Abstract
We present the first three-dimensional reconstruction of human fatty acid synthase obtained by electron cryomicroscopy and single-particle image processing. The structure shows that the synthase is composed of two monomers, arranged in an antiparallel orientation, which is consistent with biochemical data. The monomers are connected to each other at their middle by a bridge of density, a site proposed to be the combination of the interdomain regions of the two monomers. Each monomer subunit appears to be subdivided into three structural domains. With this reconstruction of the synthase, we propose a location for the enzyme's two fatty acid synthesis sites.
Obesity, a major health factor, is a measure of the fat deposited with adipose in consequence to food intake, fatty acid and triglyceride synthesis and oxidation, and energy homeostasis (1). Excess food provides not only the energy needs of the body but promotes the synthesis of fatty acids and triglycerides and storage of the latter in liver and adipose tissues. The de novo synthesis of fatty acids from acetyl-CoA and malonyl-CoA in the presence of the reducing substrate NADPH is catalyzed by the enzyme fatty acid synthase (FAS, EC 2-3-185; ref. 2). Malonyl-CoA is the donor of the C2 units required for chain elongation and is generated by acetyl-CoA carboxylase reactions, the rate-limiting step in the synthesis of fatty acids. In animal tissues including that of humans, FAS is a homodimer of a multifunctional protein (Mr 272,000; refs. 2 and 3). Each subunit protein contains seven catalytic activities plus the acyl carrier protein (ACP). The organization of the catalytic activities along the polypeptide chain from the N to the C termini is as follows: β-ketoacyl synthase, acetyl/malonyl transacylases, β-hydroxyacyl dehydratase, enoyl reductase, β-ketoacyl reductase, ACP, and thioesterase. Throughout the elongation process, the fatty acyl groups are linked as a thioester to the 4′-phosphopantetheinyl-SH, the prosthetic group of the ACP component of FAS. This prosthetic group is ≈20 Å long and is an essential flexible linker for the catalytic activities of FAS (3, 4). The organization of the partial activities, as mentioned above, is based on proteolytic mapping of the subunit and is confirmed by the predicted amino acid sequences based on the cloning and nucleotide sequences of the cDNA of the FAS component enzymes (2, 5–11). Moreover, the multifunctional human FAS and its subdomains were cloned and expressed in Escherichia coli; the recombinant proteins were catalytically active, and their orders on the FAS subunit were established (12, 13). Based on these studies, the component activities were grouped into three functional subdomains: domain I, containing β-ketoacyl synthase, acetyl/malonyl transacylases, and β-hydroxyacyl dehydratase; domain II, containing enoyl reductase, β-ketoacyl reductase, and ACP; and domain III, containing thioesterase. Moreover, an intermediate polypeptide containing 640 amino acids, located between domains I and II and designated as the interdomain, has no known catalytic activity but is essential in holding together the two FAS subunits and generating the active palmitate-synthesizing center (4, 14).
Earlier studies show that dissociation of the native FAS dimer results in the loss of palmitate synthesis and the β-ketoacyl synthase activity but not the remaining six partial activities (2). Further investigations of these observations led Stoops and Wakil (2, 15) to propose an antiparallel model for the organization of the two FAS subunits, generating two palmitate-synthesizing centers. Each center consists of domain I derived from one subunit and domains II and III derived from the second subunit. In the active palmitate-synthesizing center, the active cysteine-SH of the β-ketoacyl synthase, of domain I, where the acyl group is bound is juxtaposed within ≈2 Å from the cysteamine-SH of domain II, where the malonyl group is bound. These structural organization and domain arrangements are necessary and are prerequisites for the synthesis of long chain fatty acids (4). We present here a three-dimensional (3D) model for human FAS using electron cryomicroscopy and single-particle reconstruction and explain the structure in terms of the available biochemical data.
Materials and Methods
Enzyme Purification.
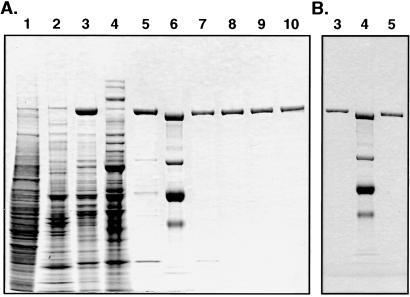
Human fatty acid synthase was purified from a breast cancer cell line, ZR75-1, as follows: the cell pellets (15 ml) were frozen in dry ice/ethanol, thawed twice, and then suspended in 100 ml of buffer (20 mM KPi, pH 7.0, 1 mM EDTA, 1 mM DTT, and 0.25 M sucrose containing protease inhibitors each at 5 μg/ml, leupeptin, antitrypsin, and aprotinin) and disrupted by using a Potter–Elvehjem homogenizer with a tight-fitting pestle. The cell lysate was sonicated three times at 50-W output for 20 sec. The suspension was clarified by centrifugation at 70,000 × g for 30 min. The supernatant was applied to a HiLoad 26/10 Q-Sepharose-FPLC column, washed with 600 ml of buffer A (20 mM KPi, pH 7.4, 1 mM EDTA, 1 mM DTT, and 5% glycerol), and the bound proteins were eluted by using a gradient of 0–30% of buffer B (buffer A + 1 M NaCl) for 240 min at a flow rate of 1 ml/min. The fractions containing FAS activity were pooled, and the enzyme was precipitated with ammonium sulfate at 50% saturation. The pellet was dissolved in 2 ml of buffer A and loaded onto a Superose 6-FPLC column. The synthase was eluted with buffer A. The active enzyme fractions were applied to an UNO-Q-6 FPLC column and washed with 20 ml of buffer A. FAS was eluted with a gradient of 0–26% buffer B for 195 min at a flow rate of 0.5 ml/min. The purified enzyme (Fig. 1B, lane 5) was stored at −80°C at a concentration of 1 mg/ml in a buffer containing 150 mM KPi, pH 7.4, 1 mM DTT, 1 mM EDTA, and 15% (vol/vol) glycerol. For electron cryomicroscopy studies, the enzyme was eluted from the UNO-Q-6 column with a gradient of 0–30% buffer B in buffer A without glycerol and used immediately.
Figure 1.
SDS/PAGE analyses of FAS purified from the breast cancer cell line ZR75–1. (A) Various fractions were subjected to SDS/PAGE analysis (4–12% gradient), and the gel then was stained with Coomassie blue. Lane 1, soluble extract (25 μg); lanes 2, 3, and 4, HiLoad 26/10 Q-Sepharose fractions 29, 30–35, and 36 (10 μg of each fraction); lane 5, Superose-6 fraction (2 μg); lane 6, protein standard (molecular masses: 217, 123, 71, and 48 kDa); lanes 7, 8, 9, and 10, UNO-Q-6 fractions 56–59 (0.6 μg each). (B) For electron cryomicroscopic examination, the purified FAS (lane 5, 0.6 μg) was applied to an UNO-Q-6 column and fraction 3 (0.6 μg) was analyzed (lane 3). Lane 4 is a protein standard.
X-Ray Scattering Studies.
X-ray scattering data of FAS was collected at the Stanford Synchrotron Radiation Laboratory on beamline BL 4-2 (16, 17). Enzyme activity was measured before and after data collection to ensure that the scattering data would not be affected by radiation damage. Scattering data were collected at different concentrations: 5, 10, 15, and 20 mg/ml of FAS in buffer A in the presence of 1 mM NADPH and 1 mM acetyl-CoA. The final composite scattering profile was a combination of the low angle data from 1/350 to 1/46 Å−1 obtained at a protein concentration of 5–10 mg/ml and the high angle data from 1/46 to 1/6 Å−1 obtained at 20 mg/ml. A tenth order polynomial was fitted to the scattering data in each of the two frequency ranges before combining the data sets. A running average across 1/46 Å−1 was applied to remove any discontinuities.
Electron Cryomicroscopy.
Glycerol was removed by applying the enzyme solution to an UNO-Q-6 FPLC column. The enzyme was eluted in buffer A. The protein concentration was adjusted to 50–70 μg/ml in freshly made buffer A containing 1 mM NADPH and 1 mM acetyl-CoA and incubated at room temperature for 30–45 min to promote dimerization of FAS (18). The enzyme was embedded in vitreous ice across holes of 400-mesh carbon-coated holey grids (19, 20). A JEOL1200EX transmission electron microscope operating at 100 kV, equipped with a Gatan cryospecimen holder, and kept at −166°C was used for imaging. A beam diameter of ≈2.5 μm was used with spot size 3 and a 100-μm condenser aperture along with a 50-μm objective lens aperture. Focal pair images with a defocus difference of 1.5 μm were recorded with 8–19 electrons per Å2 per micrograph at ×60,000 magnification. All images were recorded on Kodak SO-163 photographic film, which was developed in full-strength D-19 at 20°C for 12 min.
Image Preprocessing.
Micrographs were screened visually for proper particle concentration, ice quality, and contrast. The best focal pairs were digitized by using a Zeiss Phodis SCIA microdensitometer at 7-μm intervals; subsequently, the pixels were binned 2 × 2 to reduce the modulation transfer effects from the scanner's optical system. Particle selection was performed semiautomatically using standard procedures in BOXER, a graphical tool in the EMAN package, by combining information from the 3 × 3 median-filtered far-from-focus and close-to-focus micrographs (21). Median filtering was performed solely to aid in particle identification, and the center of the particle was determined by locating its center of mass. After the particles were located and centered, the unfiltered particle images were used for subsequent data analysis. Finally, the selected particle images were 2 × 2-averaged, yielding 4.67 Å per pixel, and high-pass filtered at ≈1/300 Å−1.
The eight parameters describing the contrast transfer function (CTF), the Fourier amplitude decay function, and the background noise function for each micrograph were determined by using CTFIT, a graphical tool in EMAN (21). The one-dimensional power spectrum of the particle images was fit by combining the eight-parameter model with the x-ray scattering intensity (22).
Image Corrections and Model-Based Refinement.
CTF corrections were performed on both amplitudes and phases by using standard methods in EMAN (23). Phase-flipping was performed immediately on each particle as a preprocessing step before refinement. Amplitude corrections were performed as particles were combined into class averages. A Wiener filter using the spectral signal/noise ratio accurately estimated from the fitting was incorporated into the amplitude correction.
Various techniques have existed for generating an initial model suitable for refinement. In our case, two featureless ellipsoids, consistent with previous neutron-scattering data (24), were generated and used as an initial model. Standard techniques in EMAN (21) were used to obtain a 3D reconstruction of FAS. Briefly, projections of a starting model were generated, followed by aligning each particle image with and comparing it to each projection. The particle images were classified with whichever projection they matched best. Next, the particle images within each class were aligned relative to each other and averaged together as CTF corrections were applied (23). Finally, the class averages were combined to produce a new 3D model. This process was iterated until convergence was achieved. Resolution was determined by using a simple two-way t test and a 0.5 Fourier shell coefficient threshold (21).
Data processing was done on multiprocessor Silicon Graphics Origin 2000 machines at Baylor College of Medicine and National Center for Supercomputing Applications (Urbana-Champaign, IL). Visualization of the 3D reconstruction was done by using VIS5D.§
Results
X-Ray Scattering and Electron Cryomicroscopy Data.
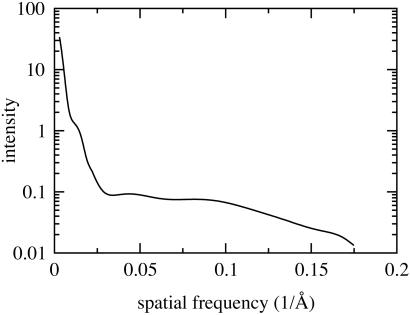
The FAS preparations had a specific activity of ≈1,500 mmol of NADPH oxidized min−1⋅mg−1 at 25°C and displayed a typical gel pattern under denaturing conditions as shown in Fig. 1. X-ray scattering intensities were collected from these preparations at different camera lengths and protein concentrations (Fig. 2). The reliability of this composite scattering curve, which covered spatial frequencies ranging from 1/300 to 1/6 Å−1, was substantiated by the small variation in the radius of gyration (Rg = 64.14 Å, σ = 0.2 Å) obtained at different sample concentrations. The scattering profile revealed a clear shoulder at 1/70 Å−1 arising from the monomer-monomer spacing in the dimeric enzyme. An equivalent feature was observed in the scattering curve obtained by using neutron scattering (24).
Figure 2.
X-ray solution scattering intensity curve of FAS collected at the Stanford Synchrotron Radiation Laboratory on beamline BL 4-2. The scattering curve exhibits a shoulder at 1/70 Å−1, which is characteristic for a dimeric particle. The curve was obtained by combining two separate scattering curves as explained in Materials and Methods.
Micrograph and Image Analysis of FAS.
A typical micrograph of FAS revealed several well defined views of the molecule, i.e., some had a distinctive dumbbell shape, whereas others looked like two parallel arcs (Fig. 3). Closer examination reveals some views other than those mentioned. Moderately hydrophobic grids yielded the thinnest ice, resulting in particle images with the highest contrast. A set of 29 micrographs was selected for use in the reconstruction. The parameters of these micrographs relevant to the imaging conditions are summarized in Table 1. A total of nearly 20,000 particle images were selected after screening out those that would most likely not correspond to a projection of a dimeric particle based on their contrast and size. In cases where this could not be assessed easily, the particles were simply left in the data set. Overall, screening removed typically less than 5% of the particles from the original data set. The experimental B factors (22) determined after fitting the observed CTF ranged from 690 to 1,000 Å2, equivalent to envelopes of 17 and 20 Å, respectively. These values are typical for the microscope used in imaging FAS.
Figure 3.
Typical micrograph of FAS embedded in vitreou ice and recorded at 2.7-μm defocus. The image shown was median-filtered by using a 3 × 3-pixel window to facilitate identification and selection of the molecules. For clarity, several particle images that were selected for image processing have been boxed out. These particles show FAS in different views (e.g. double-arched and dumbbell-shaped). The accompanying close-to-focus micrograph of this area was used to select particles from for image processing. (Bar, 500 Å.)
Table 1.
Image parameters of FAS
| Defocus group, μm
|
|||
|---|---|---|---|
| <1 | 1–2 | >2 | |
| No. of micrographs | 12 | 12 | 5 |
| No. of particles | 9378 | 6711 | 3722 |
| Average defocus, μm | 0.8 (0.2) | 1.7 (0.3) | 2.4 (0.2) |
| Average B factor, Å2 | 688 (4) | 790 (4) | 1007 (2) |
| Average envelope, Å | 16.7 (1.3) | 17.9 (1.2) | 20.2 (0.9) |
Three ranges of defocus values with the number of micrographs and particle images extracted of the micrograph sets are shown. The average defocus, experimental B factor (22), and corresponding envelope were determined using ctfit for each set of micrographs. Values in parentheses denote the standard deviation.
Image Analysis.
In earlier studies, it was suggested that FAS was a symmetric enzyme with both monomers juxtaposed head-to-tail, resulting in two active sites for fatty acid biosynthesis (2, 15, 25). Additionally, a dyad axis was implied, which would be perpendicular to a plane through both monomers (24). This dyad axis related one monomer in its entirety to the other, i.e., each active site was an exact structural duplicate of the other one. However, no direct structural evidence existed yet either to substantiate or disprove this symmetrical model. Therefore, in processing the image data, we considered two possibilities, namely FAS molecules with and without C2 symmetry.
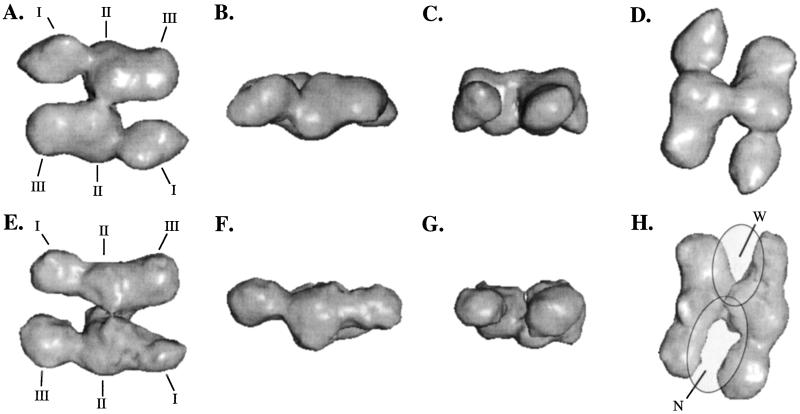
The 3D reconstruction obtained with 2-fold rotational symmetry enforced is shown in Fig. 4 A–D. The symmetry was enforced when generating 3D models from the set of class averages. This model has a resolution of ≈19 Å. Another independent reconstruction was performed starting with the same particle data and starting model but without enforcing the 2-fold rotational symmetry. As expected, the 2-fold symmetry is broken in the resulting 3D reconstruction, which has a somewhat lower resolution, namely 23 Ă (Fig. 4 E–H). The sampling of the data was adequate for the resolution of the reconstructions.
Figure 4.
3D reconstruction of FAS obtained with (A–D) and without (E–H) applying C2 symmetry. The surface renderings were done at 100% of the molecular mass assuming 1.23 Å3/Da. From left to right, different views of the molecule are shown, i.e., top view (A and E), side view (B and F), end-on view (C and G), and the view corresponding to the double-arch view visible in Fig. 3 (D and H). Individual domains of the subunits have been indicated as I–III (A and E). The far-right bottom view shows the two clefts formed by the monomers labeled as narrow (N) and wide (W). Proposed locations of the active sites in FAS are indicated by the two gray semitransparent oval regions.
In both models, FAS has similar dimensions of 180 × 130 × 75 Å with the monomers separated by a 19-Å-wide cleft. A strong bridge of density connects the two monomers at their middle. The overall dimensions and features of both models are similar except that the cleft is significantly wider on one side of the molecule in the asymmetric model. The monomers in the asymmetric model contain enough landmarks to be identified unambiguously in an antiparallel orientation, which is in agreement with previously obtained biochemical data (15, 26, 27).
Evaluation of the Validity of the Models.
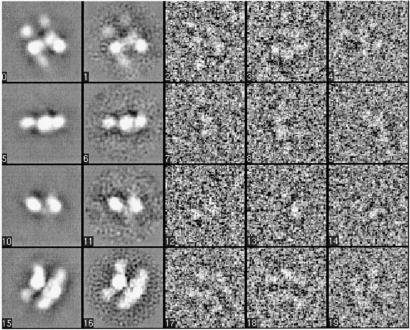
Both models satisfied criteria for a valid model. That is, both structures converged to a stable structure, both of which were reproduced when the data were split in half and independent reconstructions were performed. In addition, raw particle images quantitatively matched the class averages (Fig. 5). The agreement between the class averages and projections of the reconstructed models also was quite good, although agreement was better for the asymmetric model than for the symmetric model. These facts imply that there may be some structural heterogeneity in the data, thus the two models simply represent different amounts of averaging on the same heterogeneous population. The resolution claimed for each model is the threshold below which the heterogeneity is insignificant to cause substantial variations in the model.
Figure 5.
Excerpts from the refinement of FAS using EMAN. Shown are (from left to right) projections of the 3D model along with the corresponding class averages as well as three particle images from each class. The particle images displayed differ by an in-plane rotation. Note that during the alignment, however, this difference is taken care of. The rows show top, side, and end-on views of FAS; the Bottom shows the characteristic view of two parallel arcs as seen in Fig. 2.
In the EMAN implementation different angular views are calculated from the reference as set by the resolution that can be expected realistically. Subsequently, particles are classified and assigned to one of the views of the reference. For FAS, all classes used to cover the unit sphere had particles assigned to them. Although quite a few classes contained particle images resembling the double arcs, FAS did not exhibit a preferred orientation. Finally, nearly 80% of all particle images were used in each of the reconstructions.
Discussion
Image and Scattering Data.
Human FAS can be purified in sufficient quantity and purity (Fig. 1) to be studied by electron imaging and x-ray solution scattering. Electron imaging using cryomethods is well suited for revealing the quaternary structure of large macromolecules (28, 29). Fig. 3 demonstrates that human FAS is clearly visible in thin ice layers. The image also reveals the presence of relatively few particles, the size and molecular weight of which are clearly smaller than those of native FAS. The gel clearly attests to the purity of the FAS preparation used in this study. It thus might be possible that a small portion of FAS exists in a monomeric form. Nonetheless, despite this heterogeneity, the reconstruction is developed by using only those particle images that are consistent with each other structurally. The particle images, however, suffer from artifacts caused by the instrument's CTF (30, 31) and attenuation of the Fourier amplitudes by the various envelope functions. The latter was approximated empirically by a single Gaussian, i.e., e−Bs2, where s is the spatial frequency, and B is the experimental B factor (22) determined as part of EMAN. We made use of the x-ray solution scattering intensity of the enzyme (Fig. 2) to accurately determine the experimental and instrumental parameters of these functions for each micrograph (22). Under- or overestimating these parameters had a significant impact on the gross mass distribution in the 3D reconstruction.
One of the potential problems in obtaining reliable x-ray scattering at high resolution might be the high protein concentration required. However, the small variation in the radius of gyration calculated from scattering data, obtained at different protein concentrations, indicated that there was no aggregation as expected, presumably because the purification protocol used a chromatography step by using a size-exclusion column (Superose-6). Thus, we can state that the enzyme solution is monodisperse. Furthermore, because no significant inactivation occurred of the enzyme during these scattering experiments, the structural information gained could be interpreted without problems. By using the scattering curves, accurate corrections could be made to the images for the effects of the experimental B factor, the amplitude contrast, and the microscope's CTF. These corrections were applied during the 3D reconstruction procedure when the class averages were generated and before merging the class averages.
Starting Model.
The algorithm that EMAN used for the 3D reconstruction required an initial model against which the raw data could be refined. Earlier biophysical and biochemical experiments indicated that FAS was a dimeric particle with dimensions ≈160 × 145 × 75 Å (24, 26). The earlier studies also indicated that the monomers in FAS could be subdivided into domains (24, 26, 27). Because we did not want to introduce any structural bias into the refinement procedure, we used two featureless ellipsoids with similar dimensions as a starting model. Moreover, the subsequent refinement would converge to a new 3D model, self-consistent with the raw images, obviating the need for detail in the initial model. The fact that the initial model was featureless and that the size and shape of the monomers changed between the starting model and the final 3D reconstruction demonstrated that the initial model did indeed not bias the observed detailed features in the final 3D reconstructions.
Interpretation of Symmetry.
Although single-particle analysis can discriminate easily between, for example, 4- and 5-fold symmetries, it is a poor technique for absolutely proving whether symmetry is present at all. This is true especially in the case of pseudosymmetry where deviations from symmetry caused by subunit rearrangement are not large. Such uncertainty is analogous to x-ray crystallography where higher symmetry observed at low resolution often breaks down at higher resolution. In addition, the individual particle images used in single-particle processing are very noisy. If a given symmetry is actually present but not imposed in the reconstruction, the noise will break the symmetry, and through refinement will be strengthened iteratively, producing small but visible symmetry breaking artifacts in the model. That said, as implemented in EMAN, one clue is provided by the analysis. If symmetry is imposed in a case where it does not really exist, the class averages will not match the corresponding 3D model projections perfectly. Unfortunately, this case may occur also if the particle population is not structurally homogeneous.
For FAS, the reconstruction process converged properly both with and without the imposed 2-fold symmetry (Fig. 4). This result demonstrates that there is at least a 2-fold pseudosymmetry at the current resolution. In examining both 3D models, the overall quaternary structure arrangement is very similar. The individual class averages do show some small differences with the corresponding projections in several orientations. This observation indicates that either the symmetry may not really be present, that the particles are structurally heterogeneous, or that it may break down at higher resolution. Several tests were performed to attempt discriminating between these possibilities by simultaneously refining the data against several similar 3D models. The results of these tests were inconclusive, and thus we must conclude that we may not yet have the adequate resolution and/or number of particle images to resolve this issue unambiguously.
Several studies on human FAS and other sources have demonstrated that the enzymatically active species was dimeric (2, 3, 15, 18, 25, 32–34), with the two monomers juxtaposed head-to-tail resulting in two identical active sites for fatty acid biosynthesis (15, 24, 25). Because the active sites are completely independent functionally (35), the biochemical data supported the fidelity of the symmetric model discussed above. At the same time, however, the data did not exclude the possibility of an asymmetric molecule, especially when we consider that FAS might exist in several thermodynamically stable states. FAS is known to carry out over 50 reactions in synthesizing a single fatty acid molecule (4). It is conceivable, therefore, that the enzyme might have a large amount of structural flexibility given the fact that it is composed of two chemically identical multifunctional polypeptide chains with seven different catalytic activities. We recognize the importance of reaching a conclusive statement about the symmetry of the molecule, which would have substantial implications for interpretations of the enzyme's functions. We aim to resolve this issue by collecting data on a higher resolution instrument with a field-emission gun (36, 37). We expect that a 3D reconstruction obtained from a larger data set with higher resolution micrographs together with improved software will resolve this question.
Structural Interpretation of the 3D Models.
A prominent feature in our 3D reconstruction was the density bridge connecting both monomers (Fig. 4). The primary sequence for the FAS subunit contained a polypeptide of 649 amino acids (residues 981–1,629) separating domains I and II, designated as the interdomain region (4, 38). In the native, dimeric FAS, the interdomain regions are juxtaposed such that their interaction, presumably hydrophobic in nature (34, 39), holds the monomers together (4, 38). The observed bridge is probably where the interdomain regions combine. Recent experiments showed that this bridge likely is more than just a scaffold responsible for holding the enzyme and thus the partial activities together in a particular spatial arrangement. Labeling experiments with nanogold showed that one gold label, incorporated in the interdomain region of FAS at Cys-1225, reduced the overall activity by 50% (C.-Y.Y. and S.J.W., unpublished data). This result suggested that the interdomain region was critically important for the activity of the enzyme. Perhaps the interdomain region introduces flexibility into the structure, allowing the relevant parts of the enzyme to move around, much like the hinge region in immunoglobulins (40, 41).
Proposed Domain Assignment and Location of Active Site.
Our 3D reconstructions showed that each monomer exhibited three distinct sections: a head, a torso, and a slightly smaller foot region, designated I-III (Fig. 4 A and E). Earlier proteolytic digestion experiments on FAS revealed three fragments that could be purified while retaining all of their specific activities (3). These fragments were mapped on the primary sequence from the N to the C terminus as domains I–III, with domain I containing β-ketoacyl synthase, acetyl/malonyl transacylases, and β-hydroxyacyl dehydratase, domain II containing enoyl reductase, β-ketoacyl reductase, and ACP, and domain III containing thioesterase (2, 27). Each of these partial activities is composed of a single continuous stretch of amino acid residues connected by relatively short linking regions, the longest of which is only 25 residues. The exception is the interdomain region, which stretches from residue 981 to 1,629. Because the visual appearance of the 3D models shows a division into three parts, these structural regions (head, torso, and foot) may correspond to the functional domains seen after proteolysis (Fig. 4). Obviously, the resolution is not adequate to validate this suggestion definitively. We aim, however, to resolve this issue by obtaining a higher resolution reconstruction, possibly combined with domain localization using, for instance, antibody labeling (42, 43).
With the interdomain region and domains I, II, and III tentatively assigned in our 3D reconstruction, we propose the following location of the enzyme's active sites. In the 3D model, the walls of the clefts N and W are ≈20 and 25 Å apart, respectively (Fig. 4 D and H). This distance is close to that which the acyl chain could reach while attached to the ACP, namely ≈20 Å. Because each of the active sites is formed by two half-sites that arise from the juxtaposition of the two antiparallel monomers (15, 25), it thus seemed reasonable to propose that each cleft contained an active site (Fig. 4H). In this depiction, the partial activities would line the insides of each cleft. How the proposed subunit flexibility of FAS during turnover (4) exactly comes into play within the context of our 3D model and the proposed active sites remains to be solved by using further studies.
Acknowledgments
We acknowledge the help of Dr. Hiro Tsuruta on carrying out the x-ray solution scattering experiments at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of the U.S. Department of Energy (DOE), Office of Basic Energy Sciences (BES). The SSRL is funded by DOE BES and the SSRL Structural Biology Resource is funded by National Institutes of Health Grant P41RR012109 and the DOE Office of Biological and Environmental Research. This research has been supported by National Institutes of Health Grants P41RR02250 and GMS19091 and National Science Foundation grants through National Partnership for Advanced Computational Infrastructure–San Diego Supercomputer Center and National Center for Supercomputing Applications.
Abbreviations
- FAS
fatty acid synthase
- ACP
acyl carrier protein
- 3D
three-dimensional
- CTF
contrast transfer function
Footnotes
Available at www.ssec.wisc.edu/∼billh/vis5d.html.
References
- 1.Abu-Elheiga L, Matzuk M M, Abo-Hashema K A, Wakil S J. Science. 2001;291:2613–2616. doi: 10.1126/science.1056843. [DOI] [PubMed] [Google Scholar]
- 2.Wakil S J, Stoops J K, Joshi V C. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 3.Wakil S J. Biochemistry. 1989;28:4523–4530. doi: 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- 4.Chirala S S, Jayakumar A, Gu Z W, Wakil S J. Proc Natl Acad Sci USA. 2001;98:3104–3108. doi: 10.1073/pnas.051635998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan Z Y, Liu W, Hammes G G. Proc Natl Acad Sci USA. 1988;85:6328–6331. doi: 10.1073/pnas.85.17.6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chirala S S, Kasturi R, Pazirandeh M, Stolow D T, Huang W Y, Wakil S J. J Biol Chem. 1989;264:3750–3757. [PubMed] [Google Scholar]
- 7.Huang W Y, Chirala S S, Wakil S J. Arch Biochem Biophys. 1994;314:45–49. doi: 10.1006/abbi.1994.1410. [DOI] [PubMed] [Google Scholar]
- 8.Amy C M, Witkowski A, Naggert J, Williams B, Randhawa Z, Smith S. Proc Natl Acad Sci USA. 1989;86:3114–3118. doi: 10.1073/pnas.86.9.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schweizer M, Takabayashi K, Laux T, Beck K F, Schreglmann R. Nucleic Acids Res. 1989;17:567–586. doi: 10.1093/nar/17.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kameda K, Goodridge A G. J Biol Chem. 1991;266:419–426. [PubMed] [Google Scholar]
- 11.Paulauskis J D, Sul H S. Biochem Biophys Res Commun. 1989;158:690–695. doi: 10.1016/0006-291x(89)92776-9. [DOI] [PubMed] [Google Scholar]
- 12.Jayakumar A, Huang W Y, Raetz B, Chirala S S, Wakil S J. Proc Natl Acad Sci USA. 1996;93:14509–14514. doi: 10.1073/pnas.93.25.14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayakumar A, Tai M-H, Huang W-Y, Al-Feel W, Hsu M, Abu-Elheiga L, Chirala S S, Wakil S J. Proc Natl Acad Sci USA. 1995;92:8695–8699. doi: 10.1073/pnas.92.19.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith S. FASEB J. 1994;8:1248–1259. [PubMed] [Google Scholar]
- 15.Stoops J K, Wakil S J. J Biol Chem. 1981;256:5128–5133. [PubMed] [Google Scholar]
- 16.Tsuruta H, Brennan S, Rek Z U, Irving T C, Tompkins W H, Hodgson K O. J Appl Phys. 1998;31:672–682. [Google Scholar]
- 17.Thuman-Commike P A, Tsuruta H, Greene B, Prevelige P E, King J, Chiu W. Biophys J. 1999;76:2249–2261. doi: 10.1016/S0006-3495(99)77381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kashem M A, Hammes G G. Biochim Biophys Acta. 1988;956:39–48. doi: 10.1016/0167-4838(88)90295-6. [DOI] [PubMed] [Google Scholar]
- 19.Dubochet J, Adrian M, Chang J J, Homo J C, Lepault J, McDowall A W, Schultz P. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 20.Chiu W, Schmid M F. Curr Opin Biotechnol. 1993;4:397–402. doi: 10.1016/0958-1669(93)90004-g. [DOI] [PubMed] [Google Scholar]
- 21.Ludtke S L, Baldwin P R, Chiu W. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 22.Saad A, Ludtke S, Jakana J, Rixon F J, Tsuruta H, Chiu W. J Struct Biol. 2001;133:32–42. doi: 10.1006/jsbi.2001.4330. [DOI] [PubMed] [Google Scholar]
- 23.Ludtke S, Jakana J, Song J-L, Chuang D, Chiu W. J Mol Biol. 2001;314:241–250. doi: 10.1006/jmbi.2001.5133. [DOI] [PubMed] [Google Scholar]
- 24.Stoops J K, Wakil S J, Uberbacher E C, Bunick G J. J Biol Chem. 1987;262:10246–10251. [PubMed] [Google Scholar]
- 25.Smith S, Stern A, Randhawa Z I, Knudsen J. Eur J Biochem. 1985;152:547–555. doi: 10.1111/j.1432-1033.1985.tb09230.x. [DOI] [PubMed] [Google Scholar]
- 26.Tsukamoto Y, Wong H, Mattick J S, Wakil S J. J Biol Chem. 1983;258:15312–15322. [PubMed] [Google Scholar]
- 27.Mattick J S, Tsukamoto Y, Nickless J, Wakil S J. J Biol Chem. 1983;258:15291–15299. [PubMed] [Google Scholar]
- 28.Gabashvili I S, Agrawal R K, Spahn C M, Grassucci R A, Svergun D I, Frank J, Penczek P. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 29.Nogales E, Grigorieff N. J Cell Biol. 2001;152:F1–F10. doi: 10.1083/jcb.152.1.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erickson H P, Klug A. Philos Trans R Soc London B. 1970;261:105–118. [Google Scholar]
- 31.Thon F. Phase Contrast Electron Microscopy. New York: Academic; 1971. pp. 572–625. [Google Scholar]
- 32.Witkowski A, Joshi A, Smith S. Biochemistry. 1996;35:10569–10575. doi: 10.1021/bi960910m. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Porter J W. J Biol Chem. 1971;246:7780–7789. [PubMed] [Google Scholar]
- 34.Kumar S, Muesing R A, Porter J W. J Biol Chem. 1972;247:4749–4762. [PubMed] [Google Scholar]
- 35.Singh N, Wakil S J, Stoops J K. J Biol Chem. 1984;259:3605–3611. [PubMed] [Google Scholar]
- 36.Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann J B, Engel A, Fujiyoshi Y. Nature (London) 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 37.Fujiyoshi Y. FASEB J. 1999;13, Suppl. 2:S191–S194. doi: 10.1096/fasebj.13.9002.s191. [DOI] [PubMed] [Google Scholar]
- 38.Witkowski A, Rangan V S, Randhawa Z I, Amy C M, Smith S. Eur J Biochem. 1991;198:571–579. doi: 10.1111/j.1432-1033.1991.tb16052.x. [DOI] [PubMed] [Google Scholar]
- 39.Yun S L, Hsu R Y. J Biol Chem. 1972;247:2689–2698. [PubMed] [Google Scholar]
- 40.Kawamura S, Omoto K, Ueda S. J Mol Biol. 1990;215:201–206. doi: 10.1016/S0022-2836(05)80336-5. [DOI] [PubMed] [Google Scholar]
- 41.Smyth D S, Utsumi S. Nature (London) 1967;216:332–335. doi: 10.1038/216332a0. [DOI] [PubMed] [Google Scholar]
- 42.Lamy J, Billiald P, Taveau J C, Boisset N, Motta G. J Struct Biol. 1990;103:64–74. doi: 10.1016/1047-8477(90)90087-s. [DOI] [PubMed] [Google Scholar]
- 43.Boisset N, Penczek P, Taveau J C, Lamy J, Frank J, Lamy J. J Struct Biol. 1995;115:16–29. doi: 10.1006/jsbi.1995.1025. [DOI] [PubMed] [Google Scholar]