Abstract
Physically distinguishable microdomains associated with various functional membrane proteins are one of the major current topics in cell biology. Glycosphingolipids present in such microdomains have been used as “markers;” however, the functional role of glycosyl epitopes in microdomains has received little attention. In this review, I have tried to summarize the evidence that glycosyl epitopes in microdomains mediate cell adhesion and signal transduction events that affect cellular phenotypes. Molecular assemblies that perform such functions are hereby termed “glycosynapse” in analogy to “immunological synapse,” the membrane assembly of immunocyte adhesion and signaling. Three types of glycosynapses are so far distinguishable: (i) Glycosphingolipids organized with cytoplasmic signal transducers and proteolipid tetraspanin with or without growth factor receptors; (ii) transmembrane mucin-type glycoproteins with clustered O-linked glycoepitopes for cell adhesion and associated signal transducers at lipid domain; and (iii) N-glycosylated transmembrane adhesion receptors complexed with tetraspanin and gangliosides, as typically seen with the integrin–tetraspanin–ganglioside complex. The possibility is discussed that glycosynapses give rise to a high degree of diversity and complexity of phenotypes.
The structure and function of “microdomains” having different physical properties and specialized functions is one of the major topics of current cell biology (for review, see refs. 1–6). The development of this area has occurred over the past two decades, based on key observations such as (i) the distinctive clustering of glycosphingolipids (GSLs)† and/or glycoproteins, which has been observed by scanning electron microscopy with freeze-fracture (7). GSL clusters were observed even on liposome surfaces prepared without cholesterol (8). (ii) Detergent-insolubility of GSLs together with pericellular matrix proteins and microfilaments at fibroblast adhesion sites, termed detergent-insoluble membrane (DIM) (9, 10). (iii) Exclusive presence of “fucolipids” in villous membranes separated from intestinal mucosa (11), and enriched sphingolipids vs. glycerophospholipids in apical vs. basolateral membranes in kidney epithelial cells (12). (iv) Association of Src family kinases with glycosylphosphatidylinositol-anchored proteins found originally by Horejší and colleagues (13), and noted later in DIM or glycolipid-enriched microdomain (GEM) of many types of cells. (v) Specific membrane domains termed “caveolae,” associated with the scaffold protein caveolin (14, 15), and claimed to be involved in endocytosis and signal transduction (1). Caveolar membranes showed a composition similar to that of other low-density membranes regardless of the presence or absence of caveolin (1, 16). Various terms such as caveolae (1), caveola-like (16), DIM (10), detergent-resistant membrane (2), GEM (17–20), and most frequently “raft” (5, 6) were used to refer to similar mixtures of membranes with low density.
In most of these studies, the presence of GSLs was noted simply as a marker of low-density membrane fraction. Little attention was paid to the structural variety of glycoclusters and their potential roles in microdomains, even though carbohydrate-dependent cell adhesion and/or signaling have been well studied subjects (for reviews, see refs. 21–31).
Our studies have established novel structures of globo-series, lacto-series types 1 and 2, and hybrid types (≈30 structures in total; for review, see ref. 23). Some of the novel structures expressed highly at defined stages of development are termed “stage-specific embryonic antigens,” and some expressed highly after oncogenic transformation are identified as tumor-associated antigens (Table 1). We subsequently tried to determine the biological significance of the glycosylation changes during development or tumor progression. Certain GSLs expressed highly at defined stages of these processes have been identified as adhesion molecules recognized by two mechanisms: (i) through binding proteins, i.e., endogenous lectins—mainly selectins (e.g., refs. 32–36; for review, see refs. 26–28) and siglecs (e.g., 37–39; for review 30); and (ii) through complementary carbohydrates expressed at the target cell surface, based on carbohydrate to carbohydrate interaction (40–47; for review, see refs. 24 and 25). GSL antigens involved in cell adhesion are highlighted in Table 1, with appropriate references, and examples of these mechanisms are given in subsequent sections.
Table 1.
Major GSLs identified as developmentally regulated or tumor-associated antigens involved in cell adhesion
Each GSL class has a characteristic core structure as indicated by underlining in the table, i.e., Galα4Gal for globo-series; Galβ3GlcNAc for lacto-series type 1; Galβ4GlcNAc for lacto-series type 2; GalNAcβ4Gal and SAα3Gal for ganglio-series. In hybrid-type, “a” as indicated is ganglio-, and “b” is lacto type 1. TerCa, teratocarcinoma; RC Ca, renal cell carcinoma; SA, sialic acid; Fuc, fucose.
GSLs, particularly gangliosides, have been characterized by their ability to interact with key transmembrane receptors or signal transducers involved in cell adhesion and signaling. Examples are (i) modulation of growth factor receptors with intrinsic tyrosine kinases (48–51; for review, see ref. 31); (ii) modulation of integrin function (52) through their complex with tetraspanin CD9 (53); and (iii) interaction with and activation of cytoplasmic signal transducers such as Src family kinases and small G proteins present in microdomain (17–20).
Based on these findings, I have tried to construct a conceptual view with a focus on how the mechanistic process of carbohydrate-dependent cell adhesion/recognition is converted to signaling impulses affecting cellular phenotype—the essential theme of “cell sociology.”
Type 1 Glycosynapse:‡ GSL Clusters, Organized with Signal Transducers, Involved in Cell Adhesion Coupled with Signal Transduction
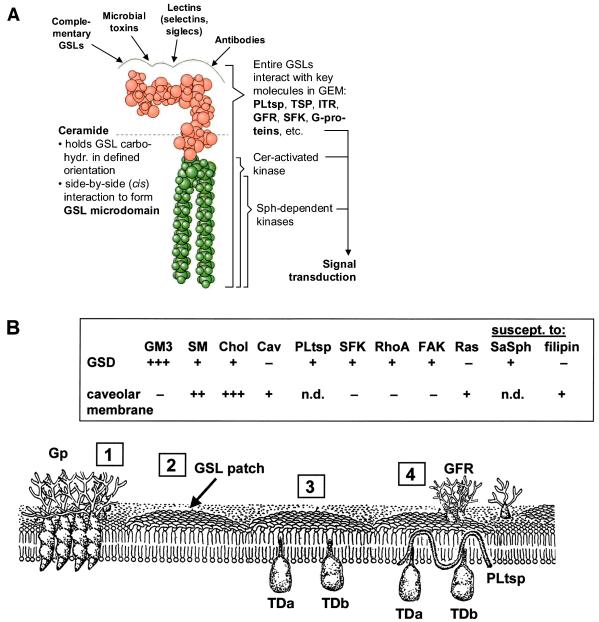
Regardless of the structural variations of the carbohydrate moieties, GSLs have a common minimum-energy conformational structure in which the oligosaccharide chain, having specific ligand-binding activity, is oriented at a defined angle to the axis of ceramide (Fig. 1A). They have the physical property to form extensive clusters (“GSL patch”), separate from glycerophospholipids and some types of glycoprotein clusters, in the plasma membrane (Fig. 1B). Although the majority of glycoproteins are separated from GSL patches, some functional glycoproteins such as growth factor receptors associated with intrinsic tyrosine kinases are often found within GSL domains and display clear interaction with and functional susceptibility to gangliosides (see below).
Figure 1.
GSL conformation and organization in membrane. (A) Minimum-energy model of GSL, Gb5 taken as an example, showing that the axis of carbohydrate moiety is perpendicular to the axis of ceramide (133, 134). The surface profile of carbohydrate provides binding sites for Abs, lectins, microbial toxins, and complementary GSLs. (Left) Role of ceramide (Cer) to form microdomain. (Right) Interaction of entire GSLs with key molecules (TSP, tetraspanin; ITR, integrin receptor; GFR, growth factor receptor; SFK, Src family kinase). These molecules, together with Cer or sphingosine (Sph) released from sphingomyelin, activate or modulate their respective kinases and are involved in various ways in control of signal transduction. (B) GSLs are clustered (“GSL patch”) and inserted via ceramide into the outer leaflet of the membrane, without (“2”) or with (“3”) signal transducers (TDa, TDb). GSL clusters organized with signal transducers, PL tetraspanin (PLtsp), and growth factor receptor (GFR) are shown in “4.” Glycoprotein (Gp) clusters (“1”) in many cases may be separated from GSL patches. (Inset) Contrasting properties of glycosignaling domain (GSD), the domain enriched in GSL, TD, and PL, separable from caveolar membrane. Data from refs. 20 and 57 and from K. Handa, D. A. Withers, and S.H. (unpublished data). Cav, caveolin; SaSph, sialyl 2 → 1 sphingosine.
We have studied the composition and functional roles of GSL glycosynapses in mouse B16 melanoma (19, 20), human teratocarcinoma 2102 (54), mouse neuroblastoma Neuro2a (55), and human renal cell carcinoma TOS-1 cell lines (56). In each case, the major GSL or ganglioside in the assembly mediates cell adhesion or displays susceptibility to exogenous gangliosides to activate cytoplasmic signal transducers. This event induces downstream signaling leading to change in transcription factors and/or consequent phenotypic changes. Well studied examples are signal transduction events resulting from GM3-dependent adhesion of B16 cells (42) and Gb4-dependent adhesion of 2102 cells (54). GM3-enriched membranes in B16 cells are separable from caveolar membranes by anti-GM3 or anti-caveolin Abs (20). Contrasting properties of GM3-enriched signaling domain vs. caveolar membranes are summarized in Fig. 1B Inset. GM3-dependent adhesion and associated activation of Src and focal adhesion kinase in B16 cells were not inhibited by cholesterol-binding reagents filipin and nystatin (20), but were inhibited by sialyl α2 → 1 sphingosine (57).
In addition to nonreceptor signal transducers, GEM contains hydrophobic chloroform/methanol-soluble proteins termed “proteolipids” (PLs), originally described by Folch and Lees (58) as PL-A, -B, and -C, which are now identified as a class of tetraspanin present at GSL glycosynapse. We found a PL-C having molecular mass of 15 kDa, present exclusively in Folch's lower phase of B16 melanoma, showing strong binding affinity to GM3. A similar PL-C, originally termed “maturation-associated lymphocyte antigen,” expressed in ldlD cells by its gene transfection, has dual affinity to GM3 and to cSrc, but not to caveolin. PL-C is considered a common basic constituent of glycosynapse or GEM (Fig. 1B). PL-C may modulate signal transduction, but it seems to block signaling from upstream to downstream transducers in the case we studied (K. Handa, D. A. Withers, and S.H., unpublished data).
How are GSLs clustered, and why are GSLs associated with sphingomyelin and cholesterol often detergent-insoluble? Biophysical approaches to answer these questions have been limited to liposome models, in studies led by D. Brown and colleagues (for review, see ref. 2). Detergent resistance of membranes has been correlated with the liquid-ordered (lo) phase based on saturated aliphatic chain, as opposed to the liquid-disordered gel phase associated with multiple olefinic bonds.
GSL clustering to form microdomains is considered a priori based on side to side (cis) interaction between GSLs, which is greater than that between glycerophospholipids. Ceramide in GSLs has a 3-O-hydroxyl group and a 2-acylamido group in sphingosine in addition to many hydroxyl groups in carbohydrate, and can therefore function more easily as hydrogen bond donor, whereas glycerophospholipids are essentially hydrogen bond acceptors (59). Ceramide with α-hydroxy fatty acid provides additional chance of hydrogen bond donors, leading to enhanced antigenicity of GSL when it contains α-hydroxy fatty acid (60). Head group interaction between GalCer and sulfatide is much higher when they contain α-hydroxy fatty acid (44). Fatty acid α-hydroxylation is unusually high in GSLs of human colonic cancer (61), human neuroblastoma (62), and Lex GSL isolated from colonic cancer (63). The last case contains exclusively 4-O-hydroxysphinganine. Thus, hydroxyl groups at fatty acid C2 or sphingosine C3 or C4 may promote cis interaction of GSLs, leading to a larger more stable GSL microdomain.
Another ceramide factor affecting GSL organization in membrane is the comparative chain length of fatty acid and sphingosine. α-Fucosylceramide from colonic cancer has C20 sphingosine and C14 fatty acid (64), and this GSL is not immunogenic. A synthetic α-fucosylceramide having C18 sphingosine and C20–24 fatty acid was strongly immunogenic (65). These findings suggest that GSLs with a sphingosine chain longer than the fatty acid chain have unstable membrane organization and vice versa.
Down-regulation of intrinsic tyrosine kinases in various growth factor receptors by gangliosides has been known for many years (48–50; for review, see ref. 31). Receptors for epidermal growth factor (EGF) and platelet-derived growth factor were found to be associated with “caveolar membrane,” or GEM (66–68). GM3 binding to EGF receptor, and GM3-dependent inhibition of receptor tyrosine kinase, were observed only when N-glycans were fully processed to complex-type structure, suggesting that GM3 interaction with N-glycosylated EGF receptor takes place in GEM and leads to down-regulation of the receptor-associated tyrosine kinase (69). GM3 may interact with a defined conformation of the receptor that depends on processed N-glycans, or it may interact directly with N-linked glycans present in EGF receptor, presumably through carbohydrate–carbohydrate interaction. In certain cases, gangliosides may up-regulate receptors. GM1 binds to TrkA (nerve growth factor receptor) in PC12 cells, activates its tyrosine kinase, promotes neuritogenesis, and inhibits apoptosis (51). GD1a, added to certain fibroblasts, followed by prolonged starvation, causes receptor kinase activation rather than down-regulation in response to EGF; this effect was claimed to be triggered by initial activation of cSrc followed by cascades of signaling (68, 70).
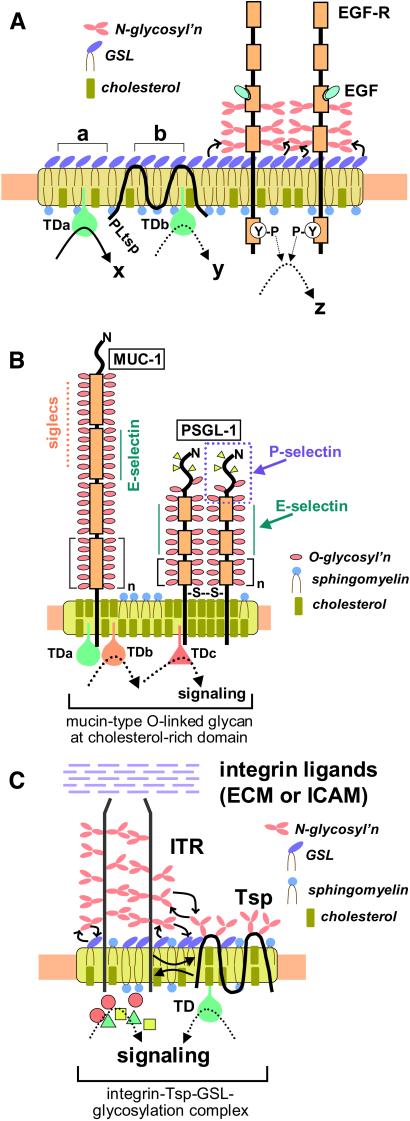
A model of GSL clusters in glycosynapse, associated with signal transducers and with growth factor receptors, is shown in Fig. 2A. The assembly plays a primary role in GSL-dependent cell adhesion (42, 54), induction of differentiation (55), and modulation of growth factor receptor function (48–51), and may lead to the dramatic phenotypic changes seen during ontogenesis and oncogenesis.
Figure 2.
Schematic models of types 1, 2, and 3 glycosynapse. (A) Type 1 glycosynapse with GSL clusters, PL tetraspanin (PLtsp), and growth factor receptor (in this example EGF-R). Clusters of GSLs are organized with signal transducer molecules (TDa, TDb). Stimulation of GSL region “a” causes strong signaling “x” through TDa (19, 20), whereas stimulation of region “b” causes weaker signaling “y” through TDb because of the presence of the blocking factor PLtsp in that region (K. Handa, D. A. Withers, and S.H., unpublished data). When EGF-R is located in a GSL-rich domain, signaling through growth factor (EGF) to activate tyrosine phosphorylation (P-Y) is blocked by the association of EGF-R with GSL (in this example, ganglioside GM3) (48). Binding of GM3 to EGF-R may result from interaction of GM3 with carbohydrate N-linked to EGF-R, as suggested by a previous study (69), and by our studies with N-glycosylation inhibitors (T. Hikita, K. Handa, and S.H., unpublished data). Note that the majority of N-glycosylation (pink oval chains) is localized at the fourth domain from the top (135), close to ganglioside clusters (purple ovals), such that interaction with gangliosides at the membrane surface is favored. (B) Type 2 glycosynapse with mucin-type transmembrane glycoprotein at cholesterol-rich membrane domain. Examples are shown for MUC-1 and PSGL-1. In MUC-1, the number of tandem repeats varies from 20 to 120, and each repeat is a 20-aa sequence. In PSGL-1, the number of tandem repeats is 15, and each repeat is a 10-aa sequence. The units, having multiple O-linked structure with glycosyl adhesion epitope, are organized with various signal transducers (TDa, TDb, TDc). In human and mouse T-cell lines, cSrc, lck56, Lyn, Fyn, and CD45 are detected (73). Both MUC-1 and PSGL-1 are associated with a membrane domain rich in cholesterol (indicated by yellow rods). Cells expressing type 2 glycosynapse are capable of binding to cells expressing P-selectin, E-selectin, or siglecs. A specific structure with three tyrosine phosphates and O-linked glycan to define P-selectin-dependent adhesion in PSGL-1 was recently elucidated (33). (C) Type 3 glycosynapse with integrin receptor (ITR) having α- and β-subunits and tetraspanin (Tsp.). N-glycosylation (pink oval chains) of ITR is essential for connection and stabilization of α5- and β1-subunits (90) and also for interaction of ITR with tetraspanin CD82 (98). Interaction of some tetraspanins (e.g., CD9) with ITR requires GM3 ganglioside (53). The complex is more stable with complete N-glycosylation and is located at a low-density domain showing resistance to β-cyclodextrin.
Type 2 Glycosynapse: O-Linked Mucin-Type Adhesion Epitopes Organized with Signal Transducers in a Cholesterol-Rich Lipid Microdomain
Mucin-type glycoproteins having tandem repeat peptides with multiple O-linked glycan are carriers of various glycoepitopes involved in cell adhesion mediated by selectins and siglecs. It was recently observed that both MUC-1 and PSGL-1 (71) are highly enriched in low-density membrane fractions (Fr. 5 and 6) of human HUT78 and mouse EL4 T-cell lines expressing MUC-1 by its gene transfection. The enrichment was observed in Brij, as well as in high-ionic alkaline conditions (500 mM Na2CO3), a drastic condition used to eliminate weak interaction (72). The association may be maintained by cholesterol, because both MUC-1 and PSGL-1 undergo solubilization and translocation from GEM to a high-density soluble fraction when cells are treated with cholesterol-binding reagent β-cyclodextrin (73). In contrast, GSLs in GEM are not affected by cholesterol-binding reagents (20).
Signal transducers characteristic of T-cells (lck56, Lyn, Fyn, and CD45) were also found to be associated with low-density lipid fraction. This finding raises an interesting possibility that binding of ligands to glycoepitopes in O-linked glycoclusters triggers change of signal transducers, leading to phenotypic modulation of cells. A new functional role of MUC-1 in T-cells was presented recently, based on observations that (i) activated but not resting human T-cells express and secrete MUC-1 (74), and that (ii) soluble MUC-1 inhibits T-cell proliferation (75, 76). These findings suggest that T-cell activation induces MUC-1 transcription, and that the activation process is inhibited by MUC-1 per se, i.e., MUC-1 is an automodulator of T-cell activation and inhibition.
O-linked glycoepitopes present in mucin-type structures include (i) ABH and Lewis blood group antigens; (ii) tumor-associated antigens T, Tn (77, 78), and sialyl-Tn (79, 80); (iii) E-selectin epitopes sialyl-Lex (32) and sialyl-Lea (81, 82) for tumor cell binding; (iv) P-selectin epitopes (33); (v) L-selectin epitopes (83); and (vi) a number of sialyl 2 → 3 or sialyl 2 → 6 epitopes, including sialyl-Tn, as targets of siglecs (84; for review, see ref. 30). The “myeloglycan” epitope (see Table 1 for structure) found originally in GSLs (35, 36) may well be present in O-linked form in PSGL-1 and other mucin-type glycoproteins. Thus, type 2 glycosynapses may play major roles in glycosylation-dependent adhesion through selectins and siglecs, and consequent changes in signaling leading to phenotypic changes, e.g., tumor cell invasiveness. A model of type 2 glycosynapse is illustrated in Fig. 2B.
Type 3 Glycosynapse: Adhesion Receptors with N-Glycosylation Complexed with Tetraspanin and Gangliosides in Microdomains
Our knowledge of cell adhesion, motility, and signaling controlled by various types of integrin receptors is comprehensive (85). Many recent studies indicate that integrins are associated with various members of the tetraspanin family [CD81, CD82, CD9, CD63, CD53 (86; for review, see ref. 87)], or tetraspanins interact with other tetraspanins or with non-tetraspanin membrane proteins (88, 89). Integrins, complexed with tetraspanin, are found at low-density membrane domain. Formation of such complexes, and their presence in microdomain, are affected by N-glycosylation of both integrin and tetraspanin, as well as by surrounding gangliosides. The association of α5 with β1 subunits in α5β1 complex is maintained by N-glycosylation (90), and optimal quantity of GM3 is necessary for maximal fibronectin-dependent adhesion through α5β1 (52). In contrast, GT1b and GD3 inhibit α5β1-dependent binding of keratinocytes. In this system, N-glycosylation of α5β1 is essential for this inhibitory effect, in which GT1b and GD3 are claimed to interact with the mannose core of N-linked glycan through carbohydrate–carbohydrate interaction (91).
Integrin α3 or α5 complexed with tetraspanin CD9 or CD82 strongly affects cell motility and may control tumor cell malignancy. CD82 was originally identified as an anti-metastatic gene product of prostate cancer (92), and its expression is down-regulated in metastatic lesions of prostate cancer (93) and lung cancer (94) as shown in clinicopathological studies. CD9, originally found as motility-regulatory protein (95), is also down-regulated in metastatic deposits of human colonic cancer (96).
To verify the effect of N-glycosylation and gangliosides on CD82 and CD9 function, we used Krieger's Chinese hamster ovary mutant deficient in UDP-Gal 4-epimerase, ldlD14 cells (97), expressing CD82 or CD9 through transfection of their genes (ldlD/CD82 and ldlD/CD9). Studies with this model system indicated that (i) CD82 with complete N-glycosylation reduces, whereas incomplete N-glycosylation or deletion of N-glycosylation enhances, the association of CD82 to integrin receptor. The motility-inhibitory effect of CD82 is strong when the association is enhanced (98). (ii) Association of CD9 with α3/α5 is not influenced by N-glycosylation, because CD9 has no N-glycosylation site. However, this association is affected by endogenously synthesized or exogenously added GM3. GM3 was identified as a cofactor for the motility-inhibitory effect of CD9, and this was also shown clearly in a few colonic and gastric cancer cell lines. Specific interaction of GM3 with CD9 was demonstrated by using photoactivatable GM3 (53). A model of type 3 glycosynapse having glycosylation complex with integrin and tetraspanin is illustrated in Fig. 2C.
Effects of N- or O-glycosylation on the function of adhesion receptors other than integrins, such as (i) cadherins, (ii) Ig family receptors, and (iii) CD44, are known. A notable example is that N-glycosylation of E-cadherin with bisecting GlcNAc enhances the cell adhesion and reduces tumor malignancy (99). In general, the presence of these receptors in low-density microdomain to form glycosynapse has not been studied.
Glycosynapse Involved in Development, Differentiation, and Tumor Progression
Phenotypic changes occurring during embryogenesis, differentiation, and oncogenic progression are all based on glycosylation-dependent cell adhesion coupled with signal transduction. These processes also take place through three types of glycosynapse as discussed in the preceding three sections. A few practical examples are briefly explained below.
1. Glycosylation-dependent adhesion of teratocarcinoma cells, coupled with changes in signal transduction: A model of “compaction process.”
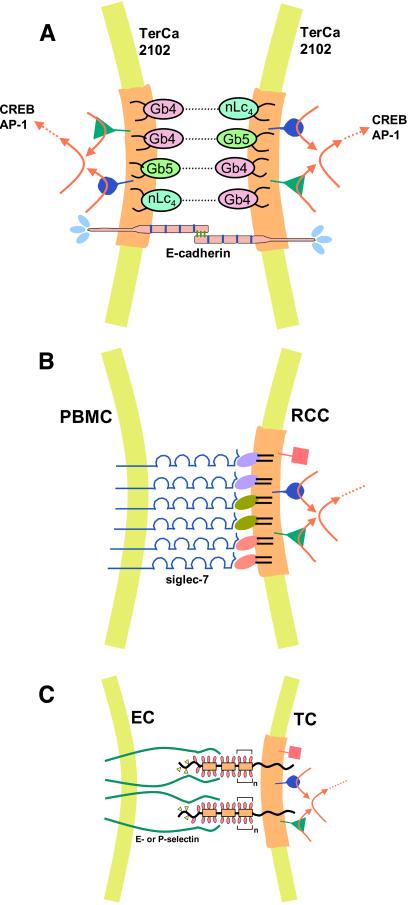
Compaction, the first cell adhesion event during embryogenesis, causes differentiation from “morula” to “blastocyst,” in which glycosynapses carrying “stage-specific” glycosyl epitopes are expressed and organized with signal transducers. At the morula stage, globo-series epitopes (100, 101) and lacto-series epitopes are coexpressed (100). Globo-epitopes in human teratocarcinoma 2102 or Tera2 cells may serve as adhesion sites, providing a model of the compaction process in human or primate embryo. All GSLs in 2102 cells, together with cSrc, RhoA, and Ras, are found in GEM (102). Autoaggregation of 2102 cells is based on interaction of Gb4 to Gb5 and Gb4 to nLc4, which induces activation of yet-unidentified signal transducers, leading to enhancement of two transcription factors, AP-1 and CREB (54). A large variety of phenotypic changes may follow as a consequence. The compaction process mediated by glycosylation may be cooperatively supported by E-cadherin, as observed originally in mouse embryo (103); this can also be applied to primate or human embryo (see Fig. 3A).
Figure 3.
Models of glycosylation-dependent cell adhesion with signaling. (A) Self-adhesion (autoaggregation) of human teratocarcinoma 2102 cells, with simultaneous signaling to activate transcription factors. The adhesion is mediated by two globo-series (Gb4, Gb5) and one lacto-series GSL (nLc4) based on Gb4–nLc4 or Gb4–Gb5 interaction in type 1 glycosynapse. Structures: Gb4, GalNAcβ3Galα4Galβ4Glcβ1Cer; nLc4, Galβ4GlcNAcβ3Galβ4Glcβ1Cer; Gb5, see Table 1. Adhesion by this process may occur in cooperation with E-cadherin-based homotypic interaction. The former process may be much faster than the latter. The glycosynapse membrane is indicated by a light brown color. Simultaneous with GSL-dependent adhesion, signal transducers (cSrc, RhoA, RasH) present in this glycosynapse may be activated, leading to activation of transcription factors AP-1 (activation protein 1) or CREB (cAMP responsive element binding protein). The adhesion process can be inhibited by Abs directed to Gb4, Gb5, or nLc4. (B) Adhesion of renal cell carcinoma (RCC) to peripheral blood mononuclear cells (PBMC), mediated by clustered disialogangliosides in type 1 glycosynapse, and binding of siglec-7 expressed at the PBMC surface. Three types of disialoganglioside in RCC, indicated by different colors, are organized with signal transducers (cSrc, RhoA, focal adhesion kinase) present in RCC glycosynapse. Disialogangliosides of RCC that bind to siglec-7 are GalNAc-disialyl-Lc4, disialyl-Lc4, and disialyl-Gb5 (see Table 1 for structures). Siglec-7 binds equally well to the three types of disialoganglioside, indicating a lack of binding specificity. Low binding specificity or lack of binding specificity in endogenous lectin is often seen in selectins, siglecs, and galectins. Adhesion of RCC to PBMC causes large-scale aggregation of these two types of cells, which may lead to microembolisms in lung. RCC adhesion to PBMC activates cSrc, followed by signaling (red arrows) to enhance motility and invasiveness. (C) Tumor cell (TC) adhesion to activated ECs, through type 2 glycosynapse in TCs, may activate transducers, followed by signaling to enhance TC motility and invasiveness. Mucin-type transmembrane glycoproteins are organized with signal transducers in glycosynapse (light brown). The majority of glycosyl epitopes in TCs involved in selectin-dependent adhesion are carried by mucin-type glycoproteins in type 2 glycosynapse. The glycosyl epitopes are SLex, SLex-Lex, and SLea, which bind to E-selectin but not to P-selectin unless they are expressed on PSGL-1, and sulfated SLex, which binds to P-selectin. This process, i.e., binding of activated ECs to type 2 glycosynapse of TCs, may activate Src family kinases, RhoA, and Ras present in the glycosynapse, leading to enhanced motility and invasiveness.
Lex-dependent adhesion of mouse embryo at compaction (104) is assumed to be mediated by Lex–Lex interaction (e.g., ref. 40) presented by “embryoglycan” (43), in cooperation with E-cadherin (103). Basigin and embigin (both Ig-like receptors) have been characterized as embryoglycan carriers (105, 106); however, their expression at morula stage and functional relationship with E-cadherin remain to be clarified.
2. GSL-induced differentiation of neuroblastoma Neuro-2a.
Mouse neuroblastoma Neuro-2a cells are highly susceptible to exogenous addition of gangliosides GM3 or GM1 to induce differentiation and neuritogenesis (107). 3H-labeled GM3/GM1, but not 3H-labeled phosphatidylcholine added exogenously, accumulates preferentially in GEM, which is characterized by the presence of >90% of cellular GSLs and signal transducers (cSrc, Csk, Lyn, RhoA, and RasH). Stimulation of GEM by GM3 induces rapid (≤5 min) cSrc activation with simultaneous 70% reduction of Csk, a physiological cSrc inhibitor, leading to activation of MAPK. A crucial question on the mechanism of Csk reduction, and the quick response to GM3/GM1 stimulation, remains to be elucidated (55).
3. Tumor cell malignancy defined by glycosynapse function.
Various mechanisms and possibilities with regard to composition and function of tumor cell glycosynapse have been indicated.
(i) Tumor-associated epitopes SLex and SLex-Lex (Table 1) have been identified as E-selectin epitopes (32) and promote tumor cell metastasis (108), particularly when carried by long glycans (109). In this case, metastasis was inhibited by peptide mimetics of SLex (110). SLex or SLex-Lex, present as ganglioside or mucin-type glycoprotein in tumor cell glycosynapse, mediates binding of tumor cells to activated endothelial cells (ECs) expressing E-selectin. The tumor-associated epitope SLea, the positional isomer of SLex, is equally active in binding to E-selectin (81, 82). The same process can be considered for promotion of tumor cell metastasis by this epitope (111). Motility and invasiveness of tumor cells may be enhanced after adhesion of tumor cells to ECs, through activation of signal transducers present in type 1 or type 2 glycosynapse.
(ii) Tumor cell adhesion to ECs through carbohydrate–carbohydrate interaction promotes tumor cell metastasis. In a typical example, adhesion of B16 melanoma cells to mouse ECs (SPE-1) is based on interaction of GM3 (expressed on B16 cells) with LacCer and Gb4 (expressed on ECs), whereby not only adhesion but also motility are strongly enhanced (42, 112). Gg3, a strong ligand of GM3, is suggested to be expressed at mouse lung microvascular ECs (112), so the Gg3–GM3 interaction may also account for B16 metastasis to lung. Liposomes containing Gg3 or GM3 inhibit this metastasis (113). These results provided a basis for further studies on GSL glycosynapse, and demonstrate that GM3-dependent adhesion is coupled with activation of cSrc, RhoA, and focal adhesion kinase (19, 20), which may promote B16 cell invasiveness.
(iii) Motility and invasiveness of colorectal cancer or bladder cancer cells is controlled by type 3 glycosynapse having a complex of N-glycosylated integrin, CD9, and GM3. For these cancers, the higher the level of CD9 and GM3, the lower the invasiveness and motility (53, 114). GM3 in bladder cancer cell lines is cryptic and not involved in GM3-dependent cell adhesion. This situation is in striking contrast to mouse melanoma cells that express high GM3 but no CD9, where the higher the level of GM3, the higher the degree of malignancy (see subsection ii above).
(iv) Aggregation of tumor cells with peripheral blood mononuclear cells is mediated by siglecs expressed on blood cells and sialoglycan expressed on tumor cells. Such aggregates may cause vascular microembolisms from which metastasis may arise. An example was shown for aggregation of renal cell carcinoma cells, expressing disialogangliosides, with peripheral blood mononuclear cells expressing siglec-7. The gangliosides involved are disialyl-Lc4, GalNAc-disialyl-Lc4, and disialyl-Gb5 (see Table 1 for structures). The aggregates may become large enough to cause microembolisms, and cSrc in tumor cell GEM may be activated, leading to enhanced invasiveness (39). Models of glycosynapses involved in tumor cell adhesion as a step in tumor progression are shown in Fig. 3 B and C.
Conclusions and Perspective
The concept that cell surface carbohydrates mediate cell adhesion, and affect gene expression (e.g., 115, 116), has been a major challenge for cell biologists working on the biological significance of cell surface glycosylation defining cell social functions. Major glycosyl structures were identified during the 1970s, and mechanisms of glycosylation and its processing were clarified during the 1980s (e.g., ref. 117). The majority of genes encoding individual glycosyltransferases were cloned and characterized during the 1990s (118). The genes essential for synthesis of glycoconjugates have been applied to test the functional role of each glycosyl residue during development or to test the effect of modified glycosylation on physiological processes, using transgenic, gene knockout, or antisense approaches (119).
We are now at the stage to answer the question of how individual glycosyl structures are assembled and organized in membranes, and how such assemblies control the basic cell social functions, adhesion/recognition coupled with signaling, to maintain or alter cellular phenotype. The assemblies are based essentially on physical interaction of the membrane components, which depend on (i) primary structure of each type of membrane component (lipids, carbohydrates, and proteins) and their quantities; (ii) N- or O-linked glycosylation status of membrane proteins, in terms of glycosyl structure and glycosylation sites; and (iii) structural variety of GSLs, their degrees of clustering, and interaction with other membrane components. Differential combinations of factors i, ii, and iii may lead to microdomains with extensive diversity of structure and function, providing a basis for great phenotypic diversity and biological complexity in organisms.
Among various functions assigned to microdomains, carbohydrate-dependent cell adhesion coupled with signaling plays a major role, and the assembly for this function is hereby termed “glycosynapse”, in analogy to “immunological synapse” controlling functional adhesion and signaling between immunocytes (120, 121). Glycosynapse is one type of assembly present in low-density membrane fraction, which may also contain other assemblies not involved in glycosylation-dependent functions. The term supplements the vague concept of “raft” (5) and the limited notion of caveolae (1).
The diversity of cellular phenotypes based on organizational diversity of membrane may provide a partial explanation for the “gap” in phenotypic and biological complexity between primitive organisms and humans. The total number of protein-coding genes in the human genome was recently estimated to be ≈30,000 (122), which is on a similar order as the number (≈20,000) found in the primitive organism Caenorhabditis elegans. The complexity “gap” may be “filled up” by various factors; mechanisms for gene activation/inhibition and for differential splicing are commonly considered. Incidence and variation of splicing and number of messengers in primitive organisms are lower than in higher animals (123), but the difference is less than expected. Diversity and complexity of membrane organization may fill up this gap as well.
Differential combinations of components may lead to large phenotypic diversity and complexity, although the physical mechanism creating different combinations of interactions is mostly unknown at this time. New biophysical or biochemical approaches are necessary to solve this problem.
The three types of glycosynapse found so far are major functional units of microdomains and provide a basis for glycosylation-dependent cell social functions. Further studies along this line are essential for understanding the functional role of glycosylation in defining not only basic mechanisms of cellular interaction but also important disease processes such as cancer, infection, inflammatory disorders, and geriatric disorders.
Acknowledgments
Our studies described in this review have been supported by National Institutes of Health/National Cancer Institute Grants CA42505 (Outstanding Investigator Grant; 1986–2000), CA80054, and CA82167 (current). I am grateful to Dr. Stuart Kornfeld for his careful review of this article. I also thank my many former and present colleagues involved in microdomain studies, particularly Naoya Kojima, Kazuko Handa, Donald A. Withers, Song Yu, Soichiro Yamamura, and Kazuhisa Iwabuchi. I thank Stephen Anderson for preparation of the manuscript and figures.
Abbreviations
- EGF
epidermal growth factor
- GEM
glycolipid-enriched microdomain
- GSL
glycosphingolipid
- PL
proteolipid
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 1, 2001.
Glycosphingolipids are abbreviated according to the recommendations of the International Union of Pure and Applied Chemistry-International Union of Biochemistry Commission on Biochemical Nomenclature (CBN) [CBN for lipids (1977) Eur. J. Biochem. 79, 11–21]; however, the suffix -OseCer is omitted. Ganglio-series gangliosides are abbreviated according to the extended version of Svennerholm's list [Holmgren, J., Svennerholm, L., Elwing, H., Fredman, P. & Strannegard, O. (1980) Proc. Natl. Acad. Sci. USA 77, 1947–1950].
The term “glycosynapse” is applied to the membrane assembly involved in glycosylation-dependent cell adhesion and signaling, in analogy to “immunological synapse,” which controls functional adhesion between immunocytes (refs. 120 and 121). The term supplements the concepts of “caveolae” (1), “raft” (5), and other terms which do not implicate glycosylation-dependent cell function.
References
- 1.Anderson R G W. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 2.Brown D A, London E. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 3.Hakomori S, Handa K, Iwabuchi K, Yamamura S, Prinetti A. Glycobiology. 1998;8:xi–xviii. doi: 10.1093/oxfordjournals.glycob.a018822. [DOI] [PubMed] [Google Scholar]
- 4.Horejsí V, Drbal K, Cebecauer M, Cerný J, Brdicka T, Angelisová P, Stockinger H. Immunol Today. 1999;20:356–361. doi: 10.1016/s0167-5699(99)01489-9. [DOI] [PubMed] [Google Scholar]
- 5.Simons K, Ikonen E. Nature (London) 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 6.Simons K, Toomre D. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 7.Tillack T W, Allietta M, Moran R E, Young W W J. Biochim Biophys Acta. 1983;733:15–24. doi: 10.1016/0005-2736(83)90086-x. [DOI] [PubMed] [Google Scholar]
- 8.Rock P, Allietta M, Young W W J, Thompson T E, Tillack T W. Biochemistry. 1991;30:19–25. doi: 10.1021/bi00215a003. [DOI] [PubMed] [Google Scholar]
- 9.Carter W G, Hakomori S. J Biol Chem. 1981;256:6953–6960. [PubMed] [Google Scholar]
- 10.Okada Y, Mugnai G, Bremer E G, Hakomori S. Exp Cell Res. 1984;155:448–456. doi: 10.1016/0014-4827(84)90205-2. [DOI] [PubMed] [Google Scholar]
- 11.Forstner G G, Wherrett J R. Biochim Biophys Acta. 1973;306:446–459. doi: 10.1016/0005-2760(73)90183-5. [DOI] [PubMed] [Google Scholar]
- 12.Simons K, van Meer G. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- 13.Stefanova I, Horejsi V, Ansotegui I J, Knapp W, Stockinger H. Science. 1991;254:1016–1019. doi: 10.1126/science.1719635. [DOI] [PubMed] [Google Scholar]
- 14.Anderson R G W, Kamen B A, Rothberg K G, Lacey S W. Science. 1992;255:410–411. doi: 10.1126/science.1310359. [DOI] [PubMed] [Google Scholar]
- 15.Sargiacomo M, Sudol M, Tang Z, Lisanti M P. J Cell Biol. 1993;122:789–807. doi: 10.1083/jcb.122.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smart E J, Graf G A, McNiven M A, Sessa W C, Engelman J A, Scherer P E, Okamoto T, Lisanti M P. Mol Cell Biol. 1999;19:7289–7304. doi: 10.1128/mcb.19.11.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorice M, Parolini I, Sansolini T, Garofalo T, Dolo V, Sargiacomo M, Tai T, Peschle C, Torrisi M R, Pavan A. J Lipid Res. 1997;38:969–980. [PubMed] [Google Scholar]
- 18.Yamamura S, Handa K, Hakomori S. Biochem Biophys Res Commun. 1997;236:218–222. doi: 10.1006/bbrc.1997.6933. [DOI] [PubMed] [Google Scholar]
- 19.Iwabuchi K, Yamamura S, Prinetti A, Handa K, Hakomori S. J Biol Chem. 1998;273:9130–9138. doi: 10.1074/jbc.273.15.9130. [DOI] [PubMed] [Google Scholar]
- 20.Iwabuchi K, Handa K, Hakomori S. J Biol Chem. 1998;273:33766–33773. doi: 10.1074/jbc.273.50.33766. [DOI] [PubMed] [Google Scholar]
- 21.Hakomori S. J Biol Chem. 1990;265:18713–18716. [PubMed] [Google Scholar]
- 22.Hakomori S, Igarashi Y. J Biochem (Tokyo) 1995;118:1091–1103. doi: 10.1093/oxfordjournals.jbchem.a124992. [DOI] [PubMed] [Google Scholar]
- 23.Hakomori S. Glycoconj J. 2000;17:143–151. doi: 10.1023/a:1026524820177. [DOI] [PubMed] [Google Scholar]
- 24.Hakomori S. Pure Appl Chem. 1991;63:473–482. [Google Scholar]
- 25.Bovin N V. In: Glycosciences: Status and Perspectives. Gabius H J, Gabius S, editors. London: Chapman & Hall; 1997. pp. 277–289. [Google Scholar]
- 26.Varki A. Proc Natl Acad Sci USA. 1994;91:7390–7397. doi: 10.1073/pnas.91.16.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe J B. Kidney Int. 1997;51:1418–1426. doi: 10.1038/ki.1997.194. [DOI] [PubMed] [Google Scholar]
- 28.Kannagi R, Kanamori A. Trends Glycosci Glycotechnol. 1999;11:329–344. [Google Scholar]
- 29.Kelm S, Schauer R. Int Rev Cytol. 1997;175:137–240. doi: 10.1016/S0074-7696(08)62127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crocker P R, Varki A. Immunology. 2001;103:137–145. doi: 10.1046/j.0019-2805.2001.01241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yates A J, Rampersaud A. In: Sphingolipids as Signaling Modulators in the Nervous System. Ledeen R W, Hakomori S, Yates A J, Schneider J S, Yu R K, editors. Vol. 845. New York: Ann. N.Y. Acad. Sci.; 1998. pp. 57–71. [Google Scholar]
- 32.Phillips M L, Nudelman E D, Gaeta F C A, Perez M, Singhal A K, Hakomori S, Paulson J C. Science. 1990;250:1130–1132. doi: 10.1126/science.1701274. [DOI] [PubMed] [Google Scholar]
- 33.Leppanen A, Mehta P, Ouyang Y, Ju T, Helin J, Moore K L, van Die I, Canfield W M, McEver R P, Cummings R D. J Biol Chem. 1999;274:24838–24848. doi: 10.1074/jbc.274.35.24838. [DOI] [PubMed] [Google Scholar]
- 34.Mitsuoka C, Ohmori K, Kimura N, Kanamori A, Komba S, Ishida H, Kiso M, Kannagi R. Proc Natl Acad Sci USA. 1999;96:1597–1602. doi: 10.1073/pnas.96.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stroud M R, Handa K, Salyan M E K, Ito K, Levery S B, Hakomori S, Reinhold B B, Reinhold V N. Biochemistry. 1996;35:770–778. doi: 10.1021/bi952461g. [DOI] [PubMed] [Google Scholar]
- 36.Handa K, Stroud M R, Hakomori S. Biochemistry. 1997;36:12412–12420. doi: 10.1021/bi971181t. [DOI] [PubMed] [Google Scholar]
- 37.Yang L J-S, Zeller C B, Shaper N L, Kiso M, Hasegawa A, Shapiro R E, Schnaar R L. Proc Natl Acad Sci USA. 1996;93:814–818. doi: 10.1073/pnas.93.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnaar R L, Collins B E, Wright L P, Kiso M, Tropak M B, Roder J C, Crocker P R. In: Sphingolipids as Signaling Modulators in the Nervous System. Ledeen R W, Hakomori S, Yates A J, Schneider J S, Yu R K, editors. Vol. 845. New York: Ann. N.Y. Acad. Sci.; 1998. pp. 92–105. [Google Scholar]
- 39.Ito A, Handa K, Withers D A, Satoh M, Hakomori S. FEBS Lett. 2001;498:116–120. doi: 10.1016/s0014-5793(01)02476-0. [DOI] [PubMed] [Google Scholar]
- 40.Eggens I, Fenderson B A, Toyokuni T, Dean B, Stroud M R, Hakomori S. J Biol Chem. 1989;264:9476–9484. [PubMed] [Google Scholar]
- 41.Misevic G N, Burger M M. J Biol Chem. 1990;265:20577–20584. [PubMed] [Google Scholar]
- 42.Kojima N, Hakomori S. J Biol Chem. 1991;266:17552–17558. [PubMed] [Google Scholar]
- 43.Kojima N, Fenderson B A, Stroud M R, Goldberg R I, Habermann R, Toyokuni T, Hakomori S. Glycoconj J. 1994;11:238–248. doi: 10.1007/BF00731224. [DOI] [PubMed] [Google Scholar]
- 44.Stewart R J, Boggs J M. Biochemistry. 1993;32:10666–10674. doi: 10.1021/bi00091a017. [DOI] [PubMed] [Google Scholar]
- 45.Matsuura K, Kitakouji H, Sawada N, Ishida H, Kiso M, Kitajima K, Kobayashi K. J Am Chem Soc. 2000;122:7406–7407. [Google Scholar]
- 46.Tromas C, Rojo J, de la Fuente J M, Barrientos A G, Garcia R, Penades S. Angew Chem Intl Ed. 2001;40:3052–3055. doi: 10.1002/1521-3773(20010817)40:16<3052::AID-ANIE3052>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 47.Haseley S R, Vermeer H J, Kamerling J P, Vliegenthart J F G. Proc Natl Acad Sci USA. 2001;98:9419–9424. doi: 10.1073/pnas.151111298. . (First Published July 17, 2001; 10.1073/pnas.151111298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bremer E G, Schlessinger J, Hakomori S. J Biol Chem. 1986;261:2434–2440. [PubMed] [Google Scholar]
- 49.Nojiri H, Stroud M R, Hakomori S. J Biol Chem. 1991;266:4531–4537. [PubMed] [Google Scholar]
- 50.Yates A J, VanBrocklyn J, Saqr H E, Guan Z, Stokes B T, O'Dorisio M S. Exp Cell Res. 1993;204:38–45. doi: 10.1006/excr.1993.1006. [DOI] [PubMed] [Google Scholar]
- 51.Mutoh T, Tokuda A, Miyada T, Hamaguchi M, Fujiki N. Proc Natl Acad Sci USA. 1995;92:5087–5091. doi: 10.1073/pnas.92.11.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng M, Fang H, Tsuruoka T, Tsuji T, Sasaki T, Hakomori S. J Biol Chem. 1993;268:2217–2222. [PubMed] [Google Scholar]
- 53.Ono M, Handa K, Sonnino S, Withers D A, Nagai H, Hakomori S. Biochemistry. 2001;40:6414–6421. doi: 10.1021/bi0101998. [DOI] [PubMed] [Google Scholar]
- 54.Yu S, Withers D A, Hakomori S. J Biol Chem. 1998;273:2517–2525. doi: 10.1074/jbc.273.5.2517. [DOI] [PubMed] [Google Scholar]
- 55.Prinetti A, Iwabuchi K, Hakomori S. J Biol Chem. 1999;274:20916–20924. doi: 10.1074/jbc.274.30.20916. [DOI] [PubMed] [Google Scholar]
- 56.Satoh M, Nejad F M, Ohtani H, Ito A, Ohyama C, Saito S, Orikasa S, Hakomori S. Int J Oncol. 2000;16:529–536. doi: 10.3892/ijo.16.3.529. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y, Iwabuchi K, Nunomura S, Hakomori S. Biochemistry. 2000;39:2459–2468. doi: 10.1021/bi991882l. [DOI] [PubMed] [Google Scholar]
- 58.Folch J, Lees M. J Biol Chem. 1951;191:807–817. [PubMed] [Google Scholar]
- 59.Pascher I. Biochim Biophys Acta. 1976;455:433–451. doi: 10.1016/0005-2736(76)90316-3. [DOI] [PubMed] [Google Scholar]
- 60.Kannagi R, Stroup R, Cochran N A, Urdal D L, Young W W J, Hakomori S. Cancer Res. 1983;43:4997–5005. [PubMed] [Google Scholar]
- 61.Hakomori S, Nudelman E D, Levery S B, Kannagi R. J Biol Chem. 1984;259:4672–4680. [PubMed] [Google Scholar]
- 62.Ladisch S, Sweeley C C, Becker H, Gage D. J Biol Chem. 1989;264:12097–12105. [PubMed] [Google Scholar]
- 63.Yang H-J, Hakomori S. J Biol Chem. 1971;246:1192–1200. [PubMed] [Google Scholar]
- 64.Watanabe K, Matsubara T, Hakomori S. J Biol Chem. 1976;251:2385–2387. [PubMed] [Google Scholar]
- 65.Yoshino T, Watanabe K, Hakomori S. Biochemistry. 1982;21:928–934. doi: 10.1021/bi00534a018. [DOI] [PubMed] [Google Scholar]
- 66.Mineo C, James G L, Smart E J, Anderson R G W. J Biol Chem. 1996;271:11930–11935. doi: 10.1074/jbc.271.20.11930. [DOI] [PubMed] [Google Scholar]
- 67.Wu C, Butz S, Ying Y, Anderson R G. J Biol Chem. 1997;272:3554–3559. doi: 10.1074/jbc.272.6.3554. [DOI] [PubMed] [Google Scholar]
- 68.Li R, Manela J, Kong Y, Ladisch S. J Biol Chem. 2000;275:34213–34223. doi: 10.1074/jbc.M906368199. [DOI] [PubMed] [Google Scholar]
- 69.Wang X-Q, Sun P, O'Gorman M, Tai T, Paller A S. Glycobiology. 2001;11:515–522. doi: 10.1093/glycob/11.7.515. [DOI] [PubMed] [Google Scholar]
- 70.Li R, Liu Y, Ladisch S. J Biol Chem. 2001;276:42782–42792. doi: 10.1074/jbc.M101481200. [DOI] [PubMed] [Google Scholar]
- 71.Sako D, Chang X-J, Barone K M, Vachino G, White H M, Shaw G, Veldman G M, Bean K M, Ahern T J, Furie B, et al. Cell. 1993;75:1179–1186. doi: 10.1016/0092-8674(93)90327-m. [DOI] [PubMed] [Google Scholar]
- 72.Song K S, Li S, Okamoto T, Quilliam L A, Sargiacomo M, Lisanti M P. J Biol Chem. 1996;271:9690–9697. doi: 10.1074/jbc.271.16.9690. [DOI] [PubMed] [Google Scholar]
- 73.Handa K, Jacobs F, Longenecker B M, Hakomori S. Biochem Biophys Res Commun. 2001;285:788–794. doi: 10.1006/bbrc.2001.5225. [DOI] [PubMed] [Google Scholar]
- 74.Agrawal B, Krantz M J, Parker J, Longenecker B M. Cancer Res. 1998;58:4079–4081. [PubMed] [Google Scholar]
- 75.Agrawal B, Gendler S J, Longenecker B M. Mol Med Today. 1998;9:397–403. doi: 10.1016/s1357-4310(98)01322-7. [DOI] [PubMed] [Google Scholar]
- 76.Agrawal B, Krantz M J, Reddish M A, Longenecker B M. Nat Med. 1998;4:43–49. doi: 10.1038/nm0198-043. [DOI] [PubMed] [Google Scholar]
- 77.Springer G F. Science. 1984;224:1198–1206. doi: 10.1126/science.6729450. [DOI] [PubMed] [Google Scholar]
- 78.Hirohashi S, Clausen H, Yamada T, Shimosato Y, Hakomori S. Proc Natl Acad Sci USA. 1985;82:7039–7043. doi: 10.1073/pnas.82.20.7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kjeldsen T B, Clausen H, Hirohashi S, Ogawa T, Iijima H, Hakomori S. Cancer Res. 1988;48:2214–2220. [PubMed] [Google Scholar]
- 80.Kurosaka A, Kitagawa H, Fukui S, Numata Y, Nakada H, Funakoshi I, Kawasaki T, Ogawa T, Iijima H, Yamashina I. J Biol Chem. 1988;263:8724–8726. [PubMed] [Google Scholar]
- 81.Takada A, Ohmori K, Takahashi N, Tsuyuoka K, Yago A, Zenita K, Hasegawa A, Kannagi R. Biochem Biophys Res Commun. 1991;179:713–719. doi: 10.1016/0006-291x(91)91875-d. [DOI] [PubMed] [Google Scholar]
- 82.Berg E L, Robinson M K, Mansson O, Butcher E C, Magnani J L. J Biol Chem. 1991;266:14869–14872. [PubMed] [Google Scholar]
- 83.Rosen S D. Am J Pathol. 1999;155:1013–1020. doi: 10.1016/S0002-9440(10)65201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brinkman-Van der Linden E C, Varki A. J Biol Chem. 2000;275:8625–8632. doi: 10.1074/jbc.275.12.8625. [DOI] [PubMed] [Google Scholar]
- 85.Hynes R O. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 86.Mannion B A, Berditchevski F, Kraeft S-K, Chen L B, Hemler M E. J Immunol. 1996;157:2039–2047. [PubMed] [Google Scholar]
- 87.Hemler M E. Curr Opin Cell Biol. 1998;10:578–585. doi: 10.1016/s0955-0674(98)80032-x. [DOI] [PubMed] [Google Scholar]
- 88.Charrin S, Le Naour F, Oualid M, Billard M, Faure G, Hanash S M, Boucheix C, Rubinstein E. J Biol Chem. 2001;276:14329–14337. doi: 10.1074/jbc.M011297200. [DOI] [PubMed] [Google Scholar]
- 89.Horvath G, Serru V, Clay D, Billard M, Boucheix C, Rubinstein E. J Biol Chem. 1998;273:30537–30543. doi: 10.1074/jbc.273.46.30537. [DOI] [PubMed] [Google Scholar]
- 90.Zheng M, Fang H, Hakomori S. J Biol Chem. 1994;269:12325–12331. [PubMed] [Google Scholar]
- 91.Wang X, Sun P, Al-Qamari A, Tai T, Kawashima I, Paller A S. J Biol Chem. 2001;276:8436–8444. doi: 10.1074/jbc.M006097200. [DOI] [PubMed] [Google Scholar]
- 92.Dong J, -T, Lamb P W, Rinker-Schaeffer C W, Vukanovic J, Ichikawa T, Isaacs J T, Barrett J C. Science. 1995;268:884–886. doi: 10.1126/science.7754374. [DOI] [PubMed] [Google Scholar]
- 93.Dong J T, Suzuki H, Pin S S, Bova S, Schalken J A, Isaacs W B, Barrett J C, Isaacs J T. Cancer Res. 1996;56:4387–4390. [PubMed] [Google Scholar]
- 94.Adachi M, Taki T, Ieki Y, Huang C, Higashiyama M, Miyake M. Cancer Res. 1996;56:1751–1755. [PubMed] [Google Scholar]
- 95.Miyake M, Koyama M, Seno M, Ikeyama S. J Exp Med. 1991;174:1347–1354. doi: 10.1084/jem.174.6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cajot J-F, Sordat I, Silvestre T, Sordat B. Cancer Res. 1997;57:2593–2597. [PubMed] [Google Scholar]
- 97.Kingsley D M, Kozarsky K F, Hobbie L, Krieger M. Cell. 1986;44:749–759. doi: 10.1016/0092-8674(86)90841-x. [DOI] [PubMed] [Google Scholar]
- 98.Ono M, Handa K, Withers D A, Hakomori S. Biochem Biophys Res Commun. 2000;279:744–750. doi: 10.1006/bbrc.2000.4030. [DOI] [PubMed] [Google Scholar]
- 99.Yoshimura M, Ihara Y, Matsuzawa Y, Taniguchi N. J Biol Chem. 1996;271:13811–13815. doi: 10.1074/jbc.271.23.13811. [DOI] [PubMed] [Google Scholar]
- 100.Fenderson B A, Andrews P W, Nudelman E D, Clausen H, Hakomori S. Dev Biol. 1987;122:21–34. doi: 10.1016/0012-1606(87)90328-9. [DOI] [PubMed] [Google Scholar]
- 101.Willison K R, Karol R A, Suzuki A, Kundu S K, Marcus D M. J Immunol. 1982;129:603–609. [PubMed] [Google Scholar]
- 102.Yu S, Withers D A, Yamamura S, Handa K, Hakomori S. Glycoconj. J. 1997. , Suppl. 14, S53. [Google Scholar]
- 103.Boubelík M, Dráberová L, Dráber P. Biochem Biophys Res Commun. 1996;224:283–288. doi: 10.1006/bbrc.1996.1022. [DOI] [PubMed] [Google Scholar]
- 104.Fenderson B A, Zehavi U, Hakomori S. J Exp Med. 1984;160:1591–1596. doi: 10.1084/jem.160.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kamada Y, Arita Y, Ogata S, Muramatsu H, Muramatsu T. Eur J Biochem. 1987;163:497–502. doi: 10.1111/j.1432-1033.1987.tb10896.x. [DOI] [PubMed] [Google Scholar]
- 106.Igakura T, Kadomatsu K, Kaname T, Muramatsu H, Fan Q W, Miyauchi T, Toyama Y, Kuno N, Yuasa S, Takahashi M, et al. Dev Biol. 1998;194:152–165. doi: 10.1006/dbio.1997.8819. [DOI] [PubMed] [Google Scholar]
- 107.Ledeen R W, Wu G, Vaswani K K, Cannella M S. In: Trophic Factors and the Nervous System. Horrocks L A, Neff N H, Yates A J, Hadjiconstantinou M, editors. NY: Raven; 1990. pp. 17–34. [Google Scholar]
- 108.Inufusa H, Kojima N, Yasutomi M, Hakomori S. Clin Exp Metastasis. 1991;9:245–257. doi: 10.1007/BF01753728. [DOI] [PubMed] [Google Scholar]
- 109.Ohyama C, Tsuboi S, Fukuda M. EMBO J. 1999;18:1516–1525. doi: 10.1093/emboj/18.6.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fukuda M N, Ohyama C, Lowitz K, Matsuo O, Pasqualini R, Ruoslahti E, Fukuda M. Cancer Res. 2000;60:450–456. [PubMed] [Google Scholar]
- 111.Kannagi R. Glycoconj J. 1997;14:577–584. doi: 10.1023/a:1018532409041. [DOI] [PubMed] [Google Scholar]
- 112.Kojima N, Shiota M, Sadahira Y, Handa K, Hakomori S. J Biol Chem. 1992;267:17264–17270. [PubMed] [Google Scholar]
- 113.Otsuji E, Park Y S, Tashiro K, Kojima N, Toyokuni T, Hakomori S. Int J Oncol. 1995;6:319–327. doi: 10.3892/ijo.6.2.319. [DOI] [PubMed] [Google Scholar]
- 114.Satoh M, Ito A, Nojiri H, Handa K, Numahata K, Ohyama C, Saito S, Hoshi S, Hakomori S. Int J Oncol. 2001;19:723–731. [PubMed] [Google Scholar]
- 115.Kalckar H M. Science. 1965;150:305–313. doi: 10.1126/science.150.3694.305. [DOI] [PubMed] [Google Scholar]
- 116.Grobstein C. In: Extracellular Matrix Influences on Gene Expression. Slavkin H C, Greulich R C, editors. New York: Academic; 1975. pp. 9–16. [Google Scholar]
- 117.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 118.Taniguchi N, Honke K, Fukuda M. Handbook of Glycosyltransferases and Their Related Genes. Tokyo: Springer; 2002. [Google Scholar]
- 119.Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J. Essentials of Glycobiology. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. [PubMed] [Google Scholar]
- 120.Ilangumaran S, He H-T, Hoessli D C. Immunol Today. 2000;21:2–7. doi: 10.1016/s0167-5699(99)01494-2. [DOI] [PubMed] [Google Scholar]
- 121.Bromley S K, Burack W R, Johnson K G, Somersalo K, Sims T N, Sumen C, Davis M M, Shaw A S, Allen P M, Dustin M L. Annu Rev Immunol. 2001;19:375–396. doi: 10.1146/annurev.immunol.19.1.375. [DOI] [PubMed] [Google Scholar]
- 122.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 123.Claverie J-M. Science. 2001;291:1255–1257. doi: 10.1126/science.1058969. [DOI] [PubMed] [Google Scholar]
- 124.Kannagi R, Levery S B, Ishigami F, Hakomori S, Shevinsky L H, Knowles B B, Solter D. J Biol Chem. 1983;258:8934–8942. [PubMed] [Google Scholar]
- 125.Andrews P W, Fenderson B A, Hakomori S. Int J Androl. 1987;10:95–104. doi: 10.1111/j.1365-2605.1987.tb00170.x. [DOI] [PubMed] [Google Scholar]
- 126.Saito S, Levery S B, Salyan M E K, Goldberg R I, Hakomori S. J Biol Chem. 1994;269:5644–5652. [PubMed] [Google Scholar]
- 127.Fukushi Y, Nudelman E D, Levery S B, Higuchi T, Hakomori S. Biochemistry. 1986;25:2859–2866. doi: 10.1021/bi00358a018. [DOI] [PubMed] [Google Scholar]
- 128.Fukushi Y, Nudelman E D, Levery S B, Rauvala H, Hakomori S. J Biol Chem. 1984;259:10511–10517. [PubMed] [Google Scholar]
- 129.Fukushi Y, Hakomori S, Shepard T. J Exp Med. 1984;159:506–520. doi: 10.1084/jem.160.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ito A, Levery S B, Saito S, Satoh M, Hakomori S. J Biol Chem. 2001;276:16695–16703. doi: 10.1074/jbc.M011791200. [DOI] [PubMed] [Google Scholar]
- 131.Saito S, Orikasa S, Satoh M, Ohyama C, Ito A, Takahashi T. Jpn J Cancer Res (GANN) 1997;88:652–659. doi: 10.1111/j.1349-7006.1997.tb00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nores G A, Dohi T, Taniguchi M, Hakomori S. J Immunol. 1987;139:3171–3176. [PubMed] [Google Scholar]
- 133.Hakomori S. Sci Am. 1986;254:44–53. doi: 10.1038/scientificamerican0586-44. [DOI] [PubMed] [Google Scholar]
- 134.Strömberg N, Nyholm P-G, Pascher I, Normark S. Proc Natl Acad Sci USA. 1991;88:9340–9344. doi: 10.1073/pnas.88.20.9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fernandes H, Cohen S, Bishayee S. J Biol Chem. 2001;276:5375–5383. doi: 10.1074/jbc.M005599200. [DOI] [PubMed] [Google Scholar]