helical stability (Fig. 2).
Table 1.
Characteristics of B2+ and B2− sequences
| Best match to ACS/11 | Helical stability | No. of As/11 | No. of Ts/11 | |
|---|---|---|---|---|
| Avg. of B2+ sequences | 7.4 ± 0.9 | 5.3 ± 1.6 Kcal/mol | 5.6 ± 1.4 | 2.8 ± 1.3 |
| Avg. of B2− sequences | 4.5 ± 1.2 | 7.9 ± 3.6 Kcal/mol | 2.5 ± 1.4 | 3.9 ± 1.5 |
| Avg. of random library sequences | 4.6 ± 1.1 | 8.2 ± 5.3 Kcal/mol | 2.7 ± 1.2 | 3.2 ± 1.5 |
| Avg. of randomly generated A-rich sequences | 6.4 ± 1.7 | 6.2 ± 1.7 Kcal/mol | 5.4 ± 1.6 | 2.3 ± 1.2 |
| P value B2+ vs. random | 3.3E-05 | 6.6E-08 | 2.9E-11 | 0.27 |
| P value B2− vs. random | 0.91 | 0.53 | 0.42 | 0.080 |
| P value B2+ vs. random A-rich | 8.8E-05 | 0.0060 | 0.52 | 0.041 |
Best match to ACS is either at the defined B2 element position or shifted one base pair to the left (based on Fig. 1). Helical stability was calculated for the 13 bp including the B2 element using the WebThermodyn program and the data sets of SantaLucia et al. (29, 30). P values are the result of two-tailed Student t tests. P values in italics are considered significant.
Figure 2.
Helical stability does not correlate with origin function of ARS1 B2 mutants. The helical stability of the mutant B2 element sequences was calculated as described in Materials and Methods. The helical stability was plotted against origin function for all B2 element mutants whose plasmid stability had been measured more than once, and a linear regression trend line and correlation value (R2) were assigned by excel (Microsoft).
As originally defined, the wild-type ARS1 B2 element is a 9/11 match to the ACS in an inverse orientation relative to the A element ACS. This result raised the possibility that a sequence similar to the ACS was important for B2 function. To test this hypothesis, we examined the incidences of inverse matches to the ACS in the B2+ sequences. There was no bias for any particular base at the first position, and the majority (≈2/3) of the sequences formed a better match to the ACS if shifted one base away from the A element to include the flanking A (Fig. 1D). Because wild-type ARS1 function is not altered by a 1-bp shift in B2 element location (8), we examined the best matches to the ACS within one base of the defined B2 element. The most frequent base at each position formed a match to the ACS at almost every position (Fig. 1E). By using this approach, the B2+ sequences included an average match to the ACS of 7.4 ± 0.9 bases, a number that is much greater than the number of matches found in the similarly shifted randomly sequenced clones (4.6 ± 1.1, Table 1).
The above analyses suggested that the selection for functional B2 element sequences identified both A-rich DNA and, more specifically, matches to the A-rich ACS. One explanation for this dual selection was that the matches to the ACS were simply due to a preference for A-rich DNA. To test this hypothesis, we computer-generated a population of random 11-bp sequences with the same percentage of As as the B2+ sequences (Table 1). We found that there was a statistically greater match to the ACS in the B2+ sequences than in the randomly generated A-rich sequences (P = 8.8 × 10−5, Table 1). Therefore, the B2+ sequences contain not simply A:T-rich DNA, but instead are selected to include a sequence similar to the ACS.
In contrast to the selection for matches to the ACS in the B2+ sequences, the B2− sequences are indistinguishable from randomly sequenced library clones by all measures examined (Table 1). These sequences have a similar composition, calculated helical stability (29), and incidence of matches to the ACS as the sequences from the starting library (Table 1).
ORC Does Not Protect B2+ Sequences from DNase I Cleavage in Vitro.
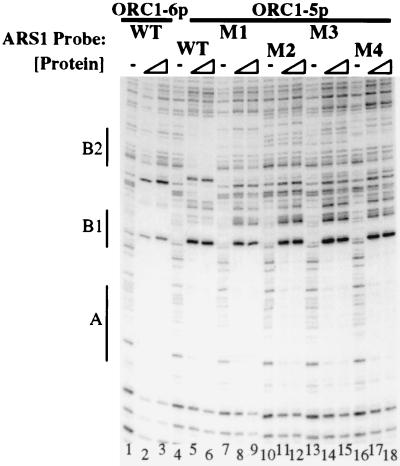
The selection for a close match to the ACS in the B2+ sequences suggested that a second inverse ORC binding site at the B2 element was important for origin function. The wild-type B2 element is a 9/11 match for the ACS, and DNase I protection assays have demonstrated that ORC can bind to this site in vitro if the A element is mutated (2). ORC will also bind to both the A and B2 elements at the same time if the smallest subunit (Orc6p), which is not required for DNA binding (32), is missing from the complex (R. Klemm and S.P.B., unpublished results, and Fig. 3 lanes 4–6). Therefore, we examined whether Orc1-5p would protect representative B2+ sequences (Table 2) from DNase I digestion. In each case, Orc1-5p strongly protected the A element. In contrast, Orc1-5p failed to protect the B2+ sequences or produce the characteristic hypersensitive site seen at the wild-type B2 element, although some small differences in the cleavage patterns at the B2+ sequences can be noted when ORC1-5p is present (Fig. 3). Likewise, wild-type ORC did not footprint over mutant B2 origin sequences when the adjacent A element was disrupted with a linker scan mutation (865–872, data not shown). This finding confirmed that the lack of Orc1-5p association with the B2+ sequences was not due to the absence of Orc6p.
Figure 3.
Orc1-5p cannot bind representative B2+ sequences in vitro. DNase I protection assays were performed with wild-type Orc1-6p (lanes 1–3), or Orc1-5p (lanes 4–18). Orc1-6p protects the A and B1 elements of wild-type ARS1 DNA (lanes 1–3), whereas Orc1-5p protects the A element and the B2 element of wild-type ARS1 DNA (lanes 4–6), with characteristic hypersensitive sites in between. In contrast, Orc1-5p does not protect the B2 elements of the B2+ ARS1 DNAs (lanes7–18). Lanes 1, 4, 7, 10, 13, and 16 contain no protein. Lanes 2, 5, 8, 11, 14, and 17 contain 50 ng of protein. Lanes 3, 6, 9, 12, 15, and 18 contain 250 ng of protein. M1–M4 refer to the B2 mutants listed in Table 2.
Table 2.
Representative B2 mutant sequences
| Sequence name | % Plasmid loss/generation | Sequence | Best match to ACS/11 | Helical stability (Kcal/mol) | No. of As/11 | No. of Ts/11 |
|---|---|---|---|---|---|---|
| WILDTYPE | 3.1 ± 1.1 | AATACTTAAAT | 9 | 2.8 | 6 | 4 |
| B2-802-10 | 7.8 ± 1.0 | CTCGAGGAAAT | 6 | 8.1 | 4 | 2 |
| B2-M1 | 2.4 ± 2.1 | AAACAAATATA | 8 | 3.4 | 8 | 2 |
| B2-M2 | 2.9 ± 0.4 | AAAATGATATA | 8 | 3.16 | 7 | 3 |
| B2-M3 | 3.0 ± 1.9 | AATTACGAAAT | 8 | 4.5 | 6 | 3 |
| B2-M4 | 4.6 ± 1.3 | ATAAACCGCCA | 7 | 8.7 | 5 | 1 |
| B2-M5 | 7.1 ± 2.2 | CTTGGCTCGCC | 3 | 12.3 | 0 | 3 |
| B2-M6 | 7.5 ± 0.6 | TTTCGTACCGC | 4 | 9.6 | 1 | 4 |
| B2-M7 | 10.4 ± 0.6 | TCTTTTAGCAT | 4 | 5.3 | 2 | 6 |
| B2-M8 | 10.7 ± 1.0 | CCAAGTTTCTT | 4 | 7.2 | 2 | 5 |
| B2-M9 | 15.0 ± 6.6 | CTTCCTGGCTT | 4 | 9.4 | 0 | 5 |
| B2-M10 | 23.1 ± 6.4 | TGATCCTCACA | 6 | 8.2 | 3 | 3 |
Plasmid loss assays were performed at least three times. Best match to the ACS and helical stability were calculated as in Table 1.
MCM Loading at the Chromosomal ARS1 Correlates with Plasmid Origin Function.
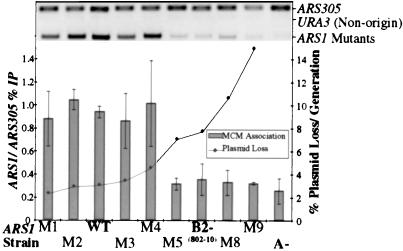
Because we could not detect an ORC interaction with the B2 element, we investigated the effect of the B2 mutants on the formation of the pre-RC in vivo. Previous reports have shown that Mcm2-7p loading is reduced in a B2 linker scan mutant in vivo when assayed by ChIP (17, 18). To determine whether B2 element function correlated with MCM loading, we integrated a series of representative mutants (Table 2) into the chromosome at ARS1 and performed ChIP with a monoclonal antibody that recognizes all six MCM proteins (A. Schwacha and S.P.B., unpublished results). B2− sequences associate with low levels of MCM proteins during G1, whereas B2+ sequences recruit Mcm2-7p to the origin at high levels (Fig. 4). By quantitation, MCM binding at the different ARS1 B2 mutants is bimodal: either bound at wild-type levels, or at levels near that of the nonfunctional A element mutation. In contrast, the MCM proteins are recruited to a wild-type origin, ARS305, in all strains, and never to a non-origin sequence, URA3 (Fig. 4). These data indicate that the plasmid stability data presented above is representative of origin function at the natural chromosomal locus and correlates with pre-RC formation.
Figure 4.
MCM origin association correlates with B2 mutant origin function. Strains containing ARS1 mutations were arrested in the G1 phase of the cell cycle with α factor and assayed for MCM association by ChIP. PCR was performed on both the immunoprecipitated (IPed) DNA and the input DNA with primers to the origins ARS305 and ARS1, and the non-origin sequence URA3. Sample IPed PCR is shown above. The graph represents the quantitation of the (ARS1 IP/ARS1 input)/(ARS305 IP/ARS305 input) for four individual experiments. Origin function is plotted on the second axis. The A element mutation is ARS1/865–872 and does not have a plasmid loss rate because plasmids containing this origin cannot transform. M1–M9 refer to the B2 mutants listed in Table 2.
Cdc6p Overexpression Rescues B2 Mutant Origins.
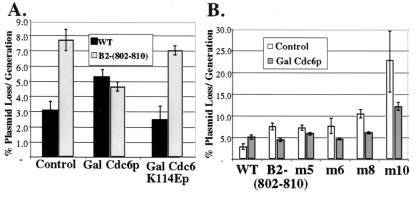
Because B2− origins have defects in MCM loading, a step requiring Cdc6p, we reasoned that they might be improved by overexpression of Cdc6p. Therefore, we introduced plasmids containing B2− origins into a strain containing an extra copy of CDC6 under control of the Gal1-10 promoter. Wild-type plasmids were lost at a slightly higher rate when the strain was grown in galactose, presumably because of titration of another factor. In contrast, the B2 linker scan mutation, and a series of other B2− mutations, were rescued to levels indistinguishable from wild type in galactose media (Fig. 5 A and B). Notably, a particularly strong B2 mutation, B2-m10, was significantly rescued, although not to wild type, suggesting that overexpression of Cdc6p cannot completely substitute for the B2 element (Fig. 5B). Overexpression of Cdc6K114Ep, which does not associate with chromatin and is nonfunctional in vivo (33–35), had no effect on either the wild-type or the mutant origins (Fig. 5A).
Figure 5.
Overexpression of wild-type but not mutant Cdc6p rescues B2 mutant origins. (A) A wild-type plasmid (pARS1/785-92) is lost at a higher rate when wild-type Cdc6p is overexpressed, whereas the plasmid loss of a B2 mutant plasmid (pARS1/802–810) decreases to the wild-type level. Plasmids were transformed into strains containing wild-type or mutant CDC6 under control of the Gal1-10 promoter, and the cells grown in media containing 2% galactose. (B) A series of representative B2 mutant plasmids are improved by galactose-induced overexpression of Cdc6p. Control, no overexpression. Gal, galactose. M5–M10 refer to the B2 mutants listed in Table 2.
Discussion
By using a genetic screen for novel B2 elements, we identified specific sequences required for the function of this element at ARS1. These sequences were found to be A-rich and specifically related to the ACS in a manner not explained by their high A-content. Analysis of these sequences suggested that the ARS1 B2 element is not a DUE, but instead requires a specific sequence. B2 element function correlates with MCM origin loading, and B2 element defects can be ameliorated by overexpressing Cdc6p. These data, combined with the observation that the pre-RC forms a footprint over the B2 element during G1 in vivo (12), suggest that B2 is a binding site for a component of the pre-RC.
Sequence and Structure of the B2 Element.
Our selection for sequences that can function in place of the B2 element provided evidence against a simple structural role, such as helical instability or DNA bending. The identified B2+ sequences could not have been selected based solely on helical instability, because they showed a strong bias for A-rich DNA, despite a starting library that was biased for Ts. The nearest neighbor base pair interactions predicted to have the lowest helical stability are 5′-TA-3′ and 5′-AT-3′, not 5′-AA-3′ (30, 36, 37). Moreover, low helical stability is not a predictor of origin function in our sequences, because we find no correlation between origin function and helical instability at ARS1 (R2 = 0.03, Fig. 2). This finding is in contrast to previous analyses at ARS307 (25). These differences may reflect differences in the two origins, but those analyses also identified outlying points that did not correlate and incorporated a smaller panel of less diverse mutations. Although A-rich DNA is often bent, the wild-type B2 element is not intrinsically bent (8, 38), nor do the sequences we isolated have high calculated curvatures (data not shown; ref. 39). Finally, the observation that the B2 element is necessary for pre-RC formation suggests a nonstructural role for this sequence because no DNA unwinding has been detected until yeast cells enter S-phase and the pre-RC is dismantled (40). Furthermore, the 11-bp ARS1 B2 element cannot itself be a DUE because it can substitute for the DUE of ARS305 only in the presence of its AT-rich flanking sequences (25).
Although the formal possibility remains that this A-rich DNA is performing an unknown structural function, our data suggest that B2 is a protein binding site related to the ACS. Matches to the ACS were more prevalent in the B2+ sequences than in random computer-generated A-rich sequences (Table 1). The parameters for this selection can be seen in Fig. 1, where it is apparent that, although there is an overall selection for As, certain positions are enriched for Ts, Gs, or Cs. Importantly, these non-A positions match the sequence of the ACS. Also, those positions with the strongest selection for As (positions 2 and 4, Fig. 1E) are those that are least tolerant of mutation within the A element at ARS307 (41). Any sequence with a 7/11 match to the ACS functions well as a B2 element, and all of the B2+ sequences contain at least a 6/11 match (Table 3, which is published as supporting information on the PNAS web site). The B2 element can be shifted slightly in either direction with no effect on origin function (8, 25), and about 2/3 of our sequences had better matches to the ACS when shifted over one base. This is not surprising because the presence of an A flanking the randomized sequences facilitated the selection of an ACS incorporating only 10 randomized bases.
The flexibility in positioning of the B2 element, the frequent occurrence of near matches to the ACS at origins, and the AT-richness of the ACS have all made uncovering of the functional importance of these sequences difficult at most origins. The presence of many more extra inverse ACS matches than would be expected given the AT-rich nature of yeast origins has been noted (42). It is striking that ARS1 is the only well-characterized origin with a single additional inverse match to the ACS greater than 7/11 bases in the region over which the pre-RC forms, and this match identifies the B2 element. Therefore, a requirement for extra inverse sequences that match the ACS may not be unique to ARS1, but simply the most easily identified.
Several prior studies addressed the role of partial matches to the ACS at other yeast origins. The H4 ARS B domain can be replaced by an unrelated easily unwound pBR322 sequence (43), and an attempt was made to remove all 8/11 or better matches to the ACS (44). However, both of these approaches left behind 7/11 and 8/11 matches to the ACS, by the current definition (41), at the approximate distance of the B2 element from the A element. A similar analysis at the rDNA ARS mutated only 9/11 matches to the ACS (45). Given the uncertainty in where an extra ACS match would have to be placed to function as a B2 element, and that many 7/11 matches were functional in our study, these studies have not fully addressed the importance of a redundant consensus similar to the ACS at other origins.
Function of the B2 Element: Pre-RC Component Binding Site?
The simplest interpretation of our sequence data is that the B2 element serves as a second binding site for ORC, because ORC binds the ACS. Multimerization of ORC during G1 would be analogous to the binding of E.coli DnaA-ATP to the 13mers embedded within the E.coli DUE during initiation to facilitate open complex formation (46, 47). The rescue of B2− origin function by Cdc6p could be mediated indirectly through improved ORC binding, because Cdc6p overexpression also rescues an Orc5-1 mutation (48). If ORC were to bind this site in vivo, it would have to be assisted by other factors and bind in a different conformation. Two purified ORCs cannot bind to the A and B2 elements at the same time (2) unless 2–3 extra base pairs are inserted between them (R. Austin and S.P.B., unpublished results), or the smallest subunit, Orc6p, is left out of the complex (R. Klemm and S.P.B., unpublished results and Fig. 3). Also, the binding of ORC to the B2+ sequences in vitro was not detected within the stringency of the Dnase I protection assay (Fig. 3). We cannot rule out the binding of a second ORC to the B2 element in vivo as a possibility, and current in vivo binding assays (e.g., ChIP) cannot detect the stoichiometry of ORC in the pre-RC.
Another candidate B2-binding protein is the Orc1p-related protein Cdc6p. The recent crystal structure of the archaeon Pyrobaculum aerophilium Cdc6p revealed a winged-helix DNA binding domain that is conserved in S. cerevisiae Cdc6p, Orc1p, and Orc4p, and in the S. pombe ortholog Cdc18p. Furthermore, this conservation in CDC6 proteins is important for function, because mutations in the putative DNA binding domain of Cdc18p cause cell cycle arrests or null phenotypes (49). This result raises the possibility that Cdc6p is recruited to the origin not solely through its interactions with ORC (50, 51) but also by binding the sequence related to the ACS at the B2 element. This finding is supported by the requirement for the B2 element in MCM loading, and by the observation that overexpressing Cdc6p ameliorates the defects in a wide range of B2 mutants. On the other hand, although DNA binding activity has been observed for purified Cdc6p, it is nonspecific and mediated by the nonessential N terminus (52). Moreover, ORC interactions may be sufficient to recruit Cdc6p to the origin, and the winged helix domain could be involved in protein–protein interactions instead of protein–DNA interactions (53). Although CDC6 appears to be a good candidate protein for binding the B2 element, we have not been able to use ChIP to directly test this model because of the low protein levels and tight cell cycle control of origin association.
Finally, it is possible that the B2 element is a binding site for another protein in the pre-RC, such as the Mcm2-7p complex, Mcm10p, or an S. cerevisiae CDT1 ortholog. It is reasonable that the MCM proteins, which are the putative eukaryotic replicative helicase, would associate with DNA embedded within the easily unwound region of the origin. Furthermore, the rescue of the B2− plasmids by Cdc6p overexpression could also have been mediated through increased MCM loading, as has previously been observed on bulk chromatin (34). Very little is known about how Mcm10p and Cdt1p associate with the origin.
By specifically randomizing the well-defined B2 element at ARS1, we have identified an ACS-related sequence requirement for this origin component. Because of redundancy at most origins, this ACS-related sequence has not previously been identified as functionally important. We also found that the ARS1 B2 element is not simply a DUE, and suggest that specific A-rich sequences like the B2 element are embedded within broad DUEs at yeast origins. We suggest that ORC, Cdc6p, or the MCMs are the most likely proteins to bind this site. In vitro eukaryotic DNA replication systems should allow the direct testing of this model, along with a clearer picture of the stoichiometry and architecture of the pre-RC.
Supplementary Material
Acknowledgments
The authors thank R. Klemm for ORC and Orc1-5p, A. Schwacha for MCM antibody, R. Austin for CDC6 plasmids, M. de Vries for assistance with statistical analyses, and T. Baker for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM 58701 and by the Howard Hughes Medical Institute.
Abbreviations
- pre-RC
pre-replicative complex
- ARS
autonomously replicating sequence
- ACS
ARS consensus sequence
- ORC
origin recognition complex
- MCM
mini-chromosome maintenance
- ChIP
chromatin immunoprecipitation
- DUE
DNA unwinding element
- IP
immunoprecipitation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.DePamphilis M L. Bioessays. 1999;21:5–16. doi: 10.1002/(SICI)1521-1878(199901)21:1<5::AID-BIES2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 2.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 3.Ogawa Y, Takahashi T, Masukata H. Mol Cell Biol. 1999;19:7228–7236. doi: 10.1128/mcb.19.10.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austin R J, Orr-Weaver T L, Bell S P. Genes Dev. 1999;13:2639–2649. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowalski D, Eddy M J. EMBO J. 1989;8:4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Natale D A, Umek R M, Kowalski D. Nucleic Acids Res. 1993;21:555–560. doi: 10.1093/nar/21.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin S, Kowalski D. J Mol Biol. 1994;235:496–507. doi: 10.1006/jmbi.1994.1009. [DOI] [PubMed] [Google Scholar]
- 8.Marahrens Y, Stillman B. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 9.Rao H, Marahrens Y, Stillman B. Mol Cell Biol. 1994;14:7643–7651. doi: 10.1128/mcb.14.11.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Theis J F, Newlon C S. Mol Cell Biol. 1994;14:7652–7659. doi: 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang R Y, Kowalski D. Nucleic Acids Res. 1996;24:816–823. doi: 10.1093/nar/24.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santocanale C, Diffley J F. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- 13.Diffley J F, Cocker J H, Dowell S J, Harwood J, Rowley A. J Cell Sci Suppl. 1995;19:67–72. doi: 10.1242/jcs.1995.supplement_19.9. [DOI] [PubMed] [Google Scholar]
- 14.Donovan S, Harwood J, Drury L S, Diffley J F. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka T, Knapp D, Nasmyth K. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 16.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 17.Zou L, Stillman B. Mol Cell Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipford J R, Bell S P. Mol Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- 19.Seki T, Diffley J F. Proc Natl Acad Sci USA. 2000;97:14115–14120. doi: 10.1073/pnas.97.26.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maiorano D, Moreau J, Mechali M. Nature (London) 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 21.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. Nature (London) 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 22.Whittaker A J, Royzman I, Orr-Weaver T L. Genes Dev. 2000;14:1765–1776. [PMC free article] [PubMed] [Google Scholar]
- 23.Huang R Y, Kowalski D. EMBO J. 1993;12:4521–4531. doi: 10.1002/j.1460-2075.1993.tb06141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto K, Ishimi Y. Mol Cell Biol. 1994;14:4624–4632. doi: 10.1128/mcb.14.7.4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin S, Kowalski D. Mol Cell Biol. 1997;17:5473–5484. doi: 10.1128/mcb.17.9.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ausubel F M. Current Protocols in Molecular Biology. New York: Greene and Wiley; 1992. [Google Scholar]
- 27.Stotz A, Linder P. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- 28.Klemm R D, Bell S P. Proc Natl Acad Sci USA. 2001;98:8361–8367. doi: 10.1073/pnas.131006898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Natale D A, Schubert A E, Kowalski D. Proc Natl Acad Sci USA. 1992;89:2654–2658. doi: 10.1073/pnas.89.7.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.SantaLucia J, Jr, Allawi H T, Seneviratne P A. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 31.Bell S P, Mitchell J, Leber J, Kobayashi R, Stillman B. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 32.Lee D G, Bell S P. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elsasser S, Lou F, Wang B, Campbell J L, Jong A. Mol Biol Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perkins G, Diffley J F. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- 35.Weinreich M, Liang C, Stillman B. Proc Natl Acad Sci USA. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breslauer K J, Frank R, Blocker H, Marky L A. Proc Natl Acad Sci USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugimoto N, Nakano S, Yoneyama M, Honda K. Nucleic Acids Res. 1996;24:4501–4505. doi: 10.1093/nar/24.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marilley M. Mol Gen Genet. 2000;263:854–866. doi: 10.1007/s004380000253. [DOI] [PubMed] [Google Scholar]
- 39.Munteanu M G, Vlahovicek K, Parthasarathy S, Simon I, Pongor S. Trends Biochem Sci. 1998;23:341–347. doi: 10.1016/s0968-0004(98)01265-1. [DOI] [PubMed] [Google Scholar]
- 40.Geraghty D S, Ding M, Heintz N H, Pederson D S. J Biol Chem. 2000;275:18011–18021. doi: 10.1074/jbc.M909787199. [DOI] [PubMed] [Google Scholar]
- 41.Van Houten J V, Newlon C S. Mol Cell Biol. 1990;10:3917–3925. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palzkill T G, Newlon C S. Cell. 1988;53:441–450. doi: 10.1016/0092-8674(88)90164-x. [DOI] [PubMed] [Google Scholar]
- 43.Umek R M, Kowalski D. Cell. 1988;52:559–567. doi: 10.1016/0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 44.Holmes S G, Smith M M. Mol Cell Biol. 1989;9:5464–5472. doi: 10.1128/mcb.9.12.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker S S, Malik A K, Eisenberg S. Nucleic Acids Res. 1991;19:6255–6262. doi: 10.1093/nar/19.22.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bramhill D, Kornberg A. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 47.Speck C, Messer W. EMBO J. 2001;20:1469–1476. doi: 10.1093/emboj/20.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang C, Weinreich M, Stillman B. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 49.Liu J, Smith C L, DeRyckere D, DeAngelis K, Martin G S, Berger J M. Mol Cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 50.Wang B, Feng L, Hu Y, Huang S H, Reynolds C P, Wu L, Jong A Y. J Biol Chem. 1999;274:8291–8298. doi: 10.1074/jbc.274.12.8291. [DOI] [PubMed] [Google Scholar]
- 51.Mizushima T, Takahashi N, Stillman B. Genes Dev. 2000;14:1631–1641. [PMC free article] [PubMed] [Google Scholar]
- 52.Feng L, Wang B, Driscoll B, Jong A. Mol Biol Cell. 2000;11:1673–1685. doi: 10.1091/mbc.11.5.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gajiwala K S, Burley S K. Curr Opin Struct Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 54.Rao H, Stillman B. Proc Natl Acad Sci USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rowley A, Cocker J H, Jarwood J, Diffley J F. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.