Abstract
Associations have been reported of the seven-repeat (7R) allele of the human dopamine receptor D4 (DRD4) gene with both attention-deficit/hyperactivity disorder and the personality trait of novelty seeking. This polymorphism occurs in a 48-bp tandem repeat in the coding region of DRD4, with the most common allele containing four repeats (4R) and rarer variants containing 2–11. Here we show by DNA resequencing/haplotyping of 600 DRD4 alleles, representing a worldwide population sample, that the origin of 2R–6R alleles can be explained by simple one-step recombination/mutation events. In contrast, the 7R allele is not simply related to the other common alleles, differing by greater than six recombinations/mutations. Strong linkage disequilibrium was found between the 7R allele and surrounding DRD4 polymorphisms, suggesting that this allele is at least 5–10-fold “younger” than the common 4R allele. Based on an observed bias toward nonsynonymous amino acid changes, the unusual DNA sequence organization, and the strong linkage disequilibrium surrounding the DRD4 7R allele, we propose that this allele originated as a rare mutational event that nevertheless increased to high frequency in human populations by positive selection.
The human dopamine receptor D4 (DRD4) gene (1), located near the telomere of chromosome 11p, is one of the most variable human genes known (2, 3). Most of this diversity is the result of length and single-nucleotide polymorphism (SNP) variation in a 48-bp tandem repeat (VNTR) in exon 3, encoding the third intracellular loop of this dopamine receptor (2, 3). Variant alleles containing two (2R) to eleven (11R) repeats are found, with the resulting proteins having 32–176 amino acids at this position. Interestingly, the frequency of these alleles varies widely. The 7R allele, for example, has an extremely low incidence in Asian populations yet a high frequency in the Americas (3).
A number of investigations have found associations between particular alleles of this highly variable gene and behavioral phenotypes (4–8). Although initial studies suggested that the 7R allele of the DRD4 gene might be associated with the personality trait of novelty seeking (7, 8), the most reproduced association is between the 7R allele and attention-deficit/hyperactivity disorder (ADHD; refs. 4–6 and 9). ADHD is the most prevalent disorder of early childhood, affecting an estimated 3% of elementary school children. As defined by DSM-IV criteria (10), ADHD consists of developmentally inappropriate inattention, impulsivity, and hyperactivity with early onset (before the age of 7). Evidence of a strong genetic component of ADHD has come from a variety of twin, adoption, and family studies (11, 12). The efficacy of methylphenidate in the treatment of ADHD indicates that genes in the dopamine pathway might play a role in the syndrome's etiology (9, 13). Initial association studies found ADHD probands to exhibit an increased frequency of DRD4 7R alleles in comparison to controls (4). Eight separate replications of this initial observation have now been reported (9). As in all association studies, however, one cannot assume that the presence of a DRD4 7R allele is either necessary or sufficient to “cause” ADHD. Further work will be required to understand the genetic/environmental factors underlying this behavior.
Nevertheless, given the likely functional importance of this change in the DRD4 protein, in a region that couples to G proteins and mediates postsynaptic effects (14), these association studies have generated considerable interest (9). In particular, this association is consistent with the common variant/common disorder hypothesis, which proposes that the high frequency of many complex genetic diseases is related to common DNA variants (15, 16). However, many questions remain as to the nature of the DRD4/ADHD association. One would like to know (i) whether particular 7R allele variants are associated with ADHD, (ii) the population distribution of variant DRD4 alleles, and/or (iii) whether the observed marker is in linkage disequilibrium (LD) with other etiologically relevant polymorphisms. Given the known high level of sequence polymorphism of this gene (2), PCR-based DNA resequencing is the most efficient and accurate method to address these questions. Here we use this approach to determine (i) the population distribution of DRD4 exon 3 haplotypes and (ii) their relative association with adjacent polymorphisms. We present haplotype data indicating that the DRD4 7R allele originated as a rare mutational event (or events) that nevertheless increased to high frequency in human populations by positive selection.
Methods
Population Samples.
Samples were obtained as reported (3, 17). The origins of the 600 alleles reported in this study, based on geographical/ethnic origin, are as follows: North and South America, 12.7% (76 alleles); Europe, 36.7% (220 alleles); Asia, 27.3% (164 alleles); Africa, 20.3% (122 alleles); and Pacific, 3.0% (18 alleles). Lymphoblastoid cell lines have been established for most of these population samples, and methods for transformation, cell culture, and DNA purification have been described (3, 17). For LD studies of the DRD4 4R-G-G SNP association, an additional 288 alleles (approximately equally derived from African, Asian, and European sources) were used. All persons gave their informed consent before their inclusion in this study, which was carried out under protocols approved by the Human Subjects Committees at the participating institutions.
PCR Amplification and DNA Sequencing.
PCR amplification of the DRD4 promoter polymorphism was conducted as described (18, 19). The program OLIGO 6.0 was used to select primer pairs for the exon 1 polymorphism (5′-TGGGCCGCCGCATTCGT-3′ and 5′-GGTGGGTGTATCGCCGAGGGA-3′; 661-nt product; ref. 20) and the exon 3 VNTR (5′-CGTACTGTGCGGCCTCAACGA-3′and 5′-GACACAGCGCCTGCGTGATGT-3′; 705-nt product for the 4R allele; ref. 2). For some amplifications of the VNTR, primers described previously were used (2). The alternative primers were chosen farther from the VNTR to minimize out-of-register hybridization during amplification. PCRs were conducted in 25-μl volumes containing 100 ng of genomic DNA, 200 μM dXTPs, 0.5 μM of each primer, 1× PCR buffer (Qiagen, Chatsworth, CA), 1× Q solution (Qiagen), and 0.625 units of Taq DNA polymerase (Qiagen). Amplification was performed by using Perkin–Elmer 9700 thermal cyclers. A 20-sec, 96°C hot start was used followed by 40 cycles of 95°C for 20 sec and 68°C for 1 min. After a 4-min chase at 72°C, excess primers were eliminated with 0.5 units of shrimp alkaline phosphatase (SAP, Amersham Pharmacia), 0.1 unit of exonuclease I (Exo I, Amersham Pharmacia), and 1× SAP buffer (Amersham Pharmacia). The SAP/Exo I reaction was carried out at 37°C for 1 h followed by a 15-min heat inactivation at 72°C. The DNA from the SAP/Exo I reaction was used directly for DNA sequencing. For most individuals, the two allelic PCR products first were separated on 1.2% agarose gels. DNA cycle sequencing was conducted by standard techniques using ABI 377 and 3700 automated sequencers (21).
Ka/Ks and Allele Age Calculations.
Ka/Ks ratios were calculated by standard methods (22, 23). Putative recombinant haplotypes were not considered independent events. Allele age calculations were conducted by standard methods as described below (24–27).
Calculated from population frequency.
E(t1) = [−2p/(1 − p)] ln(p), where E(t1) = expected age, time is measured in units of 2N generations, and p = population frequency. For DRD4, p = 19.2% for the 7R allele and 65.1% for the 4R allele. A generation time of 20–25 years and N = 10,000 were assumed [regarded as a minimum estimate of the effective population size of modern humans during the period before recent growth (24, 26)].
Calculated from intraallelic variation.
t = [1/ln(1 − c)] ln[(x(t) − y)/(1 − y)], where t = allele age, c = recombination rate, x(t) = frequency in generation t, and y = frequency on normal chromosomes. Assuming the origin of the 7R allele was on an L1L2(7R)A-C haplotype, for the (7R)A-C association c = 0.0000136 (from the average recombination rate per megabase times the VNTR-SNP distance), x(t) = 97% (the percentage of A-C SNPs associated with DRD4 7R alleles), and y = 13.9% (the percentage of A-C SNPs associated with African DRD4 4R alleles, assumed to be the “normal” allele). For the promoter polymorphism L1(7R) association, c = 0.000165, x(t) = 90.8%, and y = 61.9%.
Results and Discussion
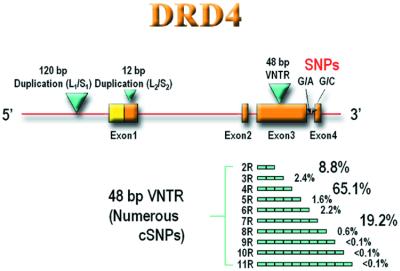
Primer sets were chosen to amplify the four exons of the highly GC-rich DRD4 gene (1) as well as the adjacent promoter region and splice junctions (Fig. 1). Initial resequencing of the entire promoter and coding region of the DRD4 gene from 20 ADHD probands (data not shown) uncovered a number of polymorphisms reported previously. These polymorphisms included two insertion/deletion polymorphisms, one in the promoter region (4.3 kb upstream of the VNTR; refs. 18 and 19) and one in exon 1 (2.7 kb upstream of the VNTR; ref. 20; see Fig. 1). In addition, a number of new coding SNPs were uncovered in the exon 3 VNTR (2) as well as two previously unreported SNPs in intron 3, 20 nt apart and ≈350 bp downstream from the center of the VNTR (Fig. 1). Given the high level of VNTR polymorphism identified in this initial sample, a more extensive PCR resequencing of 600 exon 3 VNTR alleles, obtained from a worldwide population sample (refs. 3 and 17; Table 1; Fig. 2), was conducted. This sample contained individuals representing most major geographical origins (see Methods). The majority of individuals were heterozygotes, and the two allelic PCR products could be separated by gel electrophoresis before sequencing, providing unambiguous haplotypes. Altogether, we screened over 450,000 bp of genomic DNA and 2,968 48-bp repeats.
Figure 1.
Diagrammatic representation of the human DRD4 gene region. Exon positions are indicated by blocks (yellow, noncoding; orange, coding). The approximate positions of a 120-bp promoter region duplication (blue triangle), an exon 1 12-bp duplication (blue triangle), an exon 3 VNTR (blue triangle), and two intron 3 SNPs are indicated. 2R–11R variants of the VNTR are indicated below exon 3 (blue) along with their worldwide population frequencies determined by PCR analysis (3, 17).
Table 1.
Haplotypes of 600 DRD4 exon 3 alleles
| Allele | F | N | Haplotype | Allele | F | N | Haplotype |
| 2R | 0.088 | 55 | 6R | 0.022 | 24 | ||
| 43 | 1-4 | 16 | 1-2-3-2-3-4 | ||||
| 12 | 30-4* | 2 | 1-2-6-5-2-20 | ||||
| 3R | 0.024 | 36 | 2 | 1-2-6-5-2-4 | |||
| 16 | 1-7-4 | 1 | 1-2-14-17-2-4 | ||||
| 9 | 1-2-4 | 1 | 1-6-5-2-5-4 | ||||
| 4 | 1-11-33* | 1 | 1-2-13-2-5-19 | ||||
| 3 | 1-9-4 | 1 | 24-6-5-2-5-4 | ||||
| 1 | 1-2-22 | 7R | 0.192 | 199 | |||
| 1 | 1-2-21 | 177 | 1-2-6-5-2-5-4 | ||||
| 1 | 1-2-31 | 5 | 1-2-6-5-2-5-19* | ||||
| 1 | 1-2-32 | 3 | 1-2-6-5-2-3-4 | ||||
| 4R | 0.651 | 250 | 3 | 1-2-6-5-13-5-4* | |||
| 238 | 1-2-3-4 | 2 | 1-8-25-5-2-5-4 | ||||
| 3 | 1-2-14-4 | 2 | 1-2-3-5-2-5-4 | ||||
| 2 | 1-2-13-4 | 1 | 1-2-6-5-2-13-4 | ||||
| 2 | 1-2-12-4 | 1 | 1-2-29-17-2-5-4 | ||||
| 1 | 1-17-3-4 | 1 | 1-2-6-2-2-5-4 | ||||
| 1 | 1-9-12-4 | 1 | 1-8-25-5-2-3-4 | ||||
| 1 | 1-8-3-4 | 1 | 1-2-6-16-2-3-4 | ||||
| 1 | 1-10-3-4 | 1 | 1-2-6-5-2-14-4 | ||||
| 1 | 1-9-3-4 | 1 | 1-2-3-17-2-5-4 | ||||
| 5R | 0.016 | 27 | 8R | 0.006 | 6 | ||
| 12 | 1-3-2-3-4* | 2 | 1-2-6-5-17-2-13-35* | ||||
| 4 | 1-2-13-34-4* | 1 | 1-2-6-5-2-2-5-4 | ||||
| 3 | 1-2-2-3-4 | 1 | 1-2-6-26-5-26-3-35 | ||||
| 2 | 1-2-6-5-4 | 1 | 1-2-6-26-5-26-3-4 | ||||
| 2 | 1-11-2-3-4 | 1 | 1-2-6-18-5-18-3-4 | ||||
| 1 | 1-3-2-14-4 | ||||||
| 1 | 1-2-6-23-4 | 9R | <0.001 | 1 | 1-8-25-5-2-5-2-23-4 | ||
| 1 | 1-2-3-9-4 | 10R | <0.001 | 1 | 1-2-15-6-2-6-5-2-5-4 | ||
| 1 | 1-2-3-27-4 | 11R | <0.001 | 1 | 1-2-3-27-5-23-25-5-2-5-28 |
F, observed allele frequency in 2,836 chromosomes from 37 worldwide human populations (3,17); N, allele number identified by sequence analysis in this study (non-4R alleles were oversampled by 2–3-fold); haplotype, haplotypes are indicated using the repeat motif nomenclature proposed (Fig. 2). Alleles with adjacent asterisks indicate common variants found only in a single population sample (2R 30-4, Surui; 3R 1-11-33, Nasioi; 5R 1-3-2-3-4, Chinese; 5R 1-2-13-34-4, Biaka; 7R 1-2-6-5-2-5-19, Surui; 7R 1-2-6-5-13-5-4, Nasioi; 8R 1-2-6-5-17-2-13-35, Biaka). Alleles with a single representation by definition were found in only one population.
Figure 2.
Nucleotide and amino acid sequences of VNTR motifs. The nucleotide and corresponding amino acid (red) sequences of 35 DRD4 exon 3 48-bp repeat motifs are shown. Prior nomenclature (2) for 19 of these motifs is indicated (α–ξ). The putative single-step origin of most of these motifs is indicated either as a recombination (R) or mutation (M) event. For example, the 7 motif is hypothesized to be a recombination between a 2 motif and a 3 motif (R2/3), and the 8 motif is hypothesized to be a single point mutation of a 2 motif (M2). Motifs 1–6, which account for the vast majority of observed haplotype variants (Table 1), are considered the progenitors. Motifs with no putative origin noted (for example, motif 15), have multiple possible progenitors.
In the 600 chromosomes sequenced, 56 different haplotypes were found (Table 1). These haplotypes were composed of 35 distinct 48-bp variant motifs (Fig. 2), 19 of which were reported previously (designated α–ξ in Fig. 2; ref. 2). We propose that these DRD4 48-bp variant motifs are given numbers as shown rather than the letters used previously (2), because there are not enough characters in the Greek alphabet. We propose that DRD4 exon 3 variants be designated in the format shown, i.e., the most common 4R allele being designated 4R(1-2-3-4), etc.
We intentionally over-sampled non-4R alleles approximately 2-fold, because little sequence variation was uncovered in the common 4R allele (Table 1) even though it represents 65% of the world population frequency (3, 17). Most of the haplotypes in this sample (85.7%) were found at frequencies less than 1% (Table 1). Looking at nucleotide diversity among variants defined by their VNTR number, the common 2R, 4R, and 7R alleles exhibit the least diversity, with 78.2, 95.2, and 88.9% of the alleles respectively represented by the most common 2R(1-4), 4R(1-2-3-4), and 7R(1-2-6-5-2-5-4) haplotypes (Table 1). In contrast, although the 3R, 5R, 6R, and 8R alleles are rarer, they have proportionally more variants (Table 1). This unusual pattern of allele diversity is clearly not a simple length effect, i.e., longer alleles have greater diversity. Many population-specific rare haplotypes were observed. Examples include the 2R(30-4) haplotype found only in the Surui (South America) sample and the 5R(1-3-2-3-4) haplotype found only in the Han Chinese (Asian) sample (Table 1 and Fig. 2).
The pattern of nucleotide variation observed in the VNTR haplotypes is not random (Fig. 2). Most DNA sequence variants change the amino acid sequence, sometimes quite dramatically (i.e., Gln to Pro; Fig. 2). Although many of these variants are related mutational events (below), one can account for these relationships in calculating Ka/Ks (the ratio of the number of amino acid replacements per site divided by the estimate of the number of synonymous changes). Values of Ka/Ks greater than 1 are taken usually to be a stringent indicator of positive selection at the observed DNA segment (22, 23). For a tandem repeat sequence, many assumed relationships can be inferred, and hence different Ka/Ks ratios can be calculated. For all assumed relationships of the DRD4 variants, however, Ka/Ks > 1. For example, assuming that the most abundant 1–6-variant motifs (Fig. 2) all have a common origin and that diversity was generated by both mutation and recombination (below), a Ka/Ks value of 3 is obtained. Expanding this analysis to include between-species divergence (a powerful method to improve these calculations) is not possible because of the rapid de novo generation of variation in this VNTR in primate lineages (28).
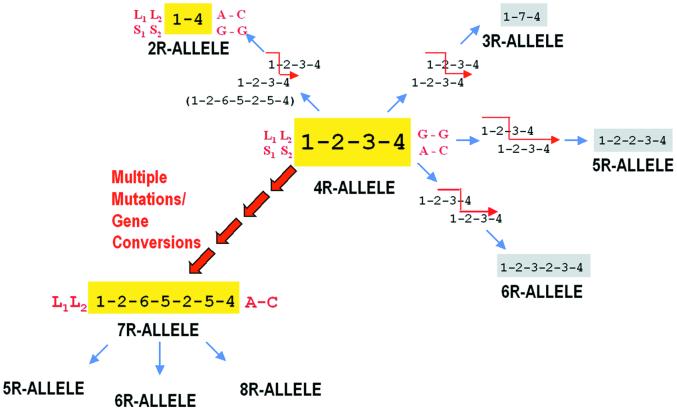
Standard approaches to defining evolutionary relationships between these haplotypes are not applicable because of the repetitive nature of the DNA sequence (23). Based on the observed DNA sequences and their nucleotide variations, however, it is straightforward to propose a simple origin for the majority of these haplotypes (Fig. 3; Table 1). One-step recombination/mutation events between the most common alleles can account for nearly all the observed variation of the 2R–6R alleles. Fig. 3 is a simplified diagram of the most common recombination events proposed. Although the inferred nucleotide sequence of an ancestral DRD4 cannot be determined, all alleles in a particular primate species seem to be derived from a relatively recent common ancestor (28). The most prevalent 4R allele is proposed as the human progenitor allele, based on (i) limited sequence data reported for primate DRD4 4R alleles (28), (ii) the lower level of LD for polymorphisms surrounding this allele (as discussed below), and (iii) the sequence motif arrangements of the non-4R alleles. Unequal recombination between two 4R(1-2-3-4) alleles would produce the observed common 2R–6R alleles (Fig. 3). The position of crossover determines the resulting sequence. For example, the most common 3R(1-7-4) and 3R(1-2-4) alleles differ only in the position of crossover either within or after the second repeat (Fig. 3; Table 1). Thus, the known high frequency of unequal recombination between tandem repeats (29) can account for most of the observed diversity of the DRD4 gene.
Figure 3.
Proposed origin of DRD4 diversity. A simplified model for exon 3 48-bp repeat sequence diversity is shown, with only major recombination events indicated (Fig. 2). The major 2R, 4R, and 7R alleles are shown in yellow, and the minor 3R, 5R, and 6R alleles are shown in gray along with their hypothesized origins by unequal recombination (red arrows). Large red arrows indicate the putative multistep origin of the 7R allele. The adjacent promoter region (L1/S1), exon 1 (L2/S2), and intron 3 (G-G/A-C) polymorphisms are indicated. The strong linkage of the L1, L2, and A-C polymorphisms with the DRD4 7R allele is noted.
In addition to unequal crossovers, single point mutations are evident in this population sample (Table 1 and Fig. 2). For example, with one exception all 2R alleles worldwide have the sequence 2R(1-4) (Table 1). All 12 2R alleles resequenced from Surui (South American) DNA were found to contain a single point mutation, the 2R(30-4) allele (Table 1 and Fig. 2). This mutation, a C to T change in the first repeat, does not alter the amino acid sequence and likely has a recent (less than 10,000–20,000-year) origin (24).
In contrast, the formation of the observed 7R and higher alleles cannot be explained by simple one-step recombination/mutation events from the 4R(1-2-3-4) haplotype (Fig. 3). The generation of a 7R allele from the most prevalent 4R allele would require at least one recombination and six mutations to arise. Even allowing for more complicated gene-conversion events, multiple low probability steps are needed to convert a 4R allele into a 7R allele (Fig. 3). For example, the central five-variant motif found in the common 7R(1-2-6-5-2-5-4) haplotype could be produced by a recombination between two 4R alleles. Recombination between the terminal four-variant motif of one 4R allele and the initial one-variant motif of the second 4R allele would yield a 7R(1-2-3-5-2-3-4) haplotype (Fig. 2). Three additional mutations of each of the two three-variant motifs in this putative 7R haplotype then are required to produce the current 7R(1-2-6-5-2-5-4) haplotype. Four of these six nucleotide changes are nonsynonymous, altering the amino acid sequence (Ser to Gly, Gln to Pro, Ala to Pro, and Ser to Gly; Fig. 2). Although gene conversion rather than mutation could be proposed as the mechanism to “insert” these nucleotide changes in a hypothetical 7R(1-2-3-5-2-3-4) allele, two unlikely events, one involving 7R-7R allele gene conversion, would be necessary (Figs. 2 and 3).
None of these putative “intermediate” 7R haplotypes were observed in this worldwide population sample. Our sample included 47 7R alleles sequenced from individuals of African origin thought to contain populations with the greatest genetic diversity and age (24). It is unlikely, then, that intermediate 7R haplotypes exist at high frequency. It is not our intention, however, to propose a specific origin of the DRD4 7R allele. Rather, we wish to emphasize that based on DNA sequence analysis, the DRD4 7R allele seems quite distinct from the common 2R–6R alleles. It is impossible to determine whether the origin of the DRD4 7R allele was a single, highly unlikely event or a series of unlikely events (Fig. 3).
Regardless of the mechanism of origin of the DRD4 7R allele, it clearly is capable of participating in recombination events with the other alleles. Most of the rare 7R haplotypes observed appear to be recombination events, mostly with the common 4R(1-2-3-4) allele (Table 1). For example, the 7R(1-2-6-5-2-3-4) haplotype appears to be a recombination between a 4R(1-2-3-4) allele and a 7R(1-2-6-5-2-5-4) allele (Table 1 and Fig. 2). This origin was confirmed by analyzing SNPs outside the recombination region (see below). Further, the origin of some of the rare 5R and 6R alleles and all the 8R and higher alleles can be explained by recombinations involving a 7R allele, because they contain the six-variant motif unique to the 7R allele (Fig. 2 and Table 1). Many of these 8R and higher alleles, however, seem to have more complicated origins based on DNA sequence analysis (Table 1 and Fig. 2).
This model (Fig. 3) explains the apparent anomaly in the observed haplotype diversity noted above (Table 1), where the most abundant (and ancient, see below) 4R allele has the lowest nucleotide diversity. If recombination is the predominant generator of diversity, then the majority of 4R-4R recombination events are predicted to have an unchanged nucleotide sequence. Such events can be inferred only by recombination of outside markers. Only when out-of-register recombination occurs will new nucleotide sequence (and length) variants be generated (Fig. 3). The observed pattern of haplotype diversity is consistent with a predominantly “2-allele” system (4R and 7R), with most of the rarer variants generated by recombination from these two haplotypes (Fig. 3).
The unusual nature of the sequence organization of the DRD4 7R allele, suggesting it arose as a rare mutational event, led us to determine whether differences in LD exist between the 4R and 7R alleles. The haplotype of two adjacent intronic SNPs (G/A-G/C; Fig. 1) could be determined directly, because they were present on the same PCR product used to amplify the 48-bpVNTR. Strong LD was found between the A-C SNP pair and the 7R allele (Fig. 3). Ninety-seven percent of 7R alleles were associated with the A-C SNP pair (66 of 68 examined). The two 7R alleles associated with G-G SNPs were 7R-4R recombinant haplotypes as determined originally from DNA sequence analysis (above). In contrast, both the G-G and A-C SNP pairs are associated with DRD4 4R alleles (487 examined alleles). However, the G-G pair is most frequent, representing 86.1% of the African sample but up to 98.6% of our Asian sample.
All African 7R alleles were associated with the A-C haplotypes, whereas only 13.9% of African 4R alleles were associated with the A-C haplotype. DNA sequence analysis of several chimp and bonobo samples (data not shown) indicates that the G-G SNP pair is likely the ancestral sequence (Fig. 3). Thus, it seems that the original DRD4 7R allele arose on this rarer A-C SNP background. A sample of 73 2R, 3R, 5R, and 6R alleles showed approximately equal association with the G-G and A-C SNPs, which is consistent with their proposed recombinational origin from both the 4R and 7R alleles (Fig. 3). Interestingly, all 26 Asian 2R allele samples examined showed association with the A-C SNPs, suggesting their origin from recombinations involving 7R alleles (Fig. 3).
Similar results were obtained for more distant promoter and exon 1 insertion/deletion polymorphisms (Fig. 1). In this case association was inferred indirectly from data obtained for our prior population studies (3, 17) and PCR analysis of a subset of the individuals used in this study. For 40 samples where parental DNA was also available and could be genotyped for these markers, phase could be inferred directly. A strong association was observed between the long (duplicated) L1 promoter polymorphism (Fig. 1) and the 7R allele (Fig. 3), with 90.8% of 7R alleles associated with L1 (607 alleles analyzed). In contrast, the L1 polymorphism is coupled with only 61.9% of 4R alleles (2,102 alleles analyzed). Although population-specific variation was observed (for example, more L1-4R coupling in Chinese than African populations), little overall L1-4R linkage was detected (Fig. 3). The closer L2 polymorphism in exon 1 (Fig. 1) was associated with 93.4% of 7R alleles and 86.4% of 4R alleles, a relative difference similar to that observed for the L1-7R and L1-4R association. The L2/S2 polymorphism is in a coding region, however, and selective constraints may be influencing allele frequency as well (30).
Standard methods of estimating coalescence time for these alleles are not applicable, given the repetitive nature of the region and high recombination frequency. However, calculations of allele age based on the relatively high worldwide population frequency of the DRD4 4R and 7R alleles suggest that these alleles are ancient (>300,000 years old; refs. 25 and 26; see Methods). On the other hand, calculations of allele age based on the observed intraallelic variability (refs. 26 and 27; see Methods) suggest that the 7R allele is 5–10-fold “younger” (30,000–50,000 years old). Such large discrepancies between allele ages calculated by these two methods usually are taken as evidence that selection has increased the frequency of the allele to higher levels than expected by random genetic drift (26). The absolute values of these estimates are greatly affected by the assumptions used in their computations, for example the assumed recombination frequency (26). We have used conservative estimates of recombination frequency based on the average observed for the terminal 20 megabases of 11p (31). Given the observed high recombination at this locus (Table 1 and Fig. 3), it is likely that the actual age of the 7R allele is even younger, and further LD analysis will refine these estimates. The important conclusion, however, is that regardless of the parameters assumed, the relative age differences for the 4R and 7R alleles calculated from intraallelic variability remains large, whereas their population frequency suggests they are both ancient.
The simplest hypothesis to account for (i) the observed bias in nucleotide changes (Ka/Ks), (ii) the unusual sequence organization of the DRD4 7R allele, and (iii) the strong LD surrounding this allele is that the 7R allele arose as a rare mutational event (or events) that nevertheless increased to high frequency by positive selection. Advantageous alleles usually take a long time to reach a frequency of 0.1, then increase rapidly to high frequencies (>0.9). Although it is possible we are observing the recent expansion of a highly advantageous 7R allele, we suggest that it is more likely that this two-allele DRD4 system (Fig. 3) is an example of balanced selection. Such selection may be more pervasive in the human genome than generally thought (24). A balanced selection model proposes that both the 4R and 7R alleles are maintained at high frequencies in human populations. A variety of mechanisms could be proposed for such balanced selection, ranging from heterozygote advantage to frequency-dependent selection (24). According to evolutionary game theory (32), the evolutionary payoff for a particular kind of personality will depend on the existing distribution of personality types. For example, high aggression may lead to high fitness if almost everyone is meek but might result in low fitness when very common, because aggressive individuals would suffer the penalties of frequent conflict. This type of frequency-dependent selection might be expected to apply to many types of psychological variation, including those associated with this particular neurotransmitter receptor (4–9).
Alternative explanations to the proposed positive selection such as recent random bottlenecks, population expansion, and/or population admixture (24) are less likely to account for the observed results. Bottlenecks certainly have occurred during human migration and evolution (33–35) and undoubtedly have influenced the current worldwide DRD4 allele frequency. Numerous population studies on other genes (24, 33, 35) have shown that an “out-of-Africa” constriction of allele diversity (and an increase in LD) likely occurred. In the present study, a greater diversity (and lower LD) was found for African DRD4 4R alleles in comparison to the remainder of our population sample, which is consistent with the out-of-Africa hypothesis (24). Although one could argue that the 7R allele frequency was increased by chance during the out-of-Africa expansion, this theory does not explain the unusual lack of diversity in African 7R alleles. The most common L1L2-7R(1-2-6-5-2-5-4)-A-C haplotype (Fig. 3) is found at frequencies comparable to those found worldwide (>85%). It is difficult to imagine what type of bottleneck could produce such results, i.e., strong worldwide LD for a single allele (DRD4 7R), yet little LD for the remaining alleles. A model that is consistent with the observed results is the “weak Garden of Eden” hypothesis (24), in which the DRD4 4R allele would be hypothesized to be ancient and present in indigenous populations, whereas the 7R allele was spread by the expansion out of (and into) Africa. In such a weak Garden of Eden hypothesis, positive selection for the DRD4 7R allele still must be proposed.
Although we suggest that a recent mutational origin and positive selection best account for the DRD4 7R allele data, another possibility cannot be ruled out. Given the highly unlikely recombination/mutation events required to generate the 7R allele from the 4R allele, a possibility worth considering is the importation of this allele from a closely related hominid lineage. What lineage that might be can only be speculated, but Neanderthal populations were present at the approximate time the 7R allele originated. Under this model, the coalescence time for the 4R and 7R alleles then would be ancient, with the importation occurring only recently, as measured by LD. Obviously, additional experimental work may clarify these speculations.
For the DRD4 locus, it is unlikely that selection for an adjacent gene can account for the proposed selection, given the distinct and unusual DNA sequence of the DRD4 7R allele itself. If the DRD4 7R allele originated ≈40,000 years ago, one might ask what was occurring at that time in human history? It is tempting to speculate that the major expansion of humans that occurred at that time, the appearance of radical new technology (the upper Paleolithic) and/or the development of agriculture (24), could be related to the increase in DRD4 7R allele frequency. Perhaps individuals with personality traits such as novelty seeking, perseverance, etc. drove the expansion (and partial replacement). The speculation that migration could account for the current 7R allele distribution has been proposed (34). In addition to such phenotypic selection, sexual selection could be operating as well. As defined originally by Darwin (36), “any advantage which certain individuals have over others of the same sex and species solely in respect of reproduction” will lead to increased offspring. If individuals with a DRD4 7R allele have personality/cognitive traits that give them an advantage (multiple sexual partners, higher probability for mate selection, etc.) then the frequency of this allele will expand rapidly depending on the cultural milieu. Perhaps cultural differences can account for some of the observed differences in DRD4 7R allele frequency (3). Obviously, determining the exact nature of the DRD4 selection and its biochemical and behavioral basis awaits further experimentation. Recent experiments indicating that individuals with ADHD and possessing this unusual DRD4 7R allele perform normally on critical neuropsychological tests of attention in comparison to other ADHD probands (6) point to but one of many areas of future investigation.
One may ask why an allele that seems to have undergone strong positive selection in human populations nevertheless is now disproportionately represented in individuals diagnosed with ADHD. The common variant/common disorder hypothesis (16) proposes that common genetic variation is related to common disease either because the disease is a product of a new environment (such that genotypes associated with the disorder were not eliminated in the past) or the disorder has small effects on fitness (because it is late onset). For early onset disorders (such as autism, ADHD, etc.) we suggest entertaining the possibility that predisposing alleles in fact are under positive selection and only result in deleterious effects when combined with other environmental/genetic factors. In this context, it is possible that prior selective constraints are no longer operating on this gene. It is possible also to speculate, however, that the very traits that may be selected for in individuals possessing a DRD4 7R allele may predispose behaviors that are deemed inappropriate in the typical classroom setting and hence diagnosed as ADHD.
Acknowledgments
This work was supported by Department of Energy Grant DE-FG03-97ER62485 and National Institute of Mental Health Grant MH60660 (to R.K.M.).
Abbreviations
- DRD4
human dopamine receptor D4
- VNTR
48-bp tandem repeat
- nR
number of repeats
- ADHD
attention-deficit/hyperactivity disorder
- LD
linkage disequilibrium
- SNP
single-nucleotide polymorphism
Footnotes
References
- 1.Van Tol H H M, Bunzow J R, Guan H-C, Sunahara R K, Seeman P, Niznik H B, Civelli O. Nature (London) 1991;350:610–614. doi: 10.1038/350610a0. [DOI] [PubMed] [Google Scholar]
- 2.Lichter J B, Barr C L, Kennedy J L, Van Tol H H M, Kidd K K, Livak K J. Hum Mol Genet. 1993;2:767–773. doi: 10.1093/hmg/2.6.767. [DOI] [PubMed] [Google Scholar]
- 3.Chang F-M, Kidd J R, Livak K J, Pakstis A J, Kidd K K. Hum Genet. 1996;98:91–101. doi: 10.1007/s004390050166. [DOI] [PubMed] [Google Scholar]
- 4.La Hoste G J, Swanson J M, Wigal S B, Glabe C, Wigal T, King N, Kennedy J L. Mol Psychiatry. 1996;1:21–24. [PubMed] [Google Scholar]
- 5.Swanson J M, Flodman P, Kennedy J, Spence M A, Moyzis R, Schuck S, Murias M, Moriarity J, Barr C, Smith M, Posner M. Neurosci Biobehav Rev. 2000;24:21–25. doi: 10.1016/s0149-7634(99)00062-7. [DOI] [PubMed] [Google Scholar]
- 6.Swanson J, Oosterlaan J, Murias M, Schuck S, Flodman, Spence M A, Wasdell M, Ding Y, Chi H-C, Smith M, et al. Proc Natl Acad Sci USA. 2000;97:4754–4759. doi: 10.1073/pnas.080070897. . (First Published April 18, 2000; 10.1073/pnas.080070897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebstein R P, Novick O, Umansky R, Priel B, Osher Y, Blaine D, Bennett E R, Nemanov L, Katz M, Belmaker R H. Nat Genet. 1996;12:78–80. doi: 10.1038/ng0196-78. [DOI] [PubMed] [Google Scholar]
- 8.Benjamin J, Li L, Patterson C, Greenberg B D, Murphy D L, Hamer D H. Nat Genet. 1996;12:81–84. doi: 10.1038/ng0196-81. [DOI] [PubMed] [Google Scholar]
- 9.Swanson J, Deutsch C, Cantwell D, Posner M, Kennedy J, Barr C, Moyzis R, Schuck S, Flodman P, Spence M A. Clin Neurosci Res. 2001;1:207–216. [Google Scholar]
- 10.American Psychiatric Association. DSM-IV: Diagnostic and Statistical Manual of Mental Disorders. 4th Ed. Washington, DC: Am. Psychiatr. Assoc.; 1994. [Google Scholar]
- 11.Faraone S V, Biederman J. Child Adolesc Psychiatr Clin North Am. 1994;3:285–291. [Google Scholar]
- 12.Thaper A, Holmes J, Poulton K, Harrington R. Br J Psychiatry. 1999;174:105–111. doi: 10.1192/bjp.174.2.105. [DOI] [PubMed] [Google Scholar]
- 13.Volkow N D, Wang G J, Fowler J S, Fischman M, Foltin R, Abumrad N N, Gatley S J, Logan J, Wang C, Gifford A, et al. Life Sci. 1999;65:PL7–PL12. doi: 10.1016/s0024-3205(99)00225-8. [DOI] [PubMed] [Google Scholar]
- 14.Asghari V, Sanyal S, Buchwaldt S, Paterson A, Jovanovic V, Von Tol H H M. J Neurochem. 1995;65:1157–1165. doi: 10.1046/j.1471-4159.1995.65031157.x. [DOI] [PubMed] [Google Scholar]
- 15.Collins F S, Guyer M S, Chakravarti A. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 16.Zwick M E, Cutler D J, Chakravarti A. Annu Rev Genomics Hum Genet. 2000;1:387–407. doi: 10.1146/annurev.genom.1.1.387. [DOI] [PubMed] [Google Scholar]
- 17.Ding Y-C, Wooding S, Harpending H C, Chi H-C, Li H-P, Fu Y-X, Pang J-F, Yao Y-G, Yu J-GX, Moyzis R, Zhang Y-P. Proc Natl Acad Sci USA. 2000;97:14003–14006. doi: 10.1073/pnas.240441297. . (First Published November 28, 2000; 10.1073/pnas.240441297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seaman M I, Fisher J B, Chang F-M, Kidd K D. Am J Med Genet. 1999;88:705–709. doi: 10.1002/(sici)1096-8628(19991215)88:6<705::aid-ajmg22>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 19.McCracken J T, Smalley S L, McGough J J, Crawford L, Del'Homme M, Cantor R M, Liu A, Nelson S F. Mol Psychiatry. 2000;5:531–536. doi: 10.1038/sj.mp.4000770. [DOI] [PubMed] [Google Scholar]
- 20.Catalano M, Nobile M, Novelli E, Nothen M M, Smeraldi E. Biol Psychiatry. 1993;34:459–464. doi: 10.1016/0006-3223(93)90236-7. [DOI] [PubMed] [Google Scholar]
- 21.Riethman H C, Xiang Z, Paul S, Morse E, Hu X-L, Flint J, Chi H-C, Grady D L, Moyzis R K. Nature (London) 2001;409:948–951. doi: 10.1038/35057180. [DOI] [PubMed] [Google Scholar]
- 22.Kimura M. Nature (London) 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 23.Kreitman M. Annu Rev Genomics Hum Genet. 2000;1:539–559. doi: 10.1146/annurev.genom.1.1.539. [DOI] [PubMed] [Google Scholar]
- 24.Harpending H, Rogers A. Annu Rev Genomics Hum Genet. 2000;1:361–385. doi: 10.1146/annurev.genom.1.1.361. [DOI] [PubMed] [Google Scholar]
- 25.Kimura M, Ohta T. Genetics. 1973;75:199–212. doi: 10.1093/genetics/75.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slatkin M, Rannala B. Annu Rev Genomics Hum Genet. 2000;1:225–249. doi: 10.1146/annurev.genom.1.1.225. [DOI] [PubMed] [Google Scholar]
- 27.Serre J L, Simon-Bouy B, Mornet E, Iaume-Roig B, Balassopoulou A, Schwartz M, Taillandier A, Boue J, Boue A. Hum Genet. 1990;84:449–454. doi: 10.1007/BF00195818. [DOI] [PubMed] [Google Scholar]
- 28.Livak K J, Rogers J, Lichter J B. Proc Natl Acad Sci USA. 1995;92:427–431. doi: 10.1073/pnas.92.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeffreys A J, Neil D L, Neumann R. EMBO J. 1998;17:4147–4157. doi: 10.1093/emboj/17.14.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seaman M I, Chang F-M, Deinard A S, Quinones A T, Kidd K K. J Exp Zool. 2000;288:32–38. [PubMed] [Google Scholar]
- 31.International Human Genome Sequencing Consortium. Nature (London) 2001;409:860–921. [Google Scholar]
- 32.Smith J M. Evolution and the Theory of Games. Cambridge, U.K.: Cambridge Univ. Press; 1982. [Google Scholar]
- 33.Tishkoff S A, Dietzsch E, Speed W, Pakstis A J, Kidd J R, Cheung K, Bonne-Tamir B, Santachiara-Benerecetti A S, Moral P, Krings M. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Burton M, Greenberger E, Dmitrieva J. Evol Hum Behav. 1999;20:309–324. [Google Scholar]
- 35.Reich D E, Cargill M, Bolk S, Ireland J, Sabeti P C, Richter D J, Lavery T, Kouyoumjian R, Farhadian S F, Ward R, Lander E S. Nature (London) 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 36.Darwin C. The Descent of Man and Selection in Relation to Sex. New York: Merrill and Baker; 1874. [Google Scholar]