Abstract
To test whether regions undergoing genomic imprinting have unique genomic characteristics, imprinted and nonimprinted human loci were compared for nucleotide and retroelement composition. Maternally and paternally expressed subgroups of imprinted genes were found to differ in terms of guanine and cytosine, CpG, and retroelement content, indicating a segregation into distinct genomic compartments. Imprinted regions have been normally permissive to L1 long interspersed transposable element retroposition during mammalian evolution but universally and significantly lack short interspersed transposable elements (SINEs). The primate-specific Alu SINEs, as well as the more ancient mammalian-wide interspersed repeat SINEs, are found at significantly low densities in imprinted regions. The latter paleogenomic signature indicates that the sequence characteristics of currently imprinted regions existed before the mammalian radiation. Transitions from imprinted to nonimprinted genomic regions in cis are characterized by a sharp inflection in SINE content, demonstrating that this genomic characteristic can help predict the presence and extent of regions undergoing imprinting. During primate evolution, SINE accumulation in imprinted regions occurred at a decreased rate compared with control loci. The constraint on SINE accumulation in imprinted regions may be mediated by an active selection process. This selection could be because of SINEs attracting and spreading methylation, as has been found at other loci. Methylation-induced silencing could lead to deleterious consequences at imprinted loci, where inactivation of one allele is already established, and expression is often essential for embryonic growth and survival.
Genomic imprinting is defined by differences in the epigenetic organization of regions dispersed throughout the genome that depend on their most recent germline exposure. These differences, which include CpG methylation (1), histone acetylation (2), and chromatin organization (3), result in a difference in potential for gene expression on the maternally and paternally derived chromosomes. The mechanism is not simply explained in terms of epigenetic differences established during gametogenesis targeted to individual promoters, as imprinted genes can cluster in megabase-sized regions, subject to coordinated regulation by elements of uncertain mechanism as distant as 1 Mb (4). Specific genomic characteristics of imprinted regions have been sought in studies limited by the relative scarcity of comprehensively sequenced imprinted loci. The association of short clustered repeat sequences with imprinted promoters (5, 6) has withstood neither subsequent comparative genomic (7) nor functional studies (8). The observation that imprinted genes have “few and small introns” (9) was made before the identification of imprinted loci such as KCNQ1 (10) and the extended SNURF-SNRPN transcript (K. Buiting, personal communication), which are unusually large. To this point, no genomic signature for imprinted regions has been found.
There are two evolutionary models that predict specific genomic characteristics to occur at imprinted loci. The first is the conflict model for genomic imprinting (11), which is based on the assumption that imprinting evolved to help balance maternal and paternal influences on offspring growth by means of genes such as the paternally expressed insulin-like growth factor 2 (Igf2) and the maternally expressed growth inhibitor Igf2r (M6pr) (12, 13). The functions of these genes are complementary (14), and the loss of either has significant effects on embryonic growth (12, 13). Among the implications of this model is the restriction of imprinting to species that are placental or at least significantly matrotrophic, for a conflict for fitness in utero to be relevant. The results of limited studies of imprinting in marsupials (15), oviparous monotremes (16), and aves (15) support this idea. One of the aims of this study was, therefore, to test whether any specific sequence patterns associated with imprinted loci could be traced by using paleogenomic techniques to a defined timepoint, such as the evolution of placental mammals. A second hypothesis addressed in this study is referred to as the host defense model, which proposes that methylation of CpGs is a genomic adaptation primarily directed at suppressing its transposable element content (17). Imprinted and X-inactivated CpG islands undergoing differential methylation are the exceptions to a major tenet of this model: that CpG islands regulating endogenous genes are never the target of methylation. As methylated cytosines are prone to deamination and transition to thymines (18), and imprinting occurs in the cells and developmental time frame in which sequence changes become fixed in the genome, one question prompting this study was whether methylation of imprinted CpG islands for half their evolutionary history is reflected by an erosion in their CpG content when compared with CpG islands that are predicted never to be methylated.
In addition to CpG content, the analyses in this study focused on the nucleotide composition and the quantities of transposable element remnants in the extended region flanking imprinted promoters. Compositional heterogeneity within the genome is manifested at the cytogenetic level as bands on chromosomes, which correlate with differences in repetitive element type (19), methylation levels (20), gene density (21), and replication timing (22). This raises the question whether regulatory influences acting on the scale of chromosomal bands might influence imprinted regions contained within them, especially as the cytogenetically apparent features of methylation (20) and replication timing (22) reflect components of the imprinting process. Before the advent of large amounts of draft genome sequence, the proposal was made that the genome heterogeneity indicated by the bands of chromosomes could be measured simply in terms of guanine and cytosine content (GC) (23). Although a correlation between such GC content “isochores” and other parameters was demonstrable for many variables, including CpG content (24), gene density (21), and repetitive sequence type (24), analysis of the draft human genome sequence indicated that the GC content is not uniform, and that the prefix “iso-” is inaccurate in implying homogeneity of GC content within a given region (25). The alternative description of isochores as “GC content domains” has been suggested as a more accurate description of this parameter of genomic compartmentalization (25).
The GC content domain characteristics of imprinted genes were described tangentially in a prior study, using the codon third position as the indicator of each gene's genomic environment. Although the sample of imprinted genes available at that time was small, one provocative finding was that the predominantly maternally expressed genes of mouse 7F/human 11p15.5 were from a high GC content domain, whereas the predominantly paternally expressed mouse 7C/human 15q11-q13 genes were located in a low GC environment (9). The present study, therefore, included the separate analysis of maternally and paternally expressed genes to see whether this preliminary observation of sequence context differences was borne out with a more comprehensive analysis. The outcome of all of these analyses defines a specific genomic signature for imprinted loci in the human genome, including a significant partitioning of maternally and paternally expressed subgroups of imprinted genes to distinct compartments within the genome, and indicates a potential evolutionary selection interplay between imprinting and short interspersed transposable elements (SINEs).
Materials and Methods
Sampling Methods.
The experimental aim was to analyze the genomic context of the regions flanking imprinted promoters. The 100 kb flanking each promoter was chosen for study, as cis-acting elements at the β-globin (26) and Igf2 (27, 28) loci, act over comparable distances. When 100-kb samples overlapped, they were treated as separate observations. Human loci were used in this study, as draft sequence is not yet widely available for sufficient imprinted loci in mouse. Some loci described as imprinted were excluded from the study if genomic sequence data were not yet available (MKRN3) or the position of the promoter is unknown (UBE3AAS) (29), conclusions about imprinting are not based on the analysis of primary tissues [HTR2A (30), ASCL2 (31)], or imprinting data are conflicting [IGF2R (32, 33)]. Two loci (WT1 and GRB10) appear to be paternally expressed in some circumstances and maternally in others (34–37) and so were not used in analyses involving the paternally or maternally expressed subgroups of imprinted genes. A list of the 31 imprinted genes used with references to their imprinting studies, their sequence accession numbers, and the genomic coordinates used are published as supporting information on the PNAS web site (www.pnas.org).
For certain imprinted loci, the promoter was contained within unassembled sequence [from bacterial artificial chromosomes averaging 178 kb in size (38)], so the flanking 100 kb is not precisely defined (COPG2, PEG10, SNURF-SNRPN, the 15q11-q13 snoRNA cluster, IPW and ATP10C). For proportional data, such as the percentage of sequence occupied by a certain characteristic or observed versus expected CpG dinucleotide ratios (see below), the result for the unassembled sequence as a whole was used to reflect the likely composition of the 100 kb around the promoter. Quantitative measurements of CpG islands or dinucleotides (see below) were adjusted to reflect the proportion per 100 kb. The promoter region was always contained within a sequence fragment of sufficient size to determine the CpG island characteristics of the 4 kb flanking the transcription start site, even in unassembled bacterial artificial chromosome sequences.
The control sample was taken from nonimprinted autosomal regions of the genome [no imprinted genes and no uniparental disomy effects (39)], where contigs of ≥2 Mb have been constructed. Characterized genes from eight contigs on separate chromosomes were numbered. A sample of 100 genes was chosen from this group randomly by using these assigned numbers, and the 100 kb flanking the 5′ end of the mRNA at that locus was sampled for each gene to represent the identical context to that for the imprinted gene sample. The frequency of sequence overlap for loci in the imprinted and control gene samples was almost identical (46 and 45%, respectively). These control loci are also described in the supporting information on the PNAS web site.
Measurement of Parameters of Sequence Context.
GC content was quantified for each sequence sample by using the base composition function of macvector (Ver. 7.0, Accelrys, Madison, WI). As CpG dinucleotides are the target of methylation signals that contribute to the epigenetic regulation of imprinted loci (1), the numbers and sizes of CpG islands throughout each sample and in the 4 kb flanking each transcription start site were determined by using the cpgplot program (http://www.uk.embnet.org/Software/EMBOSS/Apps/cpgplot.html). The CpG dinucleotide content of each sequence sample was measured as both the number of CpG dinucleotides and the observed/expected ratio within the entire 100-kb sequence sample by using the dinucleotide content function of macvector. The presence of repetitive sequences derived from transposable elements was analyzed by repeatmasker (A. F. A. Smit and P. Green, http://ftp.genome.washington.edu/RM/RepeatMasker.html) by using the slow/sensitive and primate DNA settings. The repbase update from March 31, 2001 was the database used for the repeatmasker analyses (http://www.girinst.org/). As the output of this program includes information on subfamilies of L1 long interspersed transposable elements (LINEs) and Alu transposons, for which evolutionary age information is available, these data were also collected for analysis. L1 LINEs are presumed to be of monophyletic origin, having entered the genome before the mammalian radiation and having given rise to a series of descendants that underwent active retrotransposition at different times in evolution. These assumptions are consistent with the observations that related LINEs are distributed among different mammalian lineages proportionately to their degree of sequence divergence from a consensus LINE (40). Likewise, the primate-specific Alu SINEs originated as a monomeric signal recognition particle RNA (41). The divergence of Alus at the sequence level reflects their distribution among primates and thus their timing of acquisition by an ancestral genome (42). The proportional distributions of subclasses of L1s and Alus in imprinted and control regions were analyzed to determine whether differences in the timing of acquisition of these elements exists in imprinted regions.
Linear regression analyses were performed on the control gene sample for each of the above variables using the statistical package prism (graphpad, Ver. 3, Graph Pad Software, San Diego). Values that were heterogeneously and nonrandomly distributed with respect to each other among these control loci showed a slope significantly different to zero. The large number of comparisons required an adjustment of the value at which significance could be attributed to an individual result, for which a Bonferroni correction was used. The original data from which Fig. 1 is derived are published as supporting information on the PNAS web site. The distribution of measurements for imprinted loci was compared with those for control loci using the nonparametric two-tailed Mann–Whitney test, with a Bonferroni correction to reset the threshold at which statistical significance could be attributed.
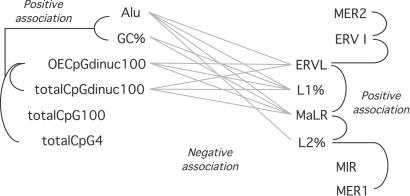
Figure 1.
Two major compartments exist within the human genome. Linear regression analyses were performed on base composition and transposon type for control genomic regions. Regressions achieving statistical significance because a positive or negative association of each parameter are illustrated. The parameters cluster into two groups, as shown. CpG dinucleotide content segregates with Alu and high GC content, defining one genomic compartment (Left) separate from that defined predominantly by LINEs and long terminal repeat transposons (Right). totalCpG100, cumulative size of CpG islands in each 100-kb sample; totalCpG4, cumulative size of CpG islands in each 4 kb flanking the transcription start site; totalCpGdinuc100, number of CpG dinucleotides per 100 kb; OECpGdinuc100, observed/expected ratio of CpG dinucleotides in the 100-kb sample. A table with the full set of results of the regression analyses is published as supporting information on the PNAS web site.
In addition to the comparison of discrete sequence samples, the two best defined regions of transition from imprinted to nonimprinted sequences in the available human genome sequence were analyzed for changes in SINE content in cis. The sizes and proportion of sequence occupied by Alus and mammalian-wide interspersed repeats (MIRs) were binned for 5-kb windows within each of these two 11p15.5 contigs (NT_009368.5 and NT_009308.5). Using excel (Microsoft), a graph was created for SINE content in a 50-kb window moving with 5-kb steps across each contig, representing known imprinted and nonimprinted sequences within each plot, annotated by using blast alignments of mRNA sequences with the sequences (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html).
Results
The control gene sample reflects the heterogeneity of the human genome, as indicated by the results of the regression analyses within this sample. The results of these analyses are summarized in Fig. 1. Genomic sequence characteristics previously found to be regionally clustered (see Introduction and refs. 19, 24, and 43) are likewise heterogenous when the genome is sampled in the manner of this study. These sequence characteristics cluster exclusively into two major groups, representing separate genomic compartments. The first group is the GC-, CpG-, and Alu-rich compartment, whereas the other compartment is GC-poor and rich in L1 LINE repeats and long terminal repeat retrotransposons. These genomic compartments are represented cytogenetically by R- and G-bands, respectively.
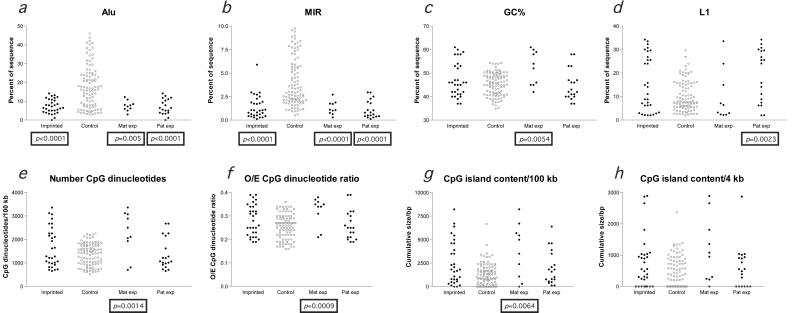
When these sequence characteristics were used to compare the imprinted with the control loci, statistically significant differences were found. The distributions of observations for imprinted and control loci are plotted in Fig. 2, with the differences of statistical significance denoted by the annotation of a P value below the imprinted sample or subsample. In the 100-kb flanking gene promoters, both major classes of SINE repeats (Alus and MIRs) are present at imprinted loci at a density much lower than at the control loci (Fig. 2 a and b). Alus are primate-specific SINEs (44), whereas MIRs were originally described to occur in all major mammalian lineages, including marsupials and monotremes (45). The exclusion of MIRs from imprinted regions therefore indicates that sequence characteristics of regions now imprinted were established before the mammalian radiation.
Figure 2.
Significant differences in sequence characteristics exist between imprinted and control loci. Individual measurements at imprinted (filled black circle) and control (open gray circle) loci for each of the parameters indicated are plotted so that the relative distributions are visually apparent. Maternally expressed and paternally expressed subgroups were separately compared with the control sample and are shown on the right of each graph. Mann–Whitney two-tailed tests that revealed differences in the distributions of measurements of imprinted loci compared with controls (remaining significant after a Bonferroni correction) are indicated by the boxed P value below that group. The major genomic characteristic of imprinted regions is the low density of Alu and MIR SINEs (a and b). The one outlying value in b, reflecting a MIR concentration of 5.90% in the 100 kb flanking its promoter, is the WT1 gene, which is paternally expressed in some tissues and maternally expressed in others (34, 35) and consequently is not included in the subgroup analyses. The maternally expressed subgroup has a significantly higher GC (c) and CpG content (e–g) than the control sample, whereas the paternally expressed subgroup has a higher L1 content, indicating that these subgroups of imprinted genes tend to segregate to the different genomic compartments illustrated in Fig. 1. No difference in the size of CpG islands in the 4 kb flanking the transcription start site at imprinted and nonimprinted loci was apparent (h).
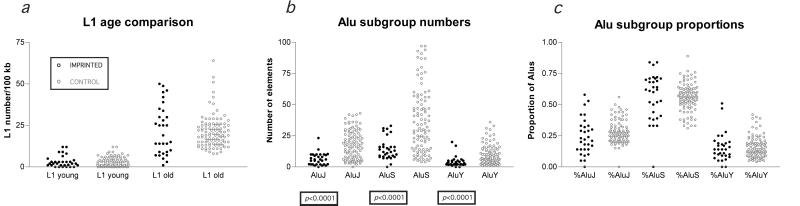
A question prompted by this finding was whether this observation could be attributed to a nonpermissiveness of imprinted regions to retroposition events in general. This would not be consistent with prior observations that imprinted regions can include retroposed endogenous genes, such as the Frat3-Ndn cluster of mouse 7C (46). As the retroposition of Alu elements appears to depend on the L1 retroposition enzyme machinery (47), the imprinted regions and control loci were compared to see whether L1 retroposition into imprinted regions has been continuous during mammalian evolution. A major enrichment of young L1s occurred in the eutherian/prosimian transition that has been proposed to have functional significance for X inactivation (48). As this enrichment occurred during the period of SINE exclusion from imprinted regions, the ongoing retroposition of L1s into imprinted regions (Fig. 2d) during SINE exclusion is demonstrable by the statistically identical sizes of the subgroups of young and old L1 transposons at imprinted and control loci (Fig. 3a). Imprinted regions have been, by this measure, normally permissive to retroposition of L1s during the period in which SINEs were being excluded.
Figure 3.
The age distributions of L1 LINE and Alu SINE repetitive sequences are compared for imprinted and control loci. L1 repetitive sequences were subcategorized into old or young, reflecting integration into the genome before or subsequent to the eutherian/prosimian transition, respectively (48). The numbers of informative L1 3′ ORFs allowing this subcategorization are plotted in a for the imprinted and control samples. No differences exist between these subgroups, indicating that imprinted regions have had ongoing integration of L1 transposons during the evolution of mammals. The Alu transposons were classified in terms of their time of integration into the primate genome, looking at the absolute numbers of each subgroup (b) or the proportion at that locus represented by each subgroup (c). Although the number of Alus in each subgroup is significantly lower than at the control loci (b), reflecting the low density of Alus in general in imprinted regions (Fig. 2a), the proportions represented by each subgroup are statistically indistinguishable at imprinted loci (c). These data indicate that Alus have continuously retroposed into imprinted regions but at a reduced rate compared with nonimprinted regions.
Imprinted regions are not absolutely exclusive of SINE elements. The presence of the primate-specific SINEs, Alu elements, indicates that imprinted regions have been subject to the continued acquisition of SINEs during the period of their relative exclusion. To determine how these new SINEs were acquired, the ages of Alus in imprinted and control regions were compared. Although the absolute numbers of the three subgroups of Alus were all significantly decreased compared with the control loci, the proportions represented by each subgroup at each locus were statistically identical (Fig. 3 b and c). This result indicates that the acquisition of SINEs by imprinted regions is a continuous process, and the local paucity of SINEs is because of a decreased rate of accumulation compared with nonimprinted regions.
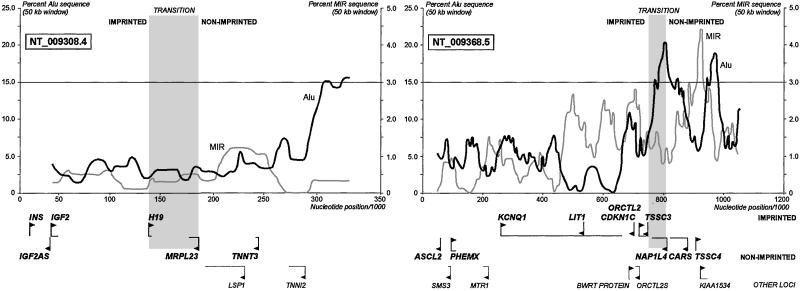
The constrained proportion of SINE content in imprinted regions is lost when the sequence transitions from imprinted to nonimprinted DNA (Fig. 4). Two well-characterized regions of transition, the H19-MRPL23 (49) and the TSSC3/NAP1L4 (50) intervals, are flanked by regions of increased SINE content. This representation illustrates clearly how the 100-kb windows used were able to detect the low SINE content of imprinted regions, which appear to maintain this sequence characteristic uniformly throughout.
Figure 4.
SINE content rises sharply in the transition between imprinted and nonimprinted regions. A window of 50 kb moving with 5-kb steps plots the content of SINEs across two well-characterized regions of transition from imprinted to nonimprinted DNA (accession numbers and gene locations shown). The approximate threshold for SINE content in the 100-kb windows of Fig. 2 a and b is represented by a line at 15% Alu/3% MIR content in these graphs. The rise in SINE content in excess of this threshold occurs immediately at the TSSC3/NAP1L4 transition, whereas the H19 locus maintains a greater buffer region of low SINE content that extends through the immediately adjacent nonimprinted flanking genes. Low SINE content is therefore a sensitive nonspecific screening tool for identifying imprinted regions.
The maternally and paternally expressed imprinted genes were separately compared with the control sample to see whether each subgroup was distinctive in terms of its genomic context. The decrease in SINE content was common to each of these subgroups (Fig. 2 a and b), but a second pattern of differences was found. The maternally expressed subgroup of imprinted loci (n = 10) has a significantly higher GC content, an increase in CpG dinucleotide content (Fig. 2e), a higher observed/expected CpG dinucleotide ratio (Fig. 2f), and a larger amount of CpG island DNA (Fig. 2g) within the 100-kb window. The paternally expressed subgroup (n = 19), on the other hand, had a significantly greater L1 content than the control sample (Fig. 2d). As these are parameters that describe two distinct genomic compartments within the human genome (Fig. 1), the results from these analyses indicate that these subgroups of imprinted genes preferentially segregate to these distinct genomic compartments.
Discussion
This study has identified two major genomic characteristics of imprinted regions. Firstly, imprinted loci in the human genome have a low density of SINE transposons. Secondly, maternally and paternally expressed imprinted genes tend to occupy the distinct genomic compartments defined previously (19, 24, 43) and by the analyses of the control genes in this study (Fig. 1). One of the starting hypotheses, that CpG islands at imprinted promoters would be more prone to mutational decay than those at nonimprinted loci, was not supported by the analyses (Fig. 2h). This may not reflect a weakness in the host defense model for genomic methylation (17) if active selection for the maintenance of CpG island size at imprinted loci is occurring. It is worth pointing out that not all CpG islands at imprinted promoters are differentially methylated [e.g., UBE3A (51)], so this question can be revisited when more is known about the germ line and early developmental methylation of a large sample of imprinted CpG islands.
Although both maternally and paternally expressed imprinted loci have a low density of SINEs, the comparison of each subgroup with the control sample revealed significant differences that indicate a partitioning of each subgroup into different genomic compartments. These results indicate that the tendency to activate each genomic compartment differs depending on each germline, the female germline more readily activating GC/CpG-rich imprinted regions, and the male germline preferentially activating AT/L1-rich regions. In other words, the imprint appears to be driven in a certain direction by the sequence context of the promoter.
Imprinted loci contain many fewer SINE transposon-derived sequences than nonimprinted loci (Fig. 2 a and b). It is possible that this difference reflects a separate unidentified variable of chromosomal context that allows imprinting but not the accumulation of SINEs. Such macrogenomic variables have become apparent when large-scale sequence analyses have been performed, revealing such phenomena as contextual biases for duplication events in the human genome (52). In the simpler model of direct interaction between SINEs and imprinting, the paucity of SINEs could be attributed either to a primary inability of SINEs to retropose into regions undergoing imprinting or to an increased rate of their removal. The latter appears to be a very infrequent event for human retroelements in general (25) and Alus in particular (53). As endogenous genes (46) and L1 LINEs (Fig. 2d) are readily capable of retroposing into imprinted regions, these regions appear to be permissive to retroposition in general. SINEs appear to use the LINE retroposition machinery for their propagation, coevolving with partner LINE elements in different species (54). Alus in particular are believed to use L1-encoded enzymes for retroposition (47). The continuous L1 retroposition into imprinted regions during mammalian evolution indicates that these regions are normally permissive to the L1 retroposition process, and that the low frequency of Alus is unlikely to be caused by a primary failure of retroposition.
SINEs of ancient (MIR) and more recent (Alu) evolutionary acquisition are both excluded from imprinted regions. If the exclusion of SINEs were absolute, Alus would not be present at all at these loci. Instead, they are present at low frequencies. The distribution of young, intermediate, and old Alus [AluY, AluS, and AluJ subgroups, respectively (55)] in imprinted regions is proportionally identical for all subgroups (Fig. 3c), indicating that Alu insertion into imprinted regions has occurred continuously but at a reduced rate compared with control loci. The L1 and Alu subgroup analyses therefore do not support a primary inability of SINEs to retropose into imprinted regions. Instead, the constraint on the rate of SINE retroposition into imprinted regions points toward a threshold of intolerance of SINEs, possibly maintained by an active selection process.
Active selection against SINEs may involve the propensity of SINEs to be targets of methylation. SINEs can be unusually CpG-rich (e.g., Alus) and constitute a major target of genomic methylation (56). Although SINEs themselves show different levels of methylation in the male and female germlines of primates (57), and this transient difference (56) was proposed to be a mediator of the imprinting signal for endogenous genes (44), the paucity of Alus in regions undergoing imprinting makes this unlikely. In somatic cells, SINEs not only attract methylation to themselves but can spread this methylation in cis to flanking nontransposon sequences in animals (58, 59) and plants (60). The methylation of SINEs is completed during embryogenesis (56), a time period during which the function of imprinted genes is critically important (61). Newly retroposed SINEs in imprinted regions that attract and spread methylation could silence imprinted promoters, which are distinctive for their sensitivity to methylation signals. The organism should be more tolerant of this process at nonimprinted loci, as a normal functional allele on the homologous chromosome would remain active. As this selection process does not require a precise site of integration for its presumed effect and can influence gene expression heterozygously, the associated genotype (SINEs in imprinted regions) would be very effectively reduced in the population. Selection against SINEs would also be expected to occur in those other regions in the genome that likewise undergo monoallelic inactivation involving methylation, such as the mammalian X chromosome (62). Statistically significant decreases in Alu content on the human X chromosome compared with autosomal regions have been shown in two separate studies (48, 63), consistent with the active selection process proposed here.
The observation that MIRs in particular are sparse in imprinted regions is interesting from the point of view of the evolution of imprinting. MIRs are similar to Alus in that they are nonautonomous retroposons with internal RNA polymerase III promoters, presumed to retropose by using enzymes encoded by LINE retroposons, with which they coevolve (54) and share 3′ sequence (64). The original description of MIRs as mammalian-wide interspersed repetitive elements was based on testing of all of the major mammalian lineages (45). Subsequent comparative sequencing confirmed the presence of MIRs in these lineages as well as similar sequences in other nonmammalian animal taxa (54). These authors proposed that a single event created an ancestral SINE precursor, and that MIRs represent a component of this CORE-SINE family of retroposons in animals (65). The exclusion of MIRs from imprinted regions links imprinting with events occurring in the genome before the evolution of placental mammals, which is unexpected in light of the evolutionary theory that is based on a requirement for imprinting to control growth in placental mammals alone (11). Although it is possible that imprinting may predate the evolution of placental mammals, and that MIRs were selected against in regions subject to imprinting just as Alus were subsequently, there are other potential reasons for this negative association. MIRs may have been preferentially lost in subsequently imprinted regions, or imprinting might have been established only in regions of the genome that lacked SINEs to start with. Genomic and imprinting analyses in different animal species should resolve these possibilities.
The low frequency of SINEs in imprinted regions may be a valuable tool in genomics-based approaches to the identification of the presence and extent of imprinted domains. The inflections of SINE content shown in Fig. 4 at the transitions from imprinted to nonimprinted DNA correlate well with the known extent of the 11p15.5 domain. Although it is obvious from the data illustrated in Fig. 2 a and b that the low abundance of SINEs in imprinted regions is not a specific feature (as many nonimprinted data points fall within the same range), a search for imprinted loci in a chromosomal region manifesting parent of origin-dependent effects can be focused using this genomic characteristic.
As more imprinted loci are identified and sequenced, the studies presented here can be expanded. Although the current sample is a fraction of expected total human imprinted gene complement [current projections suggest that over 100 genes are imprinted in mammals (66)], there is no obvious bias in ascertainment that explains why SINE-poor genes would have been identified first, so this pattern is expected to continue to hold in larger samples. The first indication that the same pattern of SINE exclusion is occurring in mouse was reported as part of the analysis of an extensive imprinted sequence on mouse 7F (Cars to Mash2). The authors described this sequence to be notable for the “absence” of SINEs throughout this imprinted region (7). The mouse genome is under continued pressure from active transposons (67) and contains fewer extremes of CpG content (68), features that will stress the ability to find significance for the variables found to be characteristic of human imprinted loci and will therefore help to test the conclusions of this study.
Supplementary Material
Acknowledgments
Drs. Rob Nicholls and Avril Friel are thanked for discussions and critical feedback. This work was supported by National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases) Grants DK02467 and DK56786.
Abbreviations
- LINE
long interspersed transposable element
- SINE
short interspersed transposable element
- MIR
mammalian-wide intespersed repeat
- GC
guanine and cytosine content
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Li E, Beard C, Jaenisch R. Nature (London) 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 2.Saitoh S, Wada T. Am J Hum Genet. 2000;66:1958–1962. doi: 10.1086/302917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schweizer J, Zynger D, Francke U. Hum Mol Genet. 1999;8:555–566. doi: 10.1093/hmg/8.4.555. [DOI] [PubMed] [Google Scholar]
- 4.Saitoh S, Buiting K, Rogan P K, Buxton J L, Driscoll D J, Arnemann J, Konig R, Malcolm S, Horsthemke B, Nicholls R D. Proc Natl Acad Sci USA. 1996;93:7811–7815. doi: 10.1073/pnas.93.15.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann B, Kubicka P, Barlow D P. Nat Genet. 1995;9:12–13. doi: 10.1038/ng0195-12. [DOI] [PubMed] [Google Scholar]
- 6.Tremblay K D, Saam J R, Ingram R S, Tilghman S M, Bartolomei M S. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 7.Engemann S, Strodicke M, Paulsen M, Franck O, Reinhardt R, Lane N, Reik W, Walter J. Hum Mol Genet. 2000;9:2691–2706. doi: 10.1093/hmg/9.18.2691. [DOI] [PubMed] [Google Scholar]
- 8.Stadnick M P, Pieracci F M, Cranston M J, Taksel E, Thorvaldsen J L, Bartolomei M S. Dev Genes Evol. 1999;209:239–248. doi: 10.1007/s004270050248. [DOI] [PubMed] [Google Scholar]
- 9.Hurst L D, McVean G, Moore T. Nat Genet. 1996;12:234–237. doi: 10.1038/ng0396-234. [DOI] [PubMed] [Google Scholar]
- 10.Lee M P, Hu R J, Johnson L A, Feinberg A P. Nat Genet. 1997;15:181–185. doi: 10.1038/ng0297-181. [DOI] [PubMed] [Google Scholar]
- 11.Moore T, Haig D. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 12.Barlow D P, Stoger R, Herrmann B G, Saito K, Schweifer N. Nature (London) 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 13.DeChiara T M, Robertson E J, Efstratiadis A. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 14.Filson A J, Louvi A, Efstratiadis A, Robertson E J. Development (Cambridge, UK) 1993;118:731–736. doi: 10.1242/dev.118.3.731. [DOI] [PubMed] [Google Scholar]
- 15.O'Neill M J, Ingram R S, Vrana P B, Tilghman S M. Dev Genes Evol. 2000;210:18–20. doi: 10.1007/pl00008182. [DOI] [PubMed] [Google Scholar]
- 16.Killian J K, Byrd J C, Jirtle J V, Munday B L, Stoskopf M K, MacDonald R G, Jirtle R L. Mol Cell. 2000;5:707–716. doi: 10.1016/s1097-2765(00)80249-x. [DOI] [PubMed] [Google Scholar]
- 17.Bestor T H. Philos Trans R Soc London B. 1990;326:179–187. doi: 10.1098/rstb.1990.0002. [DOI] [PubMed] [Google Scholar]
- 18.Duncan B K, Miller J H. Nature (London) 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- 19.Korenberg J R, Rykowski M C. Cell. 1988;53:391–400. doi: 10.1016/0092-8674(88)90159-6. [DOI] [PubMed] [Google Scholar]
- 20.Barbin A, Montpellier C, Kokalj-Vokac N, Gibaud A, Niveleau A, Malfoy B, Dutrillaux B, Bourgeois C A. Hum Genet. 1994;94:684–692. doi: 10.1007/BF00206964. [DOI] [PubMed] [Google Scholar]
- 21.Federico C, Andreozzi L, Saccone S, Bernardi G. Chromosome Res. 2000;8:737–746. doi: 10.1023/a:1026797522102. [DOI] [PubMed] [Google Scholar]
- 22.Sadoni N, Langer S, Fauth C, Bernardi G, Cremer T, Turner B M, Zink D. J Cell Biol. 1999;146:1211–1226. doi: 10.1083/jcb.146.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernardi G, Mouchiroud D, Gautier C. J Mol Evol. 1988;28:7–18. doi: 10.1007/BF02143493. [DOI] [PubMed] [Google Scholar]
- 24.Jabbari K, Bernardi G. Gene. 1998;224:123–127. doi: 10.1016/s0378-1119(98)00474-0. [DOI] [PubMed] [Google Scholar]
- 25.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature (London) 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 26.Martin D I, Fiering S, Groudine M. Curr Opin Genet Dev. 1996;6:488–495. doi: 10.1016/s0959-437x(96)80072-4. [DOI] [PubMed] [Google Scholar]
- 27.Hark A T, Schoenherr C J, Katz D J, Ingram R S, Levorse J M, Tilghman S M. Nature (London) 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 28.Bell A C, Felsenfeld G. Nature (London) 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 29.Rougeulle C, Cardoso C, Fontes M, Colleaux L, Lalande M. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 30.Kato M V, Shimizu T, Nagayoshi M, Kaneko A, Sasaki M S, Ikawa Y. Am J Hum Genet. 1996;59:1084–1090. [PMC free article] [PubMed] [Google Scholar]
- 31.Alders M, Hodges M, Hadjantonakis A K, Postmus J, van Wijk I, Bliek J, de Meulemeester M, Westerveld A, Guillemot F, Oudejans C, et al. Hum Mol Genet. 1997;6:859–867. doi: 10.1093/hmg/6.6.859. [DOI] [PubMed] [Google Scholar]
- 32.Kalscheuer V M, Mariman E C, Schepens M T, Rehder H, Ropers H H. Nat Genet. 1993;5:74–78. doi: 10.1038/ng0993-74. [DOI] [PubMed] [Google Scholar]
- 33.Xu Y, Goodyer C G, Deal C, Polychronakos C. Biochem Biophys Res Commun. 1993;197:747–754. doi: 10.1006/bbrc.1993.2542. [DOI] [PubMed] [Google Scholar]
- 34.Jinno Y, Yun K, Nishiwaki K, Kubota T, Ogawa O, Reeve A E, Niikawa N. Nat Genet. 1994;6:305–309. doi: 10.1038/ng0394-305. [DOI] [PubMed] [Google Scholar]
- 35.Mitsuya K, Sui H, Meguro M, Kugoh H, Jinno Y, Niikawa N, Oshimura M. Hum Mol Genet. 1997;6:2243–2246. doi: 10.1093/hmg/6.13.2243. [DOI] [PubMed] [Google Scholar]
- 36.Yoshihashi H, Maeyama K, Kosaki R, Ogata T, Tsukahara M, Goto Y, Hata J, Matsuo N, Smith R J, Kosaki K. Am J Hum Genet. 2000;67:476–482. doi: 10.1086/302997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blagitko N, Mergenthaler S, Schulz U, Wollmann H A, Craigen W, Eggermann T, Ropers H H, Kalscheuer V M. Hum Mol Genet. 2000;9:1587–1595. doi: 10.1093/hmg/9.11.1587. [DOI] [PubMed] [Google Scholar]
- 38.Osoegawa K, Mammoser A G, Wu C, Frengen E, Zeng C, Catanese J J, de Jong P J. Genome Res. 2001;11:483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ledbetter D H, Engel E. Hum Mol Genet. 1995;4:1757–1764. doi: 10.1093/hmg/4.suppl_1.1757. [DOI] [PubMed] [Google Scholar]
- 40.Smit A F, Toth G, Riggs A D, Jurka J. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 41.Quentin Y. Genetica. 1994;93:203–215. doi: 10.1007/BF01435252. [DOI] [PubMed] [Google Scholar]
- 42.Britten R J, Stout D B, Davidson E H. Proc Natl Acad Sci USA. 1989;86:3718–3722. doi: 10.1073/pnas.86.10.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernardi G. Gene. 2000;241:3–17. doi: 10.1016/s0378-1119(99)00485-0. [DOI] [PubMed] [Google Scholar]
- 44.Schmid C W. Nucleic Acids Res. 1998;26:4541–4550. doi: 10.1093/nar/26.20.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jurka J, Zietkiewicz E, Labuda D. Nucleic Acids Res. 1995;23:170–175. doi: 10.1093/nar/23.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chai J-H, Locke D P, Ohta T, Greally J M, Nicholls R D. Mamm Genome. 2001;12:813–821. doi: 10.1007/s00335-001-2083-1. [DOI] [PubMed] [Google Scholar]
- 47.Jurka J. Proc Natl Acad Sci USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailey J A, Carrel L, Chakravarti A, Eichler E E. Proc Natl Acad Sci USA. 2000;97:6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsang P, Gilles F, Yuan L, Kuo Y-H, Lupu F, Samara G, Moosikasuwan J, Goye A, Zelenetz A D, Selleri L, et al. Hum Mol Genet. 1995;4:1499–1507. doi: 10.1093/hmg/4.9.1499. [DOI] [PubMed] [Google Scholar]
- 50.Lee M P, Feinberg A P. Cancer Res. 1998;58:1052–1056. [PubMed] [Google Scholar]
- 51.Lossie, A. C., Whitney, M. M., Amidon, D., Chen, P., Theriaque, D., Sage, B. T., Hutson, A., Nicholls, R. D., Zori, R. T., Williams, C. A. & Driscoll, D. J. (2001) J. Med. Genet. 28, in press. [DOI] [PMC free article] [PubMed]
- 52.Eichler E E. Trends Genet. 2001;17:661–669. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- 53.Hamdi H, Nishio H, Zielinski R, Dugaiczyk A. J Mol Biol. 1999;289:861–871. doi: 10.1006/jmbi.1999.2797. [DOI] [PubMed] [Google Scholar]
- 54.Gilbert N, Labuda D. J Mol Biol. 2000;298:365–377. doi: 10.1006/jmbi.2000.3695. [DOI] [PubMed] [Google Scholar]
- 55.Batzer M A, Deininger P L, Hellmann-Blumberg U, Jurka J, Labuda D, Rubin C M, Schmid C W, Zietkiewicz E, Zuckerkandl E. J Mol Evol. 1996;42:3–6. doi: 10.1007/BF00163204. [DOI] [PubMed] [Google Scholar]
- 56.Yoder J A, Walsh C P, Bestor T H. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 57.Rubin C M, VandeVoort C A, Teplitz R L, Schmid C W. Nucleic Acids Res. 1994;22:5121–5127. doi: 10.1093/nar/22.23.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hasse A, Schulz W A. J Biol Chem. 1994;269:1821–1826. [PubMed] [Google Scholar]
- 59.Yates P A, Burman R W, Mummaneni P, Krussel S, Turker M S. J Biol Chem. 1999;274:36357–36361. doi: 10.1074/jbc.274.51.36357. [DOI] [PubMed] [Google Scholar]
- 60.Arnaud P, Goubely C, Pelissier T, Deragon J M. Mol Cell Biol. 2000;20:3434–3441. doi: 10.1128/mcb.20.10.3434-3441.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cattanach B M. J Embryol Exp Morphol. 1986;97:137–150. [PubMed] [Google Scholar]
- 62.Jaenisch R, Beard C, Lee J, Marahrens Y, Panning B. Novartis Found Symp. 1998;214:200–213. doi: 10.1002/9780470515501.ch12. [DOI] [PubMed] [Google Scholar]
- 63.Boissinot S, Entezam A, Furano A V. Mol Biol Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- 64.Okada N, Hamada M, Ogiwara I, Ohshima K. Gene. 1997;205:229–243. doi: 10.1016/s0378-1119(97)00409-5. [DOI] [PubMed] [Google Scholar]
- 65.Gilbert N, Labuda D. Proc Natl Acad Sci USA. 1999;96:2869–2874. doi: 10.1073/pnas.96.6.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reik W, Walter J. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 67.Smit A F. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 68.Antequera F, Bird A. Proc Natl Acad Sci USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.