Abstract
To study HIV-1 escape from a coreceptor antagonist, the R5 primary isolate CC1/85 was passaged in peripheral blood mononuclear cells with increasing concentrations of the CCR5-specific small molecule inhibitor, AD101. By 19 passages, an escape mutant emerged with a >20,000-fold resistance to AD101. This virus was cross-resistant to a related inhibitor, SCH-C, and partially resistant to RANTES but still sensitive to CCR5-specific mAbs. The resistant phenotype was stable; the mutant virus retained AD101 resistance during nine additional passages of culture in the absence of inhibitor. Replication of the escape mutant in peripheral blood mononuclear cells completely depended on CCR5 expression and did not occur in cells from CCR5-Δ32 homozygous individuals. The escape mutant was unable to use CXCR4 or any other tested coreceptor to enter transfected cells. Acquisition of CXCR4 use is not the dominant in vitro escape pathway for a small molecule CCR5 entry inhibitor. Instead, HIV-1 acquires the ability to use CCR5 despite the inhibitor, first by requiring lower levels of CCR5 for entry and then probably by using the drug-bound form of the receptor.
A new generation of antiviral compounds, collectively termed entry inhibitors, is presently undergoing active preclinical and clinical development as potential therapies for HIV-1 infection (1, 2). These inhibitors include the gp41-targeted peptides T20 and T1249, the gp120-targeted recombinant protein CD4-IgG2, the chemokine derivative AOP-RANTES, and several small molecules, peptides, and mAbs specific for the chemokine receptors CXCR4 and CCR5 (reviewed in refs. 1 and 2). The latter proteins act as coreceptors with CD4 during the process of HIV-1 entry (3). If the in vitro potency of the entry inhibitors can be translated successfully into clinically useful drugs, then a new group of compounds to combat HIV-1 infection would become available to supplement the existing protease and reverse transcriptase inhibitors (1, 2).
There are many hurdles to overcome in the clinical development of any compound that shows activity against HIV-1 replication in vitro. As well as the traditional issues of toxicology and pharmacology, a problem common to all HIV-1 inhibitors is the rapid development of drug resistance both in vitro and in vivo. Clinical experience has taught that HIV-1 will always mutate to escape from the selection pressure of any one inhibitor (4). Given sufficient time, it is likely that the virus also will escape from combinations of inhibitors, particularly if therapy is suboptimal. Therefore it is prudent to study the escape pathways that are adopted by HIV-1 in vitro to gain an understanding of what might happen when the same inhibitor is used clinically.
The issue of escape pathways is of particular importance with inhibitors of HIV-1 entry via CCR5 because of a well documented facet of HIV-1 pathogenesis. Almost all cases of HIV-1 transmission involve strains that use CCR5 for entry (R5 viruses); these viruses persist throughout the course of HIV-1 infection in most infected people and are pathogenic. However, in up to 50% of infected people after 5 years, on average, viruses that are able to use CXCR4 become predominant (R5X4 or X4 viruses). These strains, also known as syncytium-inducing (SI) viruses, are associated with a more rapid disease course exemplified by an accelerated rate of CD4+ T cell loss (reviewed in ref. 5). This loss may be because the ability to use CXCR4 allows the virus to better target naive CD4+ T cells and/or more effectively inhibit T cell production (6, 7).
Because of the ability of R5 viruses to undergo phenotypic evolution to acquire CXCR4 usage, there are concerns that blocking CCR5 with a specific inhibitor in vivo might force HIV-1 to evolve to use CXCR4 instead (8). This outcome would be undesirable. We therefore conducted in vitro experiments to characterize the escape pathways used by HIV-1 when replicating in peripheral blood mononuclear cells (PBMCs) under the selection pressure of a CCR5-specific small molecule inhibitor, AD101. We used an R5 virus isolate (HIV-1 CC1/85) that we knew to be capable of undergoing phenotypic evolution to CXCR4 usage. We found that the AD101 escape mutant of this virus did not use CXCR4 but instead gained the ability to use CCR5 in an AD101-insensitive manner.
Materials and Methods
Viruses and Other Reagents.
Mitogen-activated PBMCs were prepared, and CD4+ T cells were isolated and maintained as described (9), as were HeLa-CD4-CCR5 cells from D. Kabat (Oregon Health Sciences University, Portland, OR; ref. 10). GHOST-coreceptor cell lines were obtained from D. Littman (New York University, New York) and maintained as described (11). HIV-1 CC1/85 and CC2/86 isolates were from R. Connor (Aaron Diamond AIDS Research Center, New York; ref. 12). Stocks of isolates NL4-3, DH123, 92US657, DJ258, JR-CSF, and 94ZW103 were prepared as described (9). RANTES was from PeproTech (Rocky Hill, NJ), the anti-CCR5 mAb 2D7 was from PharMingen (13), and the anti-CCR5 mAb PA14 was from W. Olson (Progenics, Tarrytown, NY; ref. 14).
Generation of AD101 Escape Mutant.
HIV-1 CC1/85 (1,000 tissue culture 50% infective doses per ml) was added to 20 ml of mitogen-activated PBMCs (2 × 106/ml) with sufficient AD101 to cause >90% inhibition. Control cultures lacked AD101 but otherwise were maintained identically to the AD101-containing cultures. The cultures were passaged weekly by adding a 5-ml aliquot of supernatant and cells from each culture to 15 ml of freshly activated PBMCs, maintaining a constant density of cells throughout the experiment. On day 4 postpassage, AD101 was added to the indicated final concentration. At each passage, p24 antigen production was monitored to ensure that virus replication was occurring and to determine the extent of inhibition by AD101. Because PBMCs from a different donor were used at each passage, p24 production varies in both the AD101-treated and control cultures. The concentration of AD101 was escalated over sequential passages when viral replication began to increase in the AD101-treated cultures. At least once every 14 days culture supernatant was frozen for drug sensitivity and coreceptor utilization studies.
HIV-1 Replication.
Virus replication and the action of inhibitors were assayed in primary CD4+ T cells prepared by positive selection from freshly activated PBMCs (9); the results were comparable to those obtained by using PBMC cultures (data not shown). Individual data points within an assay were derived from duplicate wells, and the results presented represent an average of 3–6 assays. The p24 concentration in the culture supernatants was monitored as described (15) after 7 days of culture. Production of p24 antigen was calculated as a percentage of that produced in the absence of inhibitors. Assays using CCR5Δ32/Δ32 cells were performed as described above but by using PBMCs from normal or CCR5Δ32/Δ32 donors rather than CD4+ T cells. Each data point is derived from 3–9 wells, with the average value shown. Infection of GHOST indicator cells that stably express CD4 and different coreceptors and inducibly express green fluorescent protein after HIV-1 infection was performed as described (11) except that green fluorescent protein fluorescence in cell lysates prepared 3 days postinfection was measured by using a microplate fluorometer. By adding recombinant green fluorescent protein to uninfected cell lysates we found that fluorescence increases ≥5 relative fluorescence units (rfus) above background could be detected reproducibly.
Sequence Analysis of env Genes Cloned from CC1/85 and CC101.19.
Full length env genes were generated from PCR products amplified from genomic DNA purified from PBMCs infected for 4 days with the CC1/85 and CC101.19 isolates. The env genes were amplified with Pfu DNA polymerase (Stratagene) with the primers EnvF (5′-AGCAGAAGACAGTGGCAATGAGAGTGAAG-3′) and EnvR (5′-TTTTGACCACTTGCCACCCATCTTATAGC-3′). The PCR products were subcloned into pBluescriptII KS(+) (Stratagene), and individual clones were sequenced. The consensus sequences (defined as greater than 50% amino acid identity) and alignments were produced by using MACVECTOR (Oxford Molecular, UK). The predicted gp120 amino acid sequences from the available clones are published as supporting information on the PNAS web site, www.pnas.org.
Infection of HeLa-CD4-CCR5 Cells.
The single-round focal infectivity assay was performed as described (10) except that the primary antibody was an Ig fraction from HIV-1+ human sera (from J. Mascola, National Institutes of Health) at 17 μg/ml, and the secondary antibody was horseradish peroxidase-conjugated goat anti-human Ig (BioSource International, Camarillo, CA). Foci of infection were normalized to the counts derived from high CCR5 cells (clone RC.25) in the same assay. No stained cells were observed in the parental HeLa-CD4 cell line (clone HI-R).
Results
AD101: A Small Molecule Inhibitor of HIV-1 Entry via CCR5.
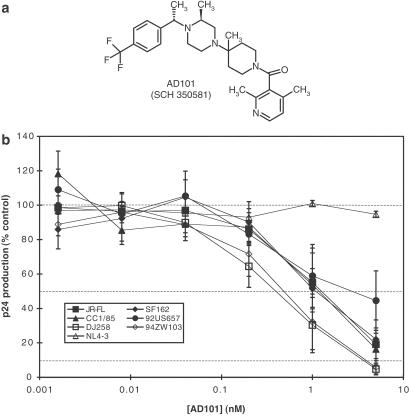
SCH-C, a small molecule CCR5 antagonist that is an effective inhibitor of HIV-1 replication in vitro, is described elsewhere (16). SCH-C is now being evaluated in phase I clinical trials. Similar to SCH-C, AD101 is a CCR5 antagonist that inhibits chemokine binding to the receptor and has potent activity against a broad range of R5 HIV-1 isolates from different genetic subtypes (Fig. 1 a and b; data not shown). The availability of AD101 early in the SCH-C development program dictated its choice for initial studies of CCR5 inhibitor escape pathways.
Figure 1.
AD101: A small molecule inhibitor of HIV-1 entry via CCR5. (a) Structure of AD101. (b) Ability of AD101 to inhibit HIV-1 replication in CD4+ T cells. The extent of virus replication is monitored by measuring p24 antigen production and is expressed as the percentage of that produced in the absence of inhibitor. Each data point is the mean (±SEM) of five experiments. The viruses used were: JRFL, SF162, CC1/85, and 92US657 (R5, subtype B); DJ258 (R5, subtype A); 94ZW103 (R5, subtype C); and NL4–3 (X4, subtype B). The mean IC50 value for the R5 isolates was ≈1.5 nM with a range of 0.53–3.5 nM.
An AD101 Escape Mutant Can Be Generated in a PBMC Culture.
In designing the escape mutant experiment we wished to mimic as closely as possible conditions relevant to the use of a CCR5 inhibitor in vivo. We therefore elected to use mitogen-stimulated PBMCs as the target cells as opposed to a CCR5-expressing cell line. We also chose to use a genetically heterogeneous HIV-1 primary isolate rather than a molecular or biological clone to allow escape mutants to be selected from a quasi-species pool as well as to be generated de novo. Moreover, we selected a primary R5 isolate, HIV-1 CC1/85 (isolated from patient case C in January 1985). At that time case C had a CD4 count of >1,000 cells per mm3 and no detectable R5X4 or X4 viruses. His viruses had the potential to evolve to use CXCR4; the February 1986 isolate from the same individual (CC2/86) had an R5X4 or X4 phenotype, although the patient's CD4 counts remained high. By July 1986, his CD4 counts had fallen to <500 cells per mm3 and then declined rapidly. Viruses with R5X4 or X4 phenotypes persisted in case C until his death from AIDS in 1989 (12).
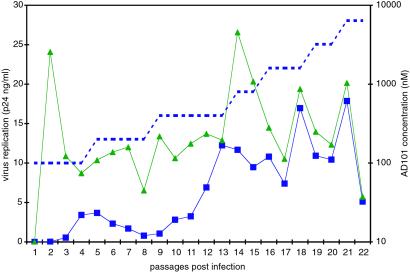
HIV-1 CC1/85 was chronically exposed to a CCR5 inhibitor by culturing it for 22 weekly passages in the presence of increasing AD101 concentrations. As a control, CC1/85 was passaged under the same conditions but without AD101 to allow us to monitor changes in the virus occurring spontaneously during prolonged PBMC passage. The p24 concentration in the culture supernatants was monitored with each passage. After six passages, partial (≈3-fold) resistance to AD101 was noted in the supernatants harvested from the AD101-treated culture, and thus the AD101 concentration was doubled. This treatment suppressed the transient peak in replication that had started after four passages with the original AD101 concentration (Fig. 2). After 16 passages, p24 production rates in the control and AD101-treated cultures were comparable. After this point, replication in the treated culture could not be suppressed further by increasing the AD101 concentration, indicating that CC1/85 had escaped from AD101 inhibition (Fig. 2). The viral supernatant from passage 19 of the AD101-treated culture, designated CC101.19, was chosen for further study, as was the supernatant from the control culture from the same time point, designated CCcon.19. Because partial resistance was apparent at earlier time points, a virus from the six-passage culture also was studied further along with its time-matched control isolate (designated CC101.6 and CCcon.6, respectively). AD101 dose escalation was terminated after 22 passages. The final isolate, CC101.22, was cultured for nine passages in the absence of AD101, yielding isolate CC101.22R9, to determine whether it had a stable phenotype or had reverted to AD101 sensitivity (see below).
Figure 2.
An escape mutant for AD101. HIV-1 CC1/85 was passaged weekly by using freshly activated PBMCs with (blue squares) or without (green triangles) the increasing concentrations of AD101 shown by the dotted line. The extent of virus replication (p24 antigen production) at each time point is shown. When replication in the control and AD101-containing cultures is comparable and replication in the latter culture can no longer be suppressed by increasing the AD101 concentration, escape from AD101 has occurred.
Phenotypic Properties of the AD101 Escape Mutant.
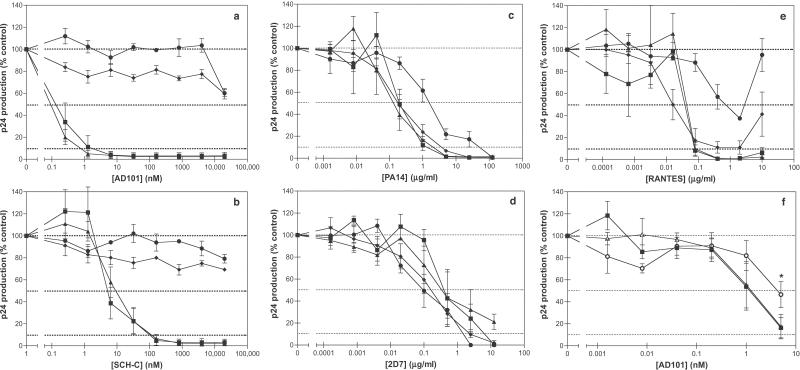
In CD4+ T cell and PBMC cultures (Fig. 3a; data not shown), the CC101.19 escape mutant was strongly (>20,000-fold) resistant to AD101, whereas CC101.6 showed partial resistance (≈3-fold; Fig. 3f), and CCcon.6 and CCcon.19 had the same sensitivity as the parental virus, CC1/85 (Fig. 3 a and f). The escape mutant phenotype was stable; the CC101.22R9 virus retained complete resistance to AD101 despite its culturing for nine passages without AD101 (Fig. 3a). The CC101.19 and CC101.22R9 viruses were cross-resistant to SCH-C, which is in the same chemical family as AD101 (Fig. 3b). The CC101.19, CCcon.19, and CC101.22R9 isolates also were less sensitive (10–20-fold) to the chemically unrelated small molecule CCR5 entry inhibitor, TAK-779 (ref. 17; data not shown). However, the parental isolate CC1/85 is only weakly inhibited by TAK-779 (IC50 = 370 nM), and all the above isolates are inhibited by >90% at TAK-779 concentrations of 25 μM (data not shown). Moreover, genetic drift in culture is a complicating factor, because the control CCcon.19 isolate also developed partial resistance to TAK-779. Thus, any cross-resistance to TAK-779 specifically generated by exposure to AD101 is marginal.
Figure 3.
Effect of CCR5 inhibitors on the AD101 escape mutant. The viruses CC1/85 (filled squares), CC101.19 (filled circles), CCcon.19 (filled triangles), and CC101.22R9 (filled diamonds) were tested for their sensitivity to AD101 (a and f), SCH-C (b), PA14 (c), 2D7 (d), and RANTES (e). (f) The isolates used were CC1/85 (filled squares), CC101.6 (open circles), and CCcon.6 (open triangles). The extent of virus replication is represented as a percentage of p24 antigen produced in the absence of any inhibitor. Each experiment was performed 3–6 times; the error bars represent the SEM. The asterisk in f indicates that the difference between the replication of CC101.6 and both CC1/85 and CCcon.6 is significant at an AD101 concentration of 5 nM, as indicated by a paired comparison t test (P < 0.05, n =5).
A modest decrease (≈10-fold) in the sensitivity of CC101.19 to the anti-CCR5 mAb, PA14, was noted. However, CC101.22R9 was as sensitive as CC1/85 and CCcon.19 to PA14, thus the partial resistance of CC101.19 to PA14 was not a stable phenotype (Fig. 3c). Both CC101.19 and CC101.22R9 retained the original sensitivity of CC1/85 to a second anti-CCR5 mAb, 2D7 (Fig. 3d). Thus, there may be subtle differences in how CC101.19 is inhibited by similar anti-CCR5 mAbs.
The CC101.19 escape mutant also was substantially cross-resistant (≈20-fold) to RANTES. However, CC101.22R9 had reverted such that its sensitivity to RANTES was comparable to CC1/85 and CCcon.19 (Fig. 3e).
The AD101 Escape Mutant Is Unable to Use CXCR4.
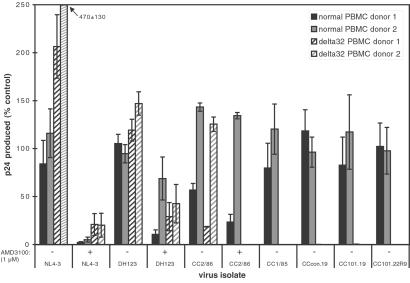
To determine whether the CC101.19 escape mutant had escaped from the AD101 selection pressure by acquiring the ability to use another coreceptor, we evaluated its ability to replicate in PBMCs that lack CCR5, i.e., cells from donors homozygous for the CCR5-Δ32 allele (18, 19). CC101.19, CCcon.19, CC101.22R9, or the parental isolate CC1/85 could replicate detectably in PBMCs from two different CCR5Δ32/Δ32 donors (Fig. 4). In contrast, the reference X4 and R5X4 viruses NL4-3 and DH123 replicated efficiently in the CCR5Δ32/Δ32 cells, as did CC2/86, a later isolate than CC1/85 from the same person (Fig. 4). Replication of CC2/86, NL4-3, and DH123 in the CCR5Δ32/Δ32 PBMCs was inhibited significantly by 1 μM AMD3100, a CXCR4-specific antagonist (ref. 20; Fig. 4). Thus, in marked contrast to the CXCR4 usage of the later in vivo variant CC2/86, replication of the CC101.19 in vitro escape mutant in PBMCs absolutely depends on CCR5 and cannot be supported by any other coreceptor expressed in these cells, including CXCR4.
Figure 4.
The AD101 escape mutant is unable to replicate in PBMCs lacking CCR5 expression. The bars represent the extent of replication of the indicated viruses in PBMCs from two donors with wild-type CCR5 alleles (solid bars) and two who are homozygous for the CCR5-Δ32 allele (hatched bars). Replication is expressed as a percentage of p24 antigen production by the same virus in control PBMC cultures. Three to nine wells were averaged from each donor for each data point; the error bars represent the SD. When 1 μM AMD3100 was added to cultures to inhibit entry via CXCR4, it is noted on the x axis.
We confirmed that CC101.19 and CC101.22R9 were unable to use CXCR4 by showing they could not replicate in GHOST-CXCR4 cells. In one experiment representative of three, green fluorescent protein fluorescence increases over the background level for uninfected GHOST-CCR5 cells (<5 rfus) was 65 rfus for CC1/85, 10 rfus for CC101.19, 7 rfus for CCcon.19, 31 rfus for CC101.22R9, and 64 rfus for CC2/86. None of the viruses caused a fluorescence increase in GHOST-CXCR4 cells except for CC2/86 (39 rfus). This infection was inhibited completely by the CXCR4 antagonist AMD3100 (1 μM). None of the viruses infected GHOST cells expressing CCR1, CCR2b, CCR3, CCR4, CCR8, CXCR6 (Bonzo), or BOB (<3 rfus) except CC2/86, which infected GHOST-CCR3 cells (7 rfus) as reported previously (12). Overall, we could find no evidence that CC101.19 or CC101.22R9 had acquired the ability to use any other coreceptor when evolving to escape from the selection pressure of AD101. We also confirmed that CC101.6 and CC101.19 still required CD4 for entry; their replication was fully inhibited by the anti-CD4 mAb RPA-T4 (data not shown).
Sequence Changes Associated with AD101 Resistance.
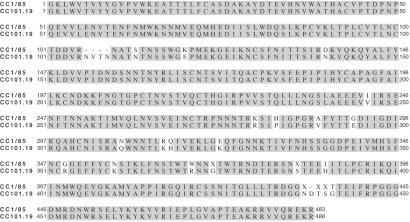
To evaluate what env sequence changes correlate with the AD101 resistance phenotype of CC101.19, eight full-length env genes from CC1/85 and six from CC101.19 were cloned and sequenced. An additional sequence of gp120 only was obtained also from each isolate. There was considerable diversity among the gp120 and gp41 coding regions of the CC1/85 clones (see the gp120 sequences, which are published as supporting information on the PNAS web site). Although there was slightly more diversity in the gp41 sequences of CC1/85 than of CC101.19, no readily discernable pattern of differences between the isolates was apparent. In contrast, the gp120 coding sequences of the CC101.19 clones showed little diversity, suggesting that the selection pressure acts on this Env subunit (S.E.K. and J.P.M., unpublished data). Alignment of the consensus amino acid sequences of the gp120 subunits of the two isolates reveals 24 differences; 22 single residue changes, one four-residue insertion in V1, and one single-residue insertion in V5. The V5 region, which is extremely diverse in the CC1/85 isolate, has little diversity in the CC101.19 clones (Fig. 5). When env genes from the CC101.19 clones were inserted into the NL4-3 provirus in the absence of any other changes, the chimeric, clonal viruses fully recapitulate the AD101 resistance of the CC101.19 isolate (S.E.K., F. Lee, and J.P.M., unpublished data).
Figure 5.
Comparison of consensus gp120 sequences from CC1/85 and CC101.19. The sequence alignment shows the consensus amino acid sequences from nine clones from the CC1/85 isolate (Upper) and seven clones from the CC101.19 isolate (Lower). Shaded amino acids are those which are identical between the two consensus sequences. Dashes indicate gaps in the consensus sequences, and Xs indicate amino acids where there was not >50% amino acid identity among the available clones.
The CC101.6 and CC101.19 Isolates Have an Increased Ability to Use Low Levels of CCR5.
One possible mechanism for CC1/85 to escape from AD101 is for the virus to develop an increased affinity for CCR5 and thus be able to “scavenge” low levels of inhibitor-free receptor. To test this theory we measured the ability of the parental (CC1/85 and CCcon.19) and escape (CC101.6 and CC101.19) isolates to infect HeLa-CD4-CCR5 clones expressing the same level of CD4 (10,000 molecules per cell) but different levels of CCR5 (10). The RC.10 clone of these cells expresses ≈7,100 CCR5 molecules per cell, and the RC.25 clone expresses ≈78,000 molecules per cell (10). As a reference virus, we used the C3 variant of HIV-1 JR-CSF, which was selected for its ability to replicate in the Molt4 T cell line and also in the SupT1 T cell line (21). The tropism expansion is the result of a single amino acid change in V1, which confers on C3 the ability to exploit the extremely low levels of CCR5 present on Molt4 and SupT1 cells, while retaining the original R5 phenotype of JR-CSF (21, 22).
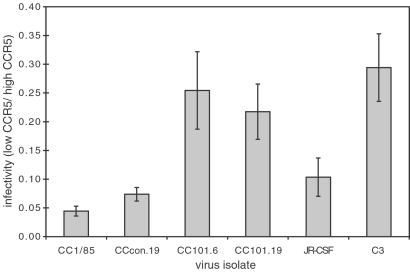
All the viruses infected the low CCR5 cells (RC.10) less efficiently than the high CCR5 cells (RC.25) in a single-round focal infectivity assay; the infectivity ratio for the two cell lines was always <1 (Fig. 6), as expected from previous studies (10). However, the C3 variant more efficiently exploited the low levels of CCR5 on RC.10 cells than did the parental JR-CSF isolate. Given the known properties of these viruses (21, 22), this observation validates the assay. Although all the tested isolates infected cells expressing high levels of CD4 and CCR5 to similar efficiencies (data not shown), the CC101.6 and CC101.19 isolates infected RC.25 cells with ≈10-fold lower efficiencies than did the CC1/85 and CCcon.19 isolates. The reason for this difference is not yet clear. However, at a constant CD4 concentration, both the early (CC101.6) and late (CC101.19) isolates used lower levels of CCR5 more efficiently than could the parental CC1/85 and CCcon.19 isolates (Fig. 6).
Figure 6.
CC101.6 and CC101.19 are better able to use low levels of CCR5. The test viruses were assayed for their ability to infect HeLa-CD4-CCR5 cells expressing high (clone RC.25, ≈78,000 CCR5 per cell) or low (clone RC.10, ≈7,100 CCR5 per cell) levels of CCR5 in a single-round focal infectivity assay. The number of infected cells was determined, and the ratio (low CCR5 cells/high CCR5 cells) was calculated (±SEM). The differences in the ratios between CC1/85 and CCcon.19 and between CC101.6 and CC101.19 were not significant. However, the differences between either of the isolates CC1/85 or CCcon.19 and the isolates CC101.6 or CC101.19 were highly significant using an unpaired t test (P < 0.03, n = 8). The difference between JR-CSF and C3 also was significant (P < 0.04, n = 4). The data shown are representative of one of three experiments.
Discussion
The principal observation made in this study is that the dominant pathway used by an HIV-1 isolate to escape from a CCR5-specific small molecule inhibitor, AD101, in vitro involves the continued use of CCR5 in an inhibitor-insensitive manner. This observation was made despite our use of PBMCs (which express CXCR4 and other potential coreceptors) as the target cells during selection and despite our choice of an HIV-1 strain that was capable of evolving the ability to use CXCR4, based on what occurred with this virus in vivo (12). Thus, although it can take only a few amino acid substitutions to convert an X4 virus into an R5 virus (23, 24) and CXCR4 was available on CD4+ T cells in the PBMC culture, HIV-1 did not follow the seemingly simple pathway of switching to CXCR4 use under the conditions of our experiment. Instead, multiple mutations accumulated in gp120 over time, creating a virus that continued to use CCR5 but was almost completely insensitive to the selecting inhibitor, AD101. This counterintuitive finding speaks to the overall efficiency gain for HIV-1 replication that is involved in continued CCR5 use, compared with a switch to use of CXCR4.
Our observations are not unique to the inhibitor and virus combination we used. An escape mutant with comparable properties was generated in response to a different small molecule CCR5 inhibitor, SCH-C, in a PBMC culture inoculated with a different R5 HIV-1 virus, the JR-FL molecular clone. Again, the SCH-C escape mutant did not switch to CXCR4 use but developed ≈50-fold resistance to SCH-C after 19 passages (S.X. and J.M.S., unpublished data). Moreover, in independent studies, the R5 HIV-1 molecular clone JR-CSF developed partial (5-fold) resistance to MIP-1α when cultured in a cell line expressing both CCR5 and CXCR4 but did not acquire CXCR4 use (25). Escape mutants with significant (>100-fold) resistance to the anti-CCR5 mAb 2D7 have been generated in both a molecular clone and a primary isolate (26). Again, coreceptor switching did not occur, although the cell line used for the selection process contains no known coreceptors other than CCR5 (26). In contrast, coreceptor switching to CXCR4 use was observed in a minority of human peripheral blood leukocyte severe combined immunodeficient mice after infection with JR-CSF in the prolonged presence of an N-terminally modified chemokine derivative, NNY-RANTES (8). Whether this reflects a specific property of this murine model is not yet known. It also may be relevant that RANTES is not only an inhibitor of R5 virus replication, but it also can actively enhance the replication of X4 viruses (27). Moreover, RANTES and its derivatives down-regulate CCR5 from cell surfaces (28), whereas AD101 and SCH-C do not (S.X., A.T., J.P.M., J.M.S., unpublished data). Hence any receptor switching in response to RANTES (or its derivatives) may represent a pathway unique to this class of compound. Overall, HIV-1 may respond differently to the selection pressure of different CCR5 inhibitors, particularly to receptor agonists such as AD101 and SCH-C and antagonists such as RANTES.
How can a CCR5 inhibitor escape mutant continue to use CCR5? We can imagine two mechanisms that are not mutually exclusive and may act sequentially. The first involves evolution of the CCR5 binding site on gp120 such that the mutant virus has a higher affinity for CCR5 and is better able to compete with AD101 or SCH-C. Under these conditions, the escape mutant would continue to use the low concentrations of free CCR5 receptors that are present despite the inhibitor, but it would do so more efficiently than the parental virus. There is indirect precedence; a single amino acid change in the V1 region of gp120 enables the R5 virus JR-CSF to exploit the nearly undetectable levels of CCR5 on the Molt4 T cell line, presumably by increasing the affinity of gp120 for CCR5 (21, 22). We have used this JR-CSF variant (C3) to validate a focal infectivity assay with HeLa-CD4-CCR5 clones expressing low and high levels of CCR5; both C3 and the CC101.6 and CC101.19 escape mutants can use low levels of CCR5 more efficiently than do their parental isolates (Fig. 6). Hence the first stage of the AD101 escape pathway may involve the acquisition of a higher CCR5 affinity associated with partial (≈3-fold) resistance, this process being complete within the first six passages. Alternatively, low levels of CCR5 may be better exploited by an escape mutant that had an increased rate of fusion once it formed the virus⋅receptor complex or required the presence of fewer CCR5 molecules in the virus⋅receptor complex for fusion to occur (29).
Between passages 6 and 19, there is no further increase in the ability of CC1/85 to use low levels of CCR5, but the escape mutant acquires full resistance to AD101. We believe this second mode of escape involves the creation of a substantially different binding site on gp120 for CCR5, such that the escape mutant can still use CCR5 even when the inhibitor is bound also to the receptor. This seems to be the mechanism by which the X4 virus HIV-1 NL4-3 escapes from the CXCR4 inhibitors AMD3100 and SDF-1α in cells that express CXCR4 but not CCR5 (30, 31). We strongly suspect that this is the dominant mechanism because of the magnitude of the eventual AD101 resistance (>20,000-fold), although we have not yet proven it.
Preliminary analyses of the sequences of viruses obtained at intermediate time points are consistent with the escape process proceeding by a two-step mechanism (S.E.K., J. Taylor, S. Wolinsky, and J.P.M., unpublished data). By passage six, 20 of the 24 differences between the consensus sequences shown in Fig. 5 dominate the gp120 sequences and apparently have been selected from among preexisting variants in the CC1/85 population. This selection is associated with partial (≈3-fold) resistance to AD101 (Fig. 3 b and f). Between passages 6 and 16, three additional single-residue changes occur in the V3 loop, probably de novo, and correlate temporally with the development of full (>20,000-fold) resistance (Fig. 3 b and f). Recently available chimeric clones of the escape viruses and specific CCR5 mutants will enable further studies of this process. If the later AD101 escape isolates do use the drug-bound form of CCR5 as a coreceptor, they also must be able to use the drug-free form efficiently, because they replicate both in the absence and presence of AD101 and SCH-C (Fig. 3 a and b).
It will be useful to determine whether the amino acid substitutions necessary to alter the gp120-CCR5 interaction in the escape mutant have created a virus with an altered sensitivity to virus-neutralizing antibodies, because these can target similar regions of the envelope glycoprotein complex (32). A CCR5 inhibitor escape mutant generated in vivo probably would not persist if it were unusually vulnerable to neutralizing antibodies.
We noted above that CCR5 inhibitors tend not to drive the phenotypic evolution of HIV-1 to CXCR4 use in vitro. A definitive answer to whether coreceptor switching in response to a CCR5 inhibitor will occur in vivo probably can be determined only by the conducting of small, carefully monitored human clinical trials or appropriately designed studies in certain animal models. However, it is worth considering why CXCR4 use is not favored in vitro and perhaps in vivo. There must be a selection pressure that limits the rate of phenotypic evolution from CCR5 to CXCR4 use in vivo. Only a few amino acid changes can convert an R5 virus to an X4 virus in vitro (23, 24), and this will happen daily given the known replication rate of HIV-1 in vivo (33). Yet X4 viruses are not abundant in many HIV-1-infected people throughout the course of disease (5). Furthermore, there are at least two examples of an early emergence of X4 viruses during primary infection followed by counterselection and the later dominance of R5 strains (34, 35). The nature of the selection pressure against X4 viruses is unresolved, but there is no convincing evidence that it is a product of acquired immunity (1). The demonstration that the burst size from HIV-1-infected primary cells is ≈10-fold greater for R5 than for X4 viruses may be highly relevant (36).
Unless a CCR5 inhibitor in some way interfered with the natural selection pressure against X4 viruses or unless there is competition, direct or otherwise, between R5 and X4 viruses for replication in a common niche in vivo, it is not obvious that suppressing the replication of R5 viruses must necessarily drive the rapid emergence of X4 viruses. Any X4 viruses that may be replicating already would presumably continue to do so in the face of a CCR5 inhibitor, but it is not clear that this would exacerbate the clinical situation that existed before the administration of the CCR5 inhibitor. Furthermore, the rate of generation of variants is a function of the extent of HIV-1 replication (33). A CCR5 inhibitor would always, in clinical practice, be combined with reverse transcriptase and protease inhibitors, further suppressing the generation of resistant viruses. Much more preclinical and clinical research needs to be performed before CCR5 inhibitors become part of the pharmacological repertoire against HIV-1 infection. Whether the evolution of HIV-1 to use CCR5 with a higher affinity, or in a different way, might have adverse consequences in vivo needs to be considered, but the present study argues against the notion that coreceptor switching to CXCR4 use must necessarily be a rapid consequence of the use of CCR5 inhibitors in HIV-1-infected people.
Supplementary Material
Acknowledgments
We thank Ruth Connor for providing HIV-1 isolates CC1/85 and CC2/86, Bill Olson for mAb PA14, Hau Truong and Fred Lee for technical assistance, and Jim Hoxie and Nelson Michael for CCR5Δ32/Δ32 blood. We are grateful to David Kabat for providing the HeLa-CD4-CCR5 cells, Paul Clapham and Robin Weiss for HIV-1 C3, and John Mascola for HIV-Ig. We thank Joann Taylor and Steve Wolinsky for preliminary sequencing data. This work was funded by RO1 AI41420, and National Institutes of Health Immunology Training Grant T32 AI07621 (to S.E.K.). J.P.M. is an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation, a Stavros S. Niarchos Scholar, and a consultant to Schering Plough. The Microbiology and Immunology Department at Weill Medical College gratefully acknowledges the support of the William Randolph Hearst Foundation.
Abbreviations
- PBMC
peripheral blood mononuclear cell
- rfu
relative fluorescence unit
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Michael N L, Moore J P. Nat Med. 1999;5:740–742. doi: 10.1038/10462. [DOI] [PubMed] [Google Scholar]
- 2.Moore J P, Stevenson M. Nat Rev Mol Cell Biol. 2000;1:40–49. doi: 10.1038/35036060. [DOI] [PubMed] [Google Scholar]
- 3.Berger E A. AIDS. 1997;11:S3–S16. [PubMed] [Google Scholar]
- 4.Perrin L, Telenti A. Science. 1998;280:1871–1873. doi: 10.1126/science.280.5371.1871. [DOI] [PubMed] [Google Scholar]
- 5.de Roda Husman A M, Schuitemaker H. Trends Microbiol. 1998;6:244–249. doi: 10.1016/s0966-842x(98)01249-9. [DOI] [PubMed] [Google Scholar]
- 6.Blaak H, van't Wout A B, Brouwer M, Hooibrink B, Hovenkamp E, Schuitemaker H. Proc Natl Acad Sci USA. 2000;97:1269–1274. doi: 10.1073/pnas.97.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maas J J, Gange S J, Schuitemaker H, Coutinho R A, van Leeuwen R, Margolick J B. AIDS. 2000;14:1155–1161. doi: 10.1097/00002030-200006160-00012. [DOI] [PubMed] [Google Scholar]
- 8.Mosier D E, Picchio G R, Gulizia R J, Sabbe R, Poignard P, Picard L, Offord R E, Thompson D A, Wilken J. J Virol. 1999;73:3544–3550. doi: 10.1128/jvi.73.5.3544-3550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trkola A, Paxton W A, Monard S P, Hoxie J A, Siani M A, Thompson D A, Wu L, Mackay C R, Horuk R, Moore J P. J Virol. 1998;72:396–404. doi: 10.1128/jvi.72.1.396-404.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mörner A, Björndal A, Albert J, KewalRamani V N, Littman D R, Inoue R, Thorstensson R, Fenyö E M, Björling E. J Virol. 1999;73:2343–2349. doi: 10.1128/jvi.73.3.2343-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L, LaRosa G, Kassam N, Gordon C J, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, et al. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson W C, Rabut G E, Nagashima K A, Tran D N, Anselma D J, Monard S P, Segal J P, Thompson D A, Kajumo F, Guo Y, et al. J Virol. 1999;73:4145–4155. doi: 10.1128/jvi.73.5.4145-4155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trkola A, Pomales A B, Yuan H, Korber B, Maddon P J, Allaway G P, Katinger H, Barbas C F, Burton D R, Ho D D, et al. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strizki J M, Xu S, Wagner N E, Wojcik L, Liu J, Hou Y, Endres M, Palani A, Shapiro S, Clader J W, et al. Proc Natl Acad Sci USA. 2001;98:12718–12723. doi: 10.1073/pnas.221375398. . (First Published October 16, 2001; 10.1073/pnas.221375398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, et al. Proc Natl Acad Sci USA. 1999;96:5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 19.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, et al. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 20.Schols D, Struyf S, Van Damme J, Esté J A, Henson G, De Clercq E. J Exp Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dejucq N, Simmons G, Clapham P R. J Virol. 1999;73:7842–7847. doi: 10.1128/jvi.73.9.7842-7847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fouchier R A, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng-Mayer C, Shioda T, Levy J A. J Virol. 1991;65:6931–6941. doi: 10.1128/jvi.65.12.6931-6941.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda Y, Foda M, Matsushita S, Harada S. J Virol. 2000;74:1787–1793. doi: 10.1128/jvi.74.4.1787-1793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aarons E J, Beddows S, Willingham T, Wu L, Koup R A. Virology. 2001;287:382–390. doi: 10.1006/viro.2001.1046. [DOI] [PubMed] [Google Scholar]
- 27.Gordon C J, Muesing M A, Proudfoot A E, Power C A, Moore J P, Trkola A. J Virol. 1999;73:684–694. doi: 10.1128/jvi.73.1.684-694.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mack M, Luckow B, Nelson P J, Cihak J, Simmons G, Clapham P R, Signoret N, Marsh M, Stangassinger M, Borlat F, et al. J Exp Med. 1998;187:1215–1224. doi: 10.1084/jem.187.8.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhmann S E, Platt E J, Kozak S L, Kabat D. J Virol. 2000;74:7005–7015. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Vreese K, Kofler-Mongold V, Leutgeb C, Weber V, Vermeire K, Schacht S, Anne J, de Clercq E, Datema R, Werner G. J Virol. 1996;70:689–696. doi: 10.1128/jvi.70.2.689-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schols D, Esté J A, Cabrera C, De Clercq E. J Virol. 1998;72:4032–4037. doi: 10.1128/jvi.72.5.4032-4037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parren P W, Moore J P, Burton D R, Sattentau Q J. AIDS. 1999;13:S137–S162. [PubMed] [Google Scholar]
- 33.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, et al. Nature (London) 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 34.Cornelissen M, Mulder-Kampinga G, Veenstra J, Zorgdrager F, Kuiken C, Hartman S, Dekker J, van der Hoek L, Sol C, Coutinho R, et al. J Virol. 1995;69:1810–1818. doi: 10.1128/jvi.69.3.1810-1818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lathey J L, Pratt R D, Spector S A. J Infect Dis. 1997;175:231–232. doi: 10.1093/infdis/175.1.231. [DOI] [PubMed] [Google Scholar]
- 36.Eckstein D A, Penn M L, Korin Y D, Scripture-Adams D D, Zack J A, Kreisberg J F, Roederer M, Sherman M P, Chin P S, Goldsmith M A. Immunity. 2001;15:671–682. doi: 10.1016/s1074-7613(01)00217-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.