Abstract
The eIF2α kinases are a family of evolutionarily conserved serine/threonine kinases that regulate stress-induced translational arrest. Here, we demonstrate that the yeast eIF2α kinase, GCN2, the target phosphorylation site of Gcn2p, Ser-51 of eIF2α, and the eIF2α-regulated transcriptional transactivator, GCN4, are essential for another fundamental stress response, starvation-induced autophagy. The mammalian IFN-inducible eIF2α kinase, PKR, rescues starvation-induced autophagy in GCN2-disrupted yeast, and pkr null and Ser-51 nonphosphorylatable mutant eIF2α murine embryonic fibroblasts are defective in autophagy triggered by herpes simplex virus infection. Furthermore, PKR and eIF2α Ser-51-dependent autophagy is antagonized by the herpes simplex virus neurovirulence protein, ICP34.5. Thus, autophagy is a novel evolutionarily conserved function of the eIF2α kinase pathway that is targeted by viral virulence gene products.
During nutrient starvation and viral infection, cells need a strategy to synthesize essential proteins in the face of a limited supply caused either by environmental depletion or intracellular parasitism. This strategy must involve not only a mechanism for strict translational regulation, but also for generating new pools of amino acids from existing proteins. Accordingly, in nutrient-limiting conditions, eukaryotic cells simultaneously decrease overall protein synthesis and increase rates of protein degradation by an autophagic pathway (1), a process involving the bulk degradation of cellular contents by autophagolysosomes (2, 3). It is not known whether stress-induced translational repression and stress-induced autophagy are regulated by a common or by genetically distinct pathways.
The phosphorylation of eukaryotic initiation factor-2-α (eIF2α) at Ser-51 by a conserved family of eIF2α protein kinases is a central mechanism in stress-induced translation regulation (4–6), but the mechanisms of stress-induced regulation of autophagy are not well understood (7). Autophagy is required for survival during amino acid starvation of eukaryotic cells (2, 3), and several yeast and mammalian APG and AUT genes have been identified that are essential for autophagy (8–13). At least in yeast, these genes act downstream of the autophagy-inhibitory target of rapamycin (TOR) signaling pathway (14). However, little is known about upstream cellular genes that are essential for initiating the process of autophagy. Furthermore, it is not known whether stimuli other than amino acid starvation such as viral infection, endoplasmic reticulum stress, or heme depletion, which trigger stress-induced translational arrest, also trigger stress-induced autophagy.
We developed the hypotheses that eIF2α kinases, well characterized regulators of stress-induced translational control programs, are also involved in the regulation of stress-induced autophagy, and that specific stress stimuli that activate eIF2α kinase-dependent translational arrest also activate eIF2α kinase-dependent autophagy. In yeast, there is only one known eIF2α kinase, GCN2, which in response to starvation inhibits global protein synthesis and stimulates specific translation of GCN4, a basic leucine zipper transcriptional transactivator of amino acid biosynthetic genes (15). In mammals, there are at least four known eIF2α kinases (reviewed in ref. 6) including GCN2 (16), PKR (17, 18), PERK (19, 20), and HRI (21), which are activated by amino acid starvation, viral infection, endoplasmic reticulum stress, and heme depletion, respectively.
To evaluate the role of eIF2α kinases in autophagy regulation, we studied autophagy in nitrogen-starved yeast cells and in virus-infected and amino acid-starved mammalian cells with genetically engineered mutations in the eIF2α kinase signaling pathway. Our results indicate that the yeast eIF2α kinase signaling pathway is essential for starvation-induced autophagy and that the mammalian eIF2α kinase signaling pathway is essential for both virus- and starvation-induced autophagy. Furthermore, we demonstrate that mammalian eIF2α kinase-dependent autophagy is antagonized by the herpes simplex virus (HSV)-encoded neurovirulence gene product, ICP34.5.
Materials and Methods
Yeast Strains, Genetic Methods, and Media.
Yeast strains used for autophagy analyses included wild-type (wt) strain SEY6210 (MATα leu2–3, 112 ura3–52 hisΔ200 trp-Δ901 lys2–801 suc2-Δ9), and Δapg6 strain JCY3000 (SEY6210 Δvps30∷HIS3), which have been previously described (22). Δgcn2 yeast (SEY6210 Δgcn2∷URA3) and Δgcn4 yeast (SEY6210 Δgcn2∷URA3) were generated by homologous recombination with PCR-generated gcn2∷URA3 and gcn4∷URA3 fragments, respectively. SUI2-S51A (SEY6210 SUI2-S51A) and SUI2-S48A (SEY6210 SUI2-S48A) yeast were generated by PCR-based allele replacement (23) by using fusion fragments containing the SUI2 mutant allele (15) and the Kluyveromyces lactis URA3 gene. GCN2 and GCN4 disruption was confirmed by PCR analysis by using forward primers that hybridize upstream of the GCN2 (positions −96 to −73) and GCN4 (positions −104 to −79) loci and a reverse primer complementary to positions 226–248 of the URA3 ORF. Allelic replacement of wt SUI2 by the mutant SUI2 alleles was confirmed by DNA sequencing. Δgcn2 yeast were transformed with empty vector, p425GPD (SEY6210 Δgcn2∷URA3 p[LEU2]), with a vector encoding human pkr (p425GPD/pkr) (SEY6210 Δgcn2∷URA3 p[pkr, LEU2]) or with a vector encoding yeast GCN2 (p3060) (provided by Thomas Dever, National Institutes of Health, Bethesda) (SEY6210 Δgcn2∷URA3 p[GCN2, LEU2]). Yeast were grown in yeast extract/peptone/dextrose (YEPD), in synthetic medium lacking uracil or leucine for transformation selection (SC-ura or SC-leu), or in SD(-N) for starvation experiments, consisting of 2% glucose and 1.7 g⋅l−1 yeast nitrogen base without amino acids and ammonium sulfate.
Mammalian Cells.
Murine embryonic fibroblasts (MEFs) were prepared and cultured by using methods previously described (24) from pkr−/− and pkr+/+ mouse embryos (mixed C57/129sv backgrounds) (25). MEFs were prepared and cultured from mutant eIF2α S51A and wt eIF2α isogenic control mouse embryos as described (26).
Virus Strains.
The HSV-1 ICP34.5 mutant 17TermA (termed here HSV-1Δ34.5) and its marker-rescued virus 17TermAR (termed here wt HSV-1) were made in the background of strain 17 of HSV-1 and have been described (27, 28). Virus stocks were grown and titered in BHK-21 cells.
Immunochemical Procedures.
Yeast cell lysates were prepared as described (29), and MEF lysates were prepared by using an Nonidet P-40 lysis buffer supplemented with 2 mg⋅ml−1 NaF, 2 mM PMSF, and 1 μg⋅ml−1 leupeptin. Yeast and mammalian lysates were separated by SDS/PAGE, immunoblotted with anti-phosphospecific eIF2α Ab (30) (1:100 dilution) (Research Genetics, Huntsville, AL) and visualized by using ECL (Amersham Pharmacia). Two hundred micrograms of protein was loaded in each lane.
Yeast Autophagy Analysis.
Yeast were grown overnight in YEPD or liquid SC-leu (final OD 1–2), resuspended at a concentration of 2 × 107 cells ml−1 in either nutrient-rich media, nutrient-rich media with 0.2 μg⋅ml−1 rapamycin, or SD(-N), and incubated for 4 h in the presence of 1 mM PMSF to facilitate autophagic body detection. For differential interference contract (DIC) microscopy, cells were visualized by using DIC optics with a Plan Neofluar 100× 1.3 NA objective and imaged with a cooled CCD Hamamatsu (Middlesex, NJ) camera and Inovision esee software (Raleigh, NC). For electron microscopy, yeast were pelleted and fixed with 1.5% KMnO4 in water and embedded in low melting point agar. The cell pellet was cut into small blocks, dehydrated in ethanol, and embedded in Epon. Thin sections were stained with uranyl acetate and lead citrate. For quantitation of autophagic bodies, 200 cell profiles containing the vacuole were screened. Profiles containing one or more autophagic bodies inside the vacuole were classified as positive.
Mammalian Cell Autophagy Analysis.
For studies of virus-induced autophagy, MEFs were pretreated with 300 units⋅ml−1 of α-IFN for 16 h and infected with HSV-1 or HSVΔ34.5 at a multiplicity of infection of five plaque-forming units cell−1 or mock-infected, and autophagy was quantitated by measurement of long-lived protein degradation at serial time points after infection and by estimation of the cellular volume density of autophagic vacuoles at 5–6 h after infection. For studies of amino acid starvation-induced autophagy, MEFs were subjected to growth in normal media [Earles Balanced Salt Solution (EBSS) + 10% serum and complete amino acids], EBSS alone, or EBSS + 10 mM 3-methyladenine, and autophagy was quantitated by measurement of long-lived protein degradation at serial time points after starvation and by estimation of the cellular volume density of autophagic vacuoles at 2 h after starvation. 3-methyladenine is a nucleotide derivative that inhibits early stages of autophagosome formation (31); it was used in amino acid starvation experiments but not in virus infection experiments because of possible inhibitory effects on viral replication.
For measurement of long-lived protein degradation, cells were labeled before infection or starvation for 72 h in media containing reduced concentrations of unlabeled leucine (0.065 mM) and [3H]leucine [1 μCi⋅ml−1 (1 Ci = 37 GBq)]. Cells were washed and then incubated for 24 h in medium containing 2 mM unlabeled leucine to allow degradation of short-lived proteins and removal of unincorporated labeled leucine, and then virus was absorbed for 2 h or cells were subjected to amino acid starvation. At 0, 2, 4, 6, and 8 h after virus absorption or 0, 2, 4, and 6 h after amino acid starvation, aliquots of the media were measured for acid-soluble radioactivity. At the end of the incubation, the trichloroacetic acid-precipitable radioactivity of the cell monolayers was measured and the percentage of long-lived protein degradation at each time point was calculated by using a described formula (32).
For estimation of the cellular volume density of autophagic vacuoles, cells were fixed with 2.5% glutaraldehyde in Sorenson's buffer, scraped off the culture dish and pelleted, postfixed in 1% OsO4 for 1 h, and embedded in Epon. The volume fraction was estimated on pellet profiles within randomly selected grid squares by using point counting (33). Point counts over cells (reference space) were obtained on photographic negatives taken at ×400 magnification, covering the whole grid square. All autophagic vacuoles found in this area were photographed at ×12,000 magnification, and point counts were obtained by using the negatives. Cell size was estimated by using the mean area of nuclear profiles and the volume density of the nucleus. The cell size in wt HSV-1 and HSV-1Δ34.5-infected MEFs was estimated to be 1.4-fold higher than in mock-infected cells. Mock-infected cells were therefore not included in the analysis.
Results and Discussion
The eIF2α Kinase, GCN2, and Its Phosphorylation Target Site, Ser-51 of eIF2α, Are Essential for Starvation-Induced Autophagy in Yeast.
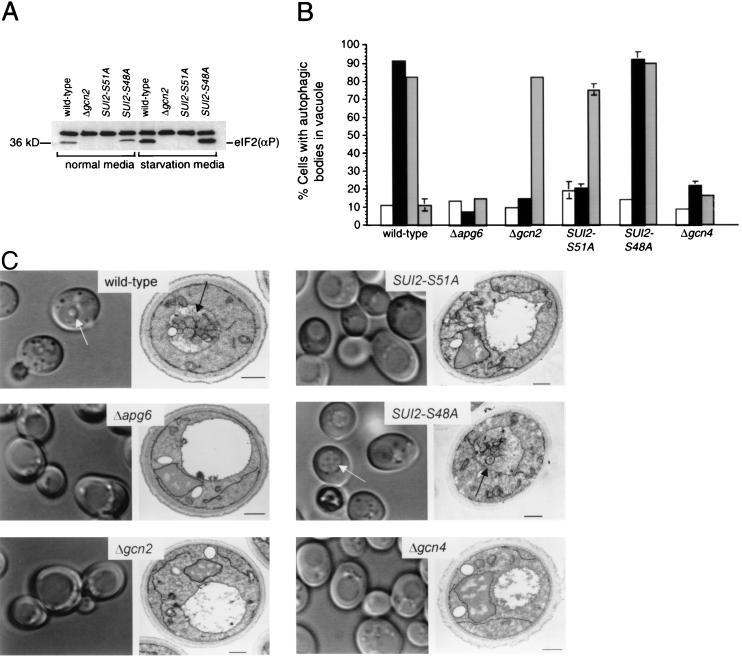
To investigate the role of the eIF2α kinase signaling pathway in nitrogen starvation-induced autophagy in yeast, we used PCR-based homologous recombination (23) to disrupt the yeast eIF2α kinase, GCN2, and to replace the yeast SUI2 gene that encodes eIF2α with two mutant alleles, including SUI2-S51A and SUI2-S48A. The SUI2-S51A allele is nonphosphorylatable by eIF2α kinases and defective in translational control during starvation, whereas the SUI2-S48A allele is phosphorylated by eIF2α kinases and phenotypically similar to wt SUI2 (15). We confirmed that the GCN2 disruption and the SUI2-S51A mutation, but not the SUI2-S48A mutation, blocked eIF2α Ser-51 phosphorylation in both normal growth conditions and in nitrogen starvation conditions by using an eIF2α Ser-51-phosphospecific Ab (Fig. 1A). Next, we measured the ability of wt yeast, yeast deleted of a known autophagy gene, APG6 (34), yeast disrupted of GCN2, and yeast with the SUI2-S48A or SUI2-S51A mutation to undergo autophagy in normal media, nitrogen deprivation media, or after treatment with rapamycin, an inhibitor of the TOR signaling pathway (refs. 35 and 36; Fig. 1 B and C). Previous studies have shown that, after nitrogen deprivation or rapamycin treatment, wt yeast but not yeast deleted of APG genes that are downstream of TOR, accumulate autophagic bodies within their central vacuole (the equivalent of mammalian lysosomes) (14, 37).
Figure 1.
Yeast with mutations in the GCN2-signaling pathway are defective in autophagy. (A) Western blot analysis of lysates from yeast grown in normal media (YEPD) or N deprivation media lacking amino acids and ammonium sulfate (SD-N), with an eIF2α Ser-51-phosphospecific Ab (1:100 dilution; Research Genetics). (B) Quantitation by DIC microscopy of autophagic body formation in yeast grown in normal growth conditions (open bars) or subjected to 4 h of nitrogen deprivation (black bars), treatment with 0.2 μg⋅ml−1 rapamycin (light gray bars), or treatment with 10 μg⋅ml−1 cycloheximide (dark gray bars). Cells with one or more autophagic bodies within the vacuole were scored as positive. A minimum of 100 cells was counted for each sample for each yeast strain. The results shown represent the mean ± SEM percentage of cells with autophagic bodies within the vacuole for triplicate samples. Similar results were obtained in five independent experiments. (C) Representative DIC micrographs (left columns of each panel) and electron micrographs (right columns of each panel) of yeast deprived of nitrogen for 4 h. Arrows in light micrographs denote representative cells which would be scored as positive in experiment shown in B. Arrows in electron micrograph denote representative autophagic bodies within the vacuole. (Scale bars, 0.5 μm.)
Using DIC microscopy (Fig. 1 B and C), we confirmed that wt yeast, but not Δapg6 yeast, had a significant increase in the percentage of cells with autophagic bodies within the vacuole following both nitrogen starvation and rapamycin treatment, as compared to basal levels observed during growth in nutrient-rich media (P < 0.001, t test). In contrast, Δgcn2 yeast and SUI2-S51A mutant yeast displayed a phenotype that differed from either wt or Δapg6 yeast. In Δgcn2 yeast and SUI2-S51A mutant yeast, there was no increase in autophagy after nitrogen starvation as compared to levels observed during normal growth conditions. However, there was a significant increase in autophagy in Δgcn2 and SUI2-S51A yeast after rapamycin treatment (P < 0.001, t test). Yeast with the control SUI2-S48A mutation were indistinguishable from wt yeast in their capacity to undergo autophagy in response to nitrogen starvation or rapamycin treatment. These data indicate that GCN2, and the Gcn2p target phosphorylation site, eIF2α Ser-51, are essential for nitrogen starvation-induced autophagy. However, neither GCN2 nor eIF2α Ser-51 are essential for rapamycin-induced autophagy, suggesting that these molecules act either upstream or in an independent pathway, rather than downstream of Tor signaling.
We performed electron microscopic analysis to confirm the defect in starvation-induced autophagy in Δgcn2 and SUI2-S51A mutant yeast (Fig. 1C). In thin sections of wt yeast and SUI2-S48A yeast, autophagic bodies were seen in 49% and 34% of cell profiles that contained the vacuole, whereas autophagic bodies were seen rarely in vacuole-containing profiles of Δgcn2 yeast (4%), SUI2-S51A mutant yeast (4%), or Δapg6 yeast (1%). Although the vacuoles of Δgcn2 and SUI2-S51A mutant yeast (like those of Δapg6 yeast) rarely contained autophagic bodies, they had a different ultrastructural appearance than the vacuoles of Δapg6 yeast; the latter were mostly empty and the former frequently contained amorphous material and electron dense particles. The molecular basis for this difference is not known but may relate to the different steps of the autophagy pathway that are blocked by the gcn2 null mutation and the SUI2-S51A mutation as compared to the apg6 null mutation. Alternatively, the amorphous material in the vacuole of Δgcn2 and SUI2-S51A mutant yeast may enter by the vacuolar protein sorting pathway, which is deficient in Δapg6 yeast (22). Regardless of the molecular basis for this difference in vacuolar ultrastructural appearance, the paucity of detectable autophagic bodies in Δgcn2 and SUI2-S51A yeast confirms the essential role for GCN2 and eIF2α Ser-51 in autophagy.
The GCN2-Dependent Transcriptional Transactivator, GCN4, but Not Translational Arrest, Is Essential for Starvation-Induced Autophagy in Yeast.
In response to starvation, eIF2α Ser-51 phosphorylation by Gcn2p both mediates a global translational arrest and activates Gcn4p (15), a transcriptional transactivator of starvation-induced genes. To address which downstream function of eIF2α kinase signaling is involved in autophagy induction, we evaluated whether pharmacological inhibition of translation triggers autophagy and/or whether the transcriptional transactivator, GCN4, is required for nitrogen starvation- and rapamycin-induced autophagy. We found that treatment of wt yeast with the translational inhibitor, cycloheximide, had no stimulatory effect on autophagy (Fig. 1B). However, disruption of GCN4 blocked nitrogen starvation-induced autophagy as well as rapamycin-induced autophagy (Fig. 1 B and C). Thus, autophagy induction in yeast requires function of the GCN4 transcriptional transactivator and is unlikely to be an indirect consequence of eIF2α kinase-dependent translational arrest. The concept that autophagy is not merely a function of eIF2α-dependent translational arrest but requires de novo protein synthesis is further supported by previous studies indicating that cycloheximide blocks later stages of autophagy, including autophagosomal expansion (38) and delivery of lysosomal enzymes (39). The recent observation that certain APG genes (e.g., APG1, APG13, and APG14) are regulated in a GCN4-dependent fashion (40) is likely to explain the requirement for GCN4 in autophagy signaling. Furthermore, GCN4, which is required for both nitrogen starvation- and rapamycin-induced autophagy, may be a convergent target of regulation by the autophagy stimulatory eIF2α kinase-signaling pathway and the autophagy inhibitory TOR-signaling pathway.
The Mammalian IFN Inducible eIF2α Kinase, PKR, Promotes Autophagy in GCN2-Disrupted Yeast.
Together, the above data demonstrate an essential role for the yeast eIF2α kinase signaling pathway and its downstream target, GCN4, in autophagy. To determine whether this pathway is evolutionarily conserved, we investigated whether PKR, one of the four identified mammalian eIF2α kinases, can functionally substitute for GCN2 in starvation-induced autophagy in yeast. Although thought to be involved primarily in translational inhibition during viral infections in mammalian cells (reviewed in refs. 17 and 18), PKR is known to functionally substitute for GCN2 in the GCN4 translational control mechanism in yeast (41). PKR overexpression in yeast results in autophosphorylation, activation of downstream targets, and a slow growth phenotype, and it has been suggested that the eIF2α kinase activity is constitutive (41), rather than dependent on endogenous activators.
To avoid constitutive eIF2α kinase activity associated with PKR overexpression, we selected for stable pkr transformants of Δgcn2 yeast that had a normal growth phenotype. We confirmed that stable pkr transformants that had a normal growth phenotype restored eIF2α kinase activity during N starvation conditions, and that these pkr-transformed Δgcn2 yeast lacked spontaneous activation of autophagy during growth in normal media (Fig. 2 A and B). After nitrogen starvation, DIC microscopic analysis indicated that autophagy levels increased in pkr-transformed Δgcn2 yeast to levels similar to those observed in GCN2-transformed Δgcn2 yeast (Fig. 2 B and C). Similarly, on electron microscopic analysis of nitrogen-starved yeast, the percentage of cell profiles with autophagic bodies within the vacuole was 50% in GCN2-transformed Δgcn2 yeast and 38% in pkr-transformed Δgcn2 yeast as compared to only 12% in Δgcn2 yeast transformed with an empty vector (see representative photomicrographs, Fig. 2C). Thus, pkr can functionally substitute for GCN2 in the control of starvation-induced autophagy in yeast, raising the possibility that autophagy may be an evolutionarily conserved function of different eIF2α kinase family members that have diverged to respond to different stress signals.
Figure 2.
Mammalian pkr promotes autophagy in GCN2-disrupted yeast. (A) Western blot analysis of lysates from N-starved Δgcn2 yeast by using an eIF2α Ser-51-phosphospecific Ab. (B) Quantitative effects of GCN2 and pkr transformation on autophagic body formation in Δgcn2 yeast grown in normal growth conditions (open bars), or subjected to 4 h of nitrogen deprivation (black bars) or treatment with 0.2 μg⋅ml−1 rapamycin (light gray bars). Cells with one or more autophagic bodies within the vacuole were scored as positive. A minimum of 100 cells was counted for each sample for each yeast strain. The results shown represent the mean ± SEM percentage of cells with autophagic bodies within the vacuole for triplicate samples. Similar results were obtained in five independent experiments. Similar results were also observed for five independent clones of pkr-transformed Δgcn2 yeast that displayed normal growth phenotypes in nutrient rich media. (C) Representative light micrographs (Left column) and electron micrographs (Right column) of GCN2- and pkr-transformed Δgcn2 yeast deprived of nitrogen for 4 h. Arrows in light micrographs denote representative cells that would be scored as positive in experiment shown in B. Arrows in electron micrographs denote representative autophagic bodies within the vacuole. (Scale bars, 0.5 μm.)
PKR Is Required for Virus-Induced Autophagy in MEFs, Which Is Blocked by the HSV-1 Encoded PKR Inhibitor and Neurovirulence Protein, ICP34.5.
We evaluated whether autophagy in mammalian cells also is regulated by the eIF2α kinase pathway and whether divergent triggers of eIF2α kinase-dependent translational control in mammalian cells also trigger autophagy. We used an HSV infection model because this virus both activates the antiviral eIF2α kinase, PKR, and encodes a neurovirulence gene product, ICP34.5, that blocks PKR translational repression by binding to protein phosphatase 1α and redirecting it to dephosphorylate eIF2α (42–44). Therefore, by comparing autophagy in cells with genetic alterations in PKR and eIF2α that are infected with either a wt strain of HSV-1 or a strain of HSV-1 lacking the PKR inhibitor, ICP34.5, we could assess whether viral infection triggers eIF2α kinase-signaling-dependent autophagy.
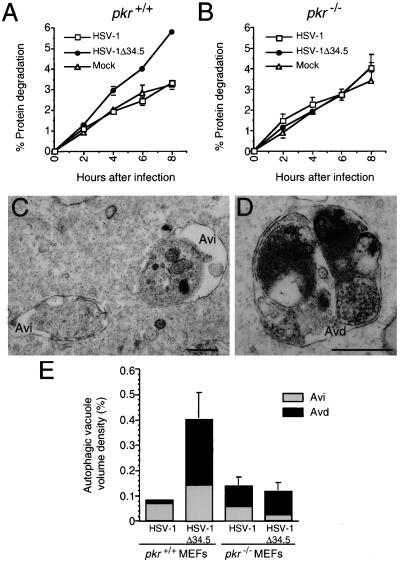
Pkr+/+ and pkr−/− MEFs were pretreated with α-IFN and infected with either a mutant strain of HSV-1 lacking ICP34.5 (HSV-1Δ34.5), or a wt marker-rescued strain of HSV-1Δ34.5 (referred to herein as wt HSV-1). α-IFN pretreatment was used to optimize induction of PKR activity in response to virus infection. Using a long-lived protein degradation assay to quantitate autophagy, we found that wt, pkr+/+ MEFs infected with HSV-1Δ34.5 had a significant increase in degradation of long-lived cellular proteins at 4, 6, and 8 h after infection (P = 0.013, 0.003, and <0.001, respectively; t test) (Fig. 3A) as compared to mock-infected pkr+/+ MEFs. Two observations suggest that this virus-induced increase in long-lived protein degradation depends on pkr function. First, wt HSV-1 that encodes ICP34.5, an antagonist of PKR, did not increase long-lived protein degradation as compared to mock infection in pkr+/+ MEFs. Second, HSV-1Δ34.5 failed to increase long-lived protein degradation in pkr−/− MEFs (Fig. 3B). Thus, HSV-1, deleted of a gene that antagonizes pkr function, increases long-lived protein degradation in pkr+/+ but not in pkr-deficient cells. These data demonstrate that viral infection can trigger increases in long-lived protein degradation in mammalian cells and that the host gene, pkr is required for this response.
Figure 3.
Pkr−/− MEFs are deficient in autophagic protein degradation and autophagic vacuole accumulation induced by α-IFN and HSV-1Δ34.5 infection. (A and B) Cumulative percentage degradation of long-lived cellular proteins in pkr+/+ and pkr−/− MEFs, respectively. Results are mean (± SEM) of triplicate wells. Similar results were obtained in five independent experiments. (C and D) Electron micrographs showing examples of early (Avi) and late (Avd) autophagic vacuoles, respectively, in pkr+/+ MEFs infected with HSV-1Δ34.5. (Scale bars, 0.5 μm.) (E) Volume density of early (Avi) and late (Avd) autophagic vacuoles in pkr−/− and pkr+/+ MEFs infected with wt HSV-1 and HSV-1Δ34.5. Error bars represent SEM of the volume densities of three to five grid squares. Similar results were obtained in two independent experiments.
To confirm that the increase in long-lived protein degradation measured in HSV-1Δ34.5-infected pkr+/+ MEFs represents an increase in autophagy, we performed quantitative electron microscopy of wt HSV-1- and HSV-1Δ34.5-infected pkr+/+ and pkr−/− MEFs (Fig. 3 C–E). Autophagic vacuoles were defined as membrane-bound 0.3 to 2 μm vesicles with clearly recognizable cytoplasmic contents, and classified as early (Avi), containing morphologically intact cytoplasm (Fig. 3C) or late (Avd), containing partially degraded but identifiable cytoplasmic material (Fig. 3D). There was a significant increase in the percentage of total cellular volume of both early autophagic vacuoles and late autophagic vacuoles in HSV-1Δ34.5-infected pkr+/+ MEFs as compared to HSV-1Δ34.5-infected pkr−/− MEFs and wt HSV-1-infected pkr+/+ and pkr−/− MEFs (P = 0.037; ANOVA; Fig. 3E). Therefore, the pkr-dependent increase in long-lived protein degradation in MEFs infected with HSV-1Δ34.5 is associated with a pkr-dependent increase in autophagic vacuole accumulation.
Together with the biochemical data, these results indicate that HSV-1 infection induces pkr-dependent autophagy in mammalian cells that is antagonized by the HSV-1 neurovirulence gene product, ICP34.5. To the best of our knowledge, these findings represent the first example of cellular autophagy induction in response to viral infection. Previous studies have shown that poliovirus infection induces the formation of double- membraned vesicles that share some similarities with autophagic vacuoles (45). However, unlike our observations in HSV-1Δ34.5-infected MEFs, these vesicles have not been shown to be associated with autophagic degradation of intracytoplasmic contents. Interestingly, the pathogenic intracellular bacterium Legionella pneumophila grows in mammalian cells in modified membranous compartments that are formed by an autophagic-like mechanism (46), and cells infected with less virulent mutant bacteria have an accumulation of lysosomal markers and degradative enzymes in these compartments (47). This suggests that wt bacterial proteins may prevent normal maturation of early autophagic vacuoles into late degradative vacuoles. Thus, virulence gene products encoded by diverse intracellular pathogens such as HSV-1 and Legionella may have devised different strategies to antagonize autophagic protein degradation.
eIF2α Ser-51 Is Required for Virus-Induced and Amino Acid Starvation-Induced Autophagy in MEFs.
In mammalian cells, there are at least four different eIF2α kinases that are specialized to respond to divergent, nonoverlapping stress stimuli (reviewed in ref. 6), including PKR, which is activated primarily by double-stranded RNA and inflammatory mediators (reviewed in refs. 17 and 18). Despite the specificity of stress stimuli involved in eIF2α kinase activation, the downstream target of the different activated eIF2α kinases, the Ser-51 residue of eIF2α, is identical. Therefore, to both assess the role of this downstream target in mammalian autophagy regulation and the role of the eIF2α kinase-signaling pathway in mammalian autophagy induced by a more classic stimulus, amino acid starvation, we performed autophagy studies in MEFs with a homozygous eIF2α S51A mutation that blocks eIF2α Ser-51 phosphorylation (Fig. 4A; ref. 26).
Figure 4.
Mutant eIF2α S51A MEFs are deficient in HSV-1Δ34.5 infection-induced and amino acid starvation-induced autophagic protein degradation and autophagic vacuole accumulation. (A) Western blot analysis of MEF lysates by using an eIF2α Ser-51-phosphospecific Ab. The phosphorylation of wt eIF2α at 0 h after starvation is presumed to result from eIF2α kinase activation that occurs during washing and cell lysis (29). (B, C, E, and F) Cumulative percentage degradation of long-lived cellular proteins in wt eIF2α MEFs (B and E) at serial time points after virus infection (B) or amino acid starvation (E), and in homozygous mutant eIF2α S51A MEFs (C and F) at serial time points after virus infection (C) or amino acid starvation (F). Results are mean (±SEM) of triplicate wells. Similar results were obtained in three independent experiments. (D and G) Volume density of autophagic vacuoles in wt and mutant eIF2α S51A MEFs subjected to 5 h of virus infection (D) or subjected to 2 h of amino acid starvation (G). Each bar represents volume density of both early (Avi) and late (Avd) autophagic vacuoles. The percentage of early (Avi) vacuoles is indicated above each bar. Error bars represent SEM of the volume densities of three to four grid squares.
We assessed whether eIF2α Ser-51, the phosphorylation site required for starvation-induced autophagy in yeast, is also required for HSV-1Δ34.5- and amino acid starvation-induced autophagy in mammalian cells. In isogenic control MEFs with wt eIF2α, we found that HSV-1Δ34.5 infection as compared to wt HSV-1 infection resulted in a significant increase in both long-lived protein degradation (P = 0.051, 0.035, and 0.003 at 4, 6, and 8 h after infection, respectively) and autophagic vacuole volume density (P = 0.002; Fig. 4 B and D). Similarly, in the control MEFs, amino acid starvation also resulted in a significant increase in 3-methyladenine-inhibitable long-lived protein degradation (P = 0.018, 0.002, and 0.005 at 2, 4, and 6 h after starvation, respectively) and autophagic vacuole volume density (P = 0.001; Fig. 4 E and G). However, neither of the stimuli, which increased autophagy in control MEFs—i.e., HSV-1Δ34.5 infection or amino acid starvation, increased long-lived protein degradation or autophagic vacuole volume density in the homozygous mutant eIF2α S51A MEFs that contain a nonphosphorylatable eIF2α (Fig. 4 C, D, F, and G).
These data demonstrate that the eIF2α kinase substrate site, eIF2α Ser-51, is required for both virus and amino acid starvation-induced autophagy in mammalian cells. Further studies are required to determine whether mammalian GCN2, like yeast GCN2, is the eIF2α kinase that acts upstream of eIF2α during starvation-induced autophagy. It is also as-of-yet unknown, whether other stimuli such as endoplasmic reticulum stress and heme deficiency, which activate the eIF2α kinases, PERK and HRI, respectively (21), induce eIF2α Ser-51-dependent autophagy in mammalian cells.
Conclusions
In summary, our data indicate a novel, evolutionarily conserved role of the yeast and mammalian eIF2α kinase pathway in the regulation of autophagy. We have shown that the yeast and human eIF2α kinases, GCN2 and pkr, the yeast and human phosphorylation site targeted by eIF2α kinases, eIF2α Ser-51, and the yeast GCN4 transcriptional transactivator, are all essential for stress-induced autophagy. Furthermore, our data demonstrate that divergent stress stimuli (e.g., nutrient deprivation and virus infection) known to stimulate eIF2α kinase-dependent translational arrest also stimulate eIF2α kinase-dependent autophagy. It is already well-established that autophagy is an important cellular defense mechanism against one of these stress stimuli, nutrient deprivation. Our observations indicating that autophagy is regulated by PKR, a well characterized antiviral molecule, and that autophagy is antagonized by the HSV-1 neurovirulence gene product suggest that autophagy is also an important cellular defense mechanism against viral infections. During nutrient starvation and viral infection, the synchronous regulation of translation and autophagy by the eIF2α kinase signaling pathway may be a fundamental mechanism that permits eukaryotic cells to successfully adapt to environmental stress. As part of their virulence strategy, viruses may subvert this host adaptive mechanism.
Acknowledgments
We thank Thomas Dever, Richard Thompson, and Bryan Williams for providing reagents, and David Ron for helpful discussions. This work was supported by National Institutes of Health Grants RO1 AI44157 and ROI CA84254 (to B.L.), ROI AR45019 and ROI AI39616 (to H.V.), ROI EY09083 (to D.L.), and AI42394 (to R.K.), and Royal Society Grant 574006.G503/21185 (to E.-L.E.). B.L. also was supported by a Irma T. Hirschl Trust Career Scientist Award and a Mallinckrodt Scholar Award.
Abbreviations
- MEFs
murine embryonic fibroblasts
- DIC
differential interference contrast
- HSV-1
herpes simplex virus type 1
- wt
wild type
- TOR
target of rapamycin
- PKR
double-stranded RNA-dependent protein kinase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mortimore G E, Poso A R. Methods Enzymol. 1988;166:461–476. doi: 10.1016/s0076-6879(88)66060-5. [DOI] [PubMed] [Google Scholar]
- 2.Dunn W A., Jr Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 3.Klionsky D J, Emr S D. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinnebusch A G. Semin Cell Biol. 1994;5:417–426. doi: 10.1006/scel.1994.1049. [DOI] [PubMed] [Google Scholar]
- 5.de Haro C, Mendez R, Santoyo J. FASEB J. 1996;10:1378–1387. doi: 10.1096/fasebj.10.12.8903508. [DOI] [PubMed] [Google Scholar]
- 6.Dever T E. Trends Biochem Sci. 1999;10:398–403. doi: 10.1016/s0968-0004(99)01457-7. [DOI] [PubMed] [Google Scholar]
- 7.van Sluijters D A, Dubbelhuis P F, Blommaart E F, Meijer A J. Biochem J. 2000;351:545–550. doi: 10.1042/0264-6021:3510545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohsumi Y. Philos Trans R Soc London B. 1999;354:1577–1581. doi: 10.1098/rstb.1999.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf D H. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 10.Liang X H, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Nature (London) 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 11.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. J Cell Biol. 2001;152:657–667. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. J Biol Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 14.Noda T, Ohsumi Y. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 15.Dever T E, Feng L, Wek R C, Cigan A M, Donahue T F, Hinnebusch A G. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 16.Berlanga J J, Santoyo J, de Haro C. Eur J Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 17.Williams B R. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 18.Gale M, Jr, Katze M G. Pharmacol Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 19.Shi Y, Vattem K, Sood R, An J, Liang J, Stramm L, Wek R C. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harding H P, Zhang Y, Ron D. Nature (London) 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 21.Chefalo P J, Oh J, Rafie-Kolpin M, Kan B, Chen J J. Eur J Biochem. 1998;258:820–830. doi: 10.1046/j.1432-1327.1998.2580820.x. [DOI] [PubMed] [Google Scholar]
- 22.Seaman M N J, Marcusson E G, Cereghino J L, Emr S D. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erdeniz N, Mortensen U H, Rothstein R. Genome Res. 1997;7:1174–1183. doi: 10.1101/gr.7.12.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollock J L, Presti R M, Paetzold S, Virgin H W, IV. Virology. 1997;227:168–179. doi: 10.1006/viro.1996.8303. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y-L, Reis L F, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams B R G, Aguet M, Weissmann C. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman R J. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 27.Bolovan C A, Sawtell N M, Thompson R L. J Virol. 1994;68:48–55. doi: 10.1128/jvi.68.1.48-55.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leib D A, Machalek M A, Williams B R G, Silverman R H, Virgin H W. Proc Natl Acad Sci USA. 2000;97:6097–6101. doi: 10.1073/pnas.100415697. . (First Published May 9, 2000; 10.1073/pnas.100415697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dever T E. Methods. 1997;11:403–417. doi: 10.1006/meth.1996.0437. [DOI] [PubMed] [Google Scholar]
- 30.DeGracia D J, Sullivan J M, Neumar R W, Alousi S S, Hikade K R, Pittman J E, White B C, Rafols J A, Krause G S. J Cereb Blood Flow Metab. 1997;17:1291–1302. doi: 10.1097/00004647-199712000-00004. [DOI] [PubMed] [Google Scholar]
- 31.Seglen P O, Gordon P B. Proc Natl Acad Sci USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gronostajski R M, Pardee A B. J Cell Physiol. 1984;119:127–132. doi: 10.1002/jcp.1041190120. [DOI] [PubMed] [Google Scholar]
- 33.Weibel E R. Int Rev Cytol. 1969;26:235–302. doi: 10.1016/s0074-7696(08)61637-x. [DOI] [PubMed] [Google Scholar]
- 34.Kametaka S, Okano T, Ohsumi M, Ohsumi Y. J Biol Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 35.Heitman J, Movva N R, Hall M N. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 36.Schmelzle T, Hall M N. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 37.Tsukada M, Ohsumi Y. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 38.Abeliovich H, Jr, Dunn W A, Jr, Kim J, Klionsky D J. J Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence B P, Brown W J. J Cell Sci. 1993;105:473–480. doi: 10.1242/jcs.105.2.473. [DOI] [PubMed] [Google Scholar]
- 40.Natarajan K, Meyer M R, Jackson B M, Slade D, Roberts C, Hinnebusch A G, Marton M J. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dever T E, Chen J-J, Barber G N, Cigan A M, Feng L, Donahue T F, London I M, Katze M G, Hinnebusch A G. Proc Natl Acad Sci USA. 1993;90:4616–4620. doi: 10.1073/pnas.90.10.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chou J, Kern E R, Whitley R J, Roizman B. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 43.Chou J, Chen J J, Gross M, Roizman B. Proc Natl Acad Sci USA. 1995;92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He B, Gross M, Roizman B. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suhy D A, Giddings T H, Jr, Kirkegaard K. J Virol. 2000;74:8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanson M S, Isberg R R. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrews H L, Vogel J P, Isberg R R. Infect Immun. 1998;66:950–958. doi: 10.1128/iai.66.3.950-958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]