Abstract
We have investigated the phylogenetic relationships among six wild and six domesticated taxa of Cucurbita using as a marker an intron region from the mitochondrial nad1 gene. Our study represents one of the first successful uses of a mtDNA gene in resolving inter- and intraspecific taxonomic relationships in Angiosperms and yields several important insights into the origins of domesticated Cucurbita. First, our data suggest at least six independent domestication events from distinct wild ancestors. Second, Cucurbita argyrosperma likely was domesticated from a wild Mexican gourd, Cucurbita sororia, probably in the same region of southwest Mexico that gave rise to maize. Third, the wild ancestor of Cucurbita moschata is still unknown, but mtDNA data combined with other sources of information suggest that it will probably be found in lowland northern South America. Fourth, Cucurbita andreana is supported as the wild progenitor of Cucurbita maxima, but humid lowland regions of Bolivia in addition to warmer temperate zones in South America from where C. andreana was originally described should possibly be considered as an area of origin for C. maxima. Fifth, our data support other molecular results that indicate two separate domestications in the Cucurbita pepo complex. The potential zone of domestication for one of the domesticated subspecies, C. pepo subsp. ovifera, includes eastern North America and should be extended to northeastern Mexico. The wild ancestor of the other domesticated subspecies, C. pepo subsp. pepo, is undiscovered but is closely related to C. pepo subsp. fraterna and possibly will be found in southern Mexico.
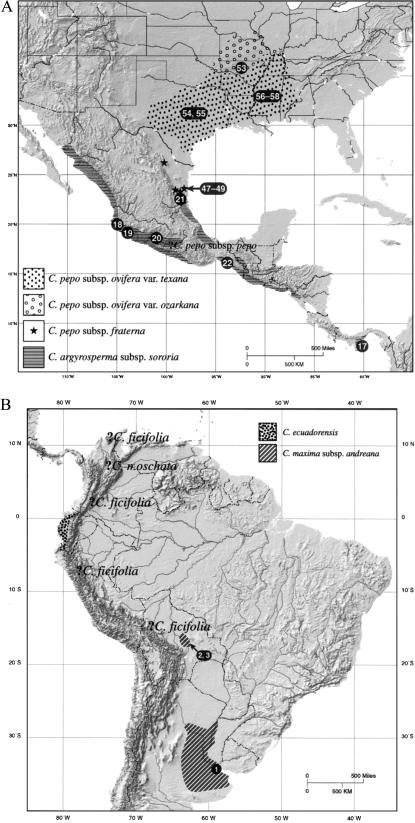
The New World genus Cucurbita (squashes, pumpkins, and yellow-flowered gourds) is composed of 12–14 species distributed from the U.S. to Argentina (1–3). At least five different species were domesticated before the European Contact, forming important sources of food in native American economies (1–4), and some of these species were among the earliest plants taken under cultivation and domesticated in the New World (Table 1) (5). Current genetic, biogeographical, and archaeological data suggest that the crop plants are not derived from a common ancestor; each species probably represents an independent domestication event. However, geographic areas of origin are not well defined for most of the crop species, and phylogenetic relationships with sympatric free-living taxa that represent possible wild progenitors are poorly understood (Fig. 1 A and B).
Table 1.
List of samples analyzed
| Individual no. | Species | Origin |
|---|---|---|
| 1–3 | C. maxima Duchesne subsp. andreana (Naud.) A.I. Filov | Argentina (1) and Bolivia (2, 3) |
| 4–10 | C. maxima | Ecuador (4–8) and U.S. (9, 10) |
| 11–16 | C. ecuadorensis Cutler & Whitaker | Ecuador |
| 17–22 | C. argyrosperma Huber subsp. sororia (L.H. Bailey) Merrick & Bates | Panama (17) and Mexico (18–22) |
| 23–24 | C. argyrosperma Huber | Mexico |
| 25–41 | C. moschata Duchesne | Puerto Rico (25), Panama (26, 31–32), Mexico (27), Colombia (28–29), Venezuela (30), Ecuador (33–36), Bolivia (37–41) |
| 42–46 | C. pepo L. subsp. pepo | Ecuador (42), Italy (43), U.S. (44), Hungary (45) |
| 47–49 | C. pepo subsp. fraterna (L.H. Bailey) Andres | Tamaulipas, Mexico |
| 50–52 | C. pepo subsp. ovifera (L.) Decker var. ovifera (L.) Decker | U.S. (acorn squash, striped pear gourd, bicolor gourd) |
| 53 | C. pepo subsp. ovifera (L.) var. ozarkana (Decker-Walters) | U.S. |
| 54–58 | C. pepo subsp. ovifera (L.) var. texana (Scheele) Decker | U.S. |
| 59–60 | C. martinezii L.H. Bailey | Mexico |
| 61–62 | C. foetidissima HBK | Mexico |
| 63–65 | C. ficifolia Bouché | U.S. (63), Mexico (64), Ecuador (65) |
| Citrullus lanatus (Thunb.) Matsum. & Nakai | Peru | |
| Sechium edule (Jacq.) Sw. | Panama |
Numbers in parentheses after countries of origin are individual numbers. Bold-printed species are domesticated.
Figure 1.
(A and B) Geographic range of putative wild ancestors of domesticated Cucurbita spp. in the U.S., Central America, and South America. Numbers on the maps are individual numbers for wild taxa listed in Table 1 and in supporting information on the PNAS web site (www.pnas.org). Question marks indicate potential areas of domestication for species that presently lack a wild ancestor.
The use of molecular markers and the deciphering of the genetically determined sequence of nucleotides in the DNA of domesticated plants and closely related wild taxa have significantly increased our understanding of crop plant evolution (6–9). In addition to providing the most accurate measure of relatedness between domesticated taxa and putative wild ancestors, these molecular systematic studies contribute essential information to archaeologists relating to the geography of plant domestication, and they serve as independent tests of hypotheses for agricultural origins posited from archaeobotanical and other kinds of research.
Previous molecular systematic work on the genus Cucurbita has been carried out by various investigators by using both nuclear and cytoplasmic markers. Wilson et al. (10) studied 15 species in the genus by using chloroplast restriction fragment length polymorphism analysis. Their results supported the separation of cultivars of Cucurbita pepo L. into two distinct lineages, C. pepo subsp. pepo and C. pepo subsp. ovifera (L.) Decker var. ovifera (L.) Decker, a division that was proposed on the basis of isozyme studies of these taxa (11, 12). The wild species C. pepo subsp. fraterna (L.H. Bailey) Andres and C. pepo subsp. ovifera var. texana (Scheele) Decker were shown by Wilson et al. to be closely aligned with cultivars of subsp. ovifera, but no wild taxon studied was considered a likely progenitor for subsp. pepo. Cucurbita ficifolia Bouché was placed as an outlier to other domesticated species of Cucurbita and was considered basal to the mesophytic species in the genus. This study could not resolve relationships among the closely related taxa Cucurbita moschata Duchesne, Cucurbita argyrosperma Huber, and C. argyrosperma subsp. sororia (L.H. Bailey) Merrick & Bates, possibly because these species have retained slow-evolving chloroplast DNA polymorphisms that were present in their ancestor, and the ancestry of Cucurbita maxima Duchesne with respect to the status of the associated free-living species C. maxima Duchesne subsp. andreana (Naud.) A.I. Filov was also left unresolved.
Jobst et al. (13) also addressed the question of evolutionary relationships in the Cucurbitaceae by using sequence data from the internal transcribed spacer nuclear region. Eleven Cucurbita species were included in this analysis. However, extensive allele sharing was observed among these species, possibly as a result of incomplete lineage sorting, which led to an inconclusive phylogenetic analysis. An inter simple sequence repeat analysis of multiple wild and domesticated plants in the C. pepo complex (14) resulted in a major division between samples of C. pepo subsp. pepo and subsp. ovifera, as reported in other studies. Cucurbita fraterna clustered with the specimens representing subsp. ovifera. A study of two different macrosatellite DNAs in nine different species of Cucurbita (15) did not result in well defined clades and thus could not provide significant phylogenetic data.
We report here the first use, to our knowledge, of a mitochondrial region to examine phylogenetic relationships among wild and domesticated plant taxa in the genus Cucurbita. Our marker displayed a surprising capacity to discriminate taxa at the specific and intraspecific levels.
Materials and Methods
Samples for this study came from four sources: (i) field collections made by various botanists, (ii) seeds obtained from the U.S. Department of Agriculture germplasm collection, (iii) specimens grown from seed by L.W.-B. at the plant breeding research station of the University of Puerto Rico in Isabela, Puerto Rico, and (iv) commercial markets [Table 1; see also supporting information on the PNAS web site (www.pnas.org) for more details on these accessions]. We analyzed a total of 65 individuals from six domesticated and six wild taxa of Cucurbita. To maximize geographic and taxonomic diversity, at least two different individuals from each species were sequenced. Only one species (C. foetidissima HBK) is considered xerophytic, whereas the rest are mesophytic. Two other members of the Cucurbitaceae, Citrullus lanatus (Thunb.) Matsum. & Nakai (watermelon) and Sechium edule (Jacq.) Sw (chayote), were used as outgroups.
We amplified and sequenced the entire mitochondrial nad1 dehydrogenase intron 2 region between exons B and C. This region is about 1.6 kb long. DNA was extracted from either leaves or seeds by using the Qiagen Miniprep Plant DNA extraction kit (Qiagen, Chatsworth, CA). PCR amplification of this region was performed by using the primers nad1 exon B and nad1 exon C (as described in ref. 16). Each 50-μl PCR reaction contained 2 units of Taq Polymerase (Applied Biosystems), 5 μl of 10× Taq buffer, 2 mM MgCl, 200 μM of each dNTP, 0.3 μM of each primer, and 15–20 ng of genomic DNA. Cycling conditions were: 94°C (4 min); 15 cycles of 92°C (45 sec), 45 + 5°C per cycle (30 sec), and 72°C (3 min); then 20 cycles of 92°C (45 sec), 50°C (30 sec), and 72°C (3 min), and finally 72°C (10 min). The PCR products were separated in a 1.5% low melting agarose gel; bands were isolated and purified by using the Qiaquick Gel Purification kit (Qiagen). DNA sequencing was performed by using the dRhodamine chemistry (Applied Biosystems) following the manufacturer's protocol and run in an automated DNA Sequencer (Applied Biosystems). To sequence the entire 1.6-kb region, we designed two internal primers, nad1-BF2 (5′-GGAGGCAAGAACCATGCTTTCA-3′) and nad1-BF3 (5′-GAAAGGGCTGTAGGTGATGGTG-3′). Sequences have been deposited in GenBank (accession nos. AF453584–AF453650).
Sequences were aligned by eye by using the program se-al Ver. 1.0 a1 (A. Rambaut, http://evolve.zoo.ox.ac.uk/software/Se-Al/Se-Al.html) [see supporting information on the PNAS web site (www.pnas.org) for alignment of the sequences). We used the program arlequin (ref. 17; http://lgb.unige.ch/arlequin) to perform the following tests: (i) linkage disequilibrium analyses to test for recombination across this region, (ii) analysis of molecular variance to determine genetic differentiation among populations and species, and (iii) to construct a minimum spanning tree. An additional test for recombination was performed by using the method described by Awadalla et al. (18). This method detects recombination by considering the correlation between linkage disequilibrium (expressed as r2) and distance. We used three measures of distance: distance including all sites, and distances excluding sites with alleles segregating at frequencies greater than 0.05 and 0.01.
We obtained the best evolutionary model of nucleotide substitution that fit the data by using the likelihood ratio test (program Model Test Ver. 3.0.6) (ref. 19). Results from these analyses showed that the best fit for our data was model F81 + I + G (ref. 20) (I, proportion of invariable sites; G, γ correction). The proportion of invariable sites (I) was determined to be 0.7898, and the γ distribution shape parameter (G) was 1.5447. All phylogenetic analyses were performed by using the program paup* 4.0b8 (ref. 21). We used distance, maximum parsimony (MP), and maximum likelihood (ML) criteria to analyze our data. We performed bootstrap analyses (under distance and MP criteria) and puzzling tests (under the ML criterion) to assess the robustness of the branches in the tree.
Results and Discussion
Molecular Variation of the nad1 Intron 2 Region.
Because of their low substitution rates (22) and high levels of gene rearrangements (23), plant mitochondrial genes have been regarded as less useful for systematic studies than animal mtDNA. However, the mitochondrial nad1 intron 2 has been successfully used in studying population structure and phylogenetic relationships among closely related taxa in conifers (24, 25). Our results showed that this mitochondrial region possesses sufficient sequence variation to distinguish among Cucurbita species. The intron sequence had conserved regions among all of the species studied, but it also showed a region (≈194 bp long) with high levels of polymorphism, mainly because of the presence of tandem repeats. This variable region was difficult to align among species and was excluded from the analysis. It is important to point out, however, that the tandem repeats within this region are potentially useful for the development of population markers. In addition to tandem repeats, this mitchondrial region also presented indels (1–11 bp long) that were coded as a fifth character for some of the phylogenetic analyses.
Results from the linkage disequilibrium test rejected the null hypothesis of no association among sites. Correlation analyses between linkage disequilibrium (LD) and distance showed that these values were not significantly different from zero, meaning that no apparent correlation was found between LD and distance. Therefore, no evidence of recombination was found. The analysis of molecular variance (AMOVA) analysis showed that 97.32% of the variation occurred among species, whereas 2.68% is because of intraspecific variation. AMOVA analysis thus supports the use of this mitochondrial marker to distinguish among species. The level of variation found within species also suggests that this intron region has the potential to be used as a marker for population studies in Angiosperms.
We found 16 distinct haplotypes among the 12 species and 65 individuals sequenced. To obtain a more clear picture of haplotype distribution among species, a minimum spanning tree was constructed by using all of the haplotypes [see supporting information on the haplotype tree on the PNAS web site (www.pnas.org)]. Each of the Cucurbita species had distinct haplotypes, in concordance with results from our analysis of molecular variance that showed high levels of variation between species. Domesticated/wild species pairs that have been proposed on the basis of isozyme, crossing studies, and other evidence (below) (e.g., C. maxima/C. andreana; C. argyrosperma/C. sororia) shared the same haplotypes, whereas domesticated species for which no wild progenitors have been proposed (e.g., C. ficifolia, C. moschata) had unique haplotypes. Of the 16 haplotypes defined, four occurred among individuals of C. sororia, three among individuals of C. moschata, and two among specimens of Cucurbita ecuadorensis Cutler & Whitaker thus demonstrating some level of population variation within species.
Phylogenetic Relationships Among Wild and Domesticated Cucurbita spp. and Crop Plant Evolution.
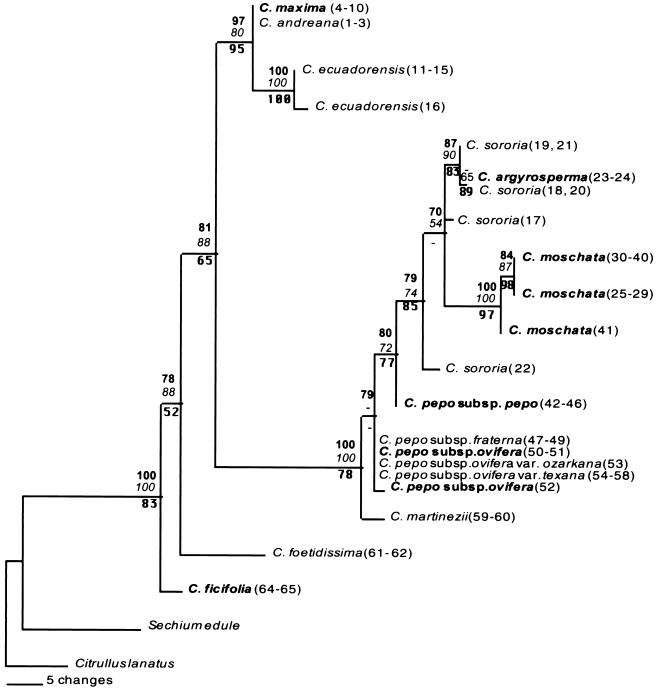
Analyses of the data under the three different phylogenetic criteria of distance, maximum parsimony, and ML showed no differences among the topologies of the trees obtained. Fig. 2 shows a ML tree for the 12 taxa of Cucurbita analyzed. Of the 1,530 characters used for the analyses, 1,423 were constant, and 75 were parsimony informative sites. Three polymorphic sites were homoplasious, meaning that the same mutational change occurred independently more than once in the tree. All of these sites involve single-base substitutions. The low level of homoplasy found in our data further indicates that recombination has not occurred in this mitochondrial region. Parsimony analyses using gaps as a fifth character and excluding gaps from the data set showed no differences among these trees, but the tree was two steps longer when using gaps as a fifth character (123 vs. 121 steps). Levels of sequence variation within each species estimated under the F81 model ranged between 0 and 0.5%, whereas sequence divergence between species ranged from 0 to 4% [see supporting information on the PNAS web site (www.pnas.org) for more information on genetic distances among species].
Figure 2.
ML tree for 11 species of Cucurbita. Individual numbers in parentheses are those listed in Table 1 and in supporting information on the PNAS web site (www.pnas.org). Taxa in bold print are domesticated species. Bootstrap values above the branches: parsimony in bold (100 bootstraps); distance in italics (1,000 bootstraps). Values below the branches represent ML steps (50,000 puzzle steps).
The mtDNA data provide important new insights into affinities between wild and domesticated taxa in Cucurbita, while leaving unresolved certain questions concerning phylogenetic relationships in the genus. When the new data are combined with existing molecular, botanical, and archaeological evidence concerning the origins of domesticated species within the genus, a clearer picture often emerges. The following relationships are suggested.
Cucurbita ? → C. ficifolia.
C. ficifolia (fig-leaf gourd; black-seeded squash), a domesticated species with a strong preference for cool, high-elevation ecological zones, was placed basal to other Cucurbita species in our analysis (Fig. 2). It has no associated wild ancestor in this or other studies. The position of this species in our tree is not robustly supported by the bootstrap-based confidence values, however. In the tree based on chloroplast restriction fragment length polymorphism data (10), the xerophytic, perennial species of Cucurbita (C. digitata A. Gray, C. foetidissima, and C. pedatifolia L.H. Bailey) occupy basal positions in the genus, whereas C. ficifolia is basal to all other mesophytic taxa. More work is needed to decipher the relationship of C. ficifolia to the xerophytic species in the genus, but here, as elsewhere (10), it has been shown to be an outlier and is probably basal to the other mesophytic Cucurbita.
C. ficifolia is often thought to be of Mesoamerican origin, but intensive searches for a wild progenitor in Mexico have not produced evidence of any such populations (2). The only reliable archaeological records come from South America (26), which we consider to be its probable area of origin.
C. andreana → C. maxima.
Specimens representing the other annual mesophytic species formed two well-defined clades in our analysis. One clade included three species of Cucurbita from South America, C. maxima, C. andreana, and C. ecuadorensis comprising one domesticated and two free-living species. This clade was supported with high bootstrap values, indicating a robust phylogenetic relationship (Fig. 2). C. maxima and C. andreana shared the same haplotypes (that is, we found no base pair differences in the sequences of these two species), supporting their assignment based on ecological and morphological evidence as a wild/domesticated species pair (1). It has been assumed that C. maxima's ancestor will be found among those fairly well-described populations of C. andreana that grow in temperate areas of Argentina (1). Fruits of free-growing C. andreana recently described from lowland central Bolivia by Michael Nee also share their haplotypes with C. maxima, perhaps extending the potential zone of domestication into this region. Historical information on the plant in Bolivia is limited, however, and thus the time depth and circumstances of its occurrence there (e.g., long-term wild species vs. a naturalized escape from cultivation) are not known. Currently available archaeological data cannot help us with this problem, because early sites in the pertinent regions are rare and poorly investigated. C. maxima was grown on the Peruvian coast by ≈4000 B.P. but apparently never left its continent of origin during the pre-Columbian era (4, 26).
Wild C. ecuadorensis ? → Semidomesticated C. ecuadorensis.
As its name implies, C. ecuadorensis is endemic to western Ecuador today. The plant is not typically counted among the domesticated species in the genus because its modern-day usage is limited and not well understood. There is considerable evidence, however, that, as first suggested by Nee (1), C. ecuadorensis was semidomesticated during the pre-Columbian era and then nearly lost from use. For example, some free-living populations have fruits with nonbitter flesh and larger sizes (≈11–13 cm) than are typical of wild gourds (1), fruit color and pattern vary considerably, and fruits with nonbitter flesh and sizes indicating a domesticated species (14 cm long × 18 cm in breadth) have recently been collected from house gardens and from slash-and-burn agricultural plots (T.C.A., unpublished data). Moreover, archaeological phytolith evidence from a preceramic (10,000–6600 B.P.) site in southwest Ecuador points to an ancient pattern of utilization and cultivation of the plant (27, 28).
There has been some question regarding the relationship of C. ecuadorensis to C. andreana and C. maxima (10). Our mtDNA data suggest that C. andreana has a haplotype that is ancestral to C. ecuadorensis, raising the possibility that C. maxima and C. ecuadorensis are both derived from C. andreana. Other existing lines of evidence support an interpretation that free-living populations of C. ecuadorensis from Ecuador gave rise to the semidomesticated forms of this species. First, the distribution of C. andreana, even with the new collections from central Bolivia, is decidedly southern South American and east of the Andes (Fig. 1B), and although the domesticated product of C. andreana, C. maxima, is recorded archaeologically and ethnohistorically in intervening areas, C. ecuadorensis is not (26). Second, intensive searches for a wild gourd in Ecuador have been unsuccessful, except for C. ecuadorensis, many populations of which exhibit all of the attributes of a wild species of Cucurbita (e.g., small fruits with bitter flesh and unvarying color and pattern) (ref. 1; T.C.A., unpublished data). Third, archaeological phytolith data from southwest Ecuador indicate the presence of a wild species of Cucurbita in contexts dated to between ≈11,000 B.P. and 10,000 B.P., before cultivated varieties are present; these phytoliths bear close size and morphological affinities to those from modern, small-fruited C. ecuadorensis but are unlike those from C. andreana (ref. 28; D.R.P., unpublished data).
C. pepo subsp. fraterna/C. pepo subsp. ovifera var. texana/C. pepo subsp. ovifera var. ozarkana → C. pepo subsp. ovifera.
A second well-defined clade in the mtDNA tree included wild and domesticated species of Cucurbita ranging from the U.S. to South America (e.g., C. pepo, C. sororia, C. argyrosperma, C. moschata) (Fig. 2). Of the taxa in this group, the C. pepo complex (pumpkins, zucchinis, acorns, crooknecks), which dominated pre-Columbian squash agriculture in the U.S. and cooler parts of Mexico, is probably the best known. How many times and in which regions of the Americas domestication occurred in C. pepo are important questions for archaeological and genetic research. Isozyme, inter simple sequence repeat, and chloroplast restriction fragment length polymorphism data (10–12, 14) all indicate that two lineages of domesticated taxa exist in the C. pepo complex, and that, whereas wild taxa constituting suitable wild progenitors are linked to one of these lineages (C. pepo subsp. ovifera), the Mexican lineage of cultivated squashes (subsp. pepo) lacks an associated wild ancestor. Our results are concordant with these analyses and further support an interpretation of two separate domestications in the species. C. pepo subsp. ovifera, C. pepo subsp. fraterna, C. pepo subsp. ovifera var. texana, and C. pepo subsp. ovifera var. ozarkana, consisting of one domesticated and three wild taxa (12), formed a group and shared the same haplotype (Fig. 2). The other domesticated species in the C. pepo complex, C. pepo subsp. pepo, showed a distinct mtDNA haplotype with no associated wild species.
Our results deviate in part from those of another study (12) regarding the identity and geographic location of the wild taxon ancestral to subsp. ovifera, the lineage of squashes considered by some scholars to have originated in eastern North America (12). On the basis of allozyme studies, Decker-Walters et al. proposed the classification of C. pepo subsp. fraterna, subsp. ovifera var. texana, and subsp. ovifera var. ozarkana as three distinct wild taxa, and argued that they had diverged long before the domestication of subsp. ovifera (12). In our analysis, all specimens of subsp. fraterna, subsp. ovifera var. texana, and subsp. ovifera var. ozarkana, together with two specimens of domesticated subsp. ovifera, shared identical mtDNA sequences. The other specimen of cultivated subsp. ovifera was tightly linked to these, differing by only 1 bp (Fig. 2). These factors suggest that revisions to the taxonomy of Decker-Walters et al. (12) are probably needed.
It was also proposed from the isozyme results that var. ozarkana was the wild progenitor of C. pepo subsp. ovifera (12). This conclusion was based on analyses of principal components and Nei's genetic identity values, but the principal component results indicated close genetic affinities between subsp. fraterna and domesticated subsp. ovifera (12). In addition, no statistical analyses were performed to test for the significance of Nei's identity values among the taxa studied. Some of the discordance between Decker's isozyme data and our mitochondrial data may also be explained by the fact that isozymes are nuclear markers; hence, postdomestication hybridization events and introgression of alleles among closely related populations can mask ancestral/derived relationships.
Our mtDNA data indicate that the three wild taxa—subsp. fraterna, subsp. ovifera var. texana, and subsp. ovifera var. ozarkana—should all be considered as candidates for the progenitor of subsp. ovifera, although we agree with Decker-Walters et al. (12) that the distinctive isozyme patterns in var. texana probably make it a less valid ancestor for subsp. ovifera than the other two. If true, the potential zone of domestication of C. pepo subsp. ovifera should be extended to northeastern Mexico, the natural habitat of C. fraterna (Fig. 1A).
Existing archaeological data do not help to resolve the question of where the domestication of subsp. ovifera occurred, because assigning ancient seed and peduncle specimens belonging to the C. pepo complex to subspecies on the basis of currently recognized physical attributes is often a difficult task. Seeds thought to be from domesticated C. pepo subsp. ovifera are known archaeologically by ≈4300 B.P. in a site from western Missouri located within the range of C. pepo subsp. ovifera var. ozarkana (29, 30). Archaeological sites in Tamaulipas state, northeastern Mexico, located less than 80 km from extant populations of C. pepo subsp. fraterna, have produced domesticated-type seeds and peduncles from unclassified subspecies of C. pepo dated to ≈5500 B.P. (31). Identification of the specimens from these sites should be pursued with alternative methods to determine whether they are from subsp. pepo or early varieties of subsp. ovifera. Additional studies using different molecular markers may help elucidate this question further.
Cucurbita ? → C. pepo subsp. pepo.
With regard to the other lineage of C. pepo, subsp. pepo, differences at only three adjacent base pairs (GAC) separate it from the subsp. ovifera group of taxa (TTA). A predomestication divergence of subsp. pepo's wild ancestor from that of subsp. ovifera is suggested by the fact that GAC is a derived character also present in the sororia and C. moschata groups. Therefore, it was probably present in the progenitor of subsp. pepo. Genetic evidence has often invalidated theories that plant taxa were domesticated more than once (6). Nevertheless, nuclear (11, 12, 14), chloroplast (10), and mtDNA data all appear to support such a scenario for C. pepo.
The close genetic relationship between C. pepo subsp. pepo and the Mexican wild species C. pepo subsp. fraterna indicated by our work may have importance in elucidating the wild ancestry of subsp. pepo. As Andres has, in part, suggested (32), it seems possible that past populations of subsp. fraterna, like today's, were small and semiisolated and somewhat genetically divergent, so that still extant but uncollected populations may represent the wild progenitors of C. pepo subsp. pepo. Existing specimens of C. fraterna from Tamaulipas and Nuevo Leon states, northeastern Mexico, derive from just four different populations, and they are completely interfertile with C. pepo subsp. pepo (1, 32).
Although any of the three wild taxa in the subsp. ovifera group are, on mtDNA grounds, equally likely to be closely related to the progenitor of subsp. pepo, geographical and archaeological information support the association advanced above between the Mexican wild taxon C. fraterna and subsp. pepo. For example, the earliest known remains of domesticated subsp. pepo occur at Guilá Naquitz Cave, a site in the Oaxaca Valley, southern Mexico, at ≈9000 B.P. (about 10,000 calendar years ago) (5), whereas domesticated forms of C. pepo are not reported in the United States until ≈4300 B.P. (29, 30). Additional collections of C. fraterna from northeastern Mexico, together with searches for this species in more southerly parts of Mexico, are sorely needed.
C. sororia → C. argyrosperma.
On the basis of isozyme, morphological, and ecological data, C. sororia has been classified as a subspecies and the presumed wild progenitor of C. argyrosperma (silverseeded gourd; green-striped cushaw) (1, 3, 33). C. argyrosperma and two of the C. sororia samples studied, from Michoacán (no. 20) and Jalisco (no. 18) states in Mexico, share the same haplotypes, strongly supporting this view. Southern Mexico is also supported as a likely area of domestication. We found four different haplotypes of C. sororia within the six individuals sequenced from six different regions of Central America ranging from Mexico to Panama, indicating that this species has high levels of diversity. Levels of sequence divergence reached 0.55% among C. sororia sequences from different localities, a level of variation comparable to that found between some species [e.g., C. andreana and C. ecuadorensis = 0.57%; see supporting information on genetic distances on the PNAS web site (www.pnas.org)].
Molecular systematic and isozyme studies have indicated that the Central Balsas River Valley of Mexico, including the region of Michoacán where our C. sororia sample (no. 20) analyzed here was collected, is the most probable origin locale for the creation of maize from its wild ancestor, teosinte (Zea mays subsp. parviglumis H.H. Iltis and Doebley), and that the region of Jalisco that provided our C. sororia sample (no. 18) is the second most likely hearth of maize (6). Hence, it is noteworthy that the four other C. sororia individuals in our study, three of which are from three other locales within Mexico and one from within Panama, are more distant from C. argyrosperma than are the Michoacán and Jalisco individuals. Maize and C. argyrosperma may have been taken under cultivation and domesticated together. At present, however, the earliest archaeological evidence for C. argyrosperma, dated to 4450 14C yr. B.P. (about 5,100 calendar years ago), comes from a cave in highland Tamaulipas, Mexico, which is considerably outside its area of origin (31). This situation highlights the paucity of archaeological research that has been carried out in the Balsas River Valley and other tropical regions of Mexico.
Cucurbita ? → C. moschata.
C. moschata (butternut squash; golden cushaw) is adapted to high temperatures and humidity, is the least cold tolerant of the domesticated plants and was probably brought under domestication somewhere in the humid tropical lowlands. That C. moschata has a greater affinity to the Sororia Group taxa than to other Cucurbita species has been previously argued from floral, seed, and ecological similarities, and the fact that these species are partly interfertile (3, 33). C. moschata formed a group with C. sororia and C. argyrosperma, supporting a close evolutionary relationship (Fig. 2). Our data suggest that C. moschata's wild progenitor was derived from a wild taxon closely related to C. sororia, but we agree with other investigators (1, 3, 33, 34) that this wild species is not a likely progenitor of C. moschata. There is no wild specimen with a closely aligned haplotype in this analysis, and previous isozyme and crossing studies indicated distinctive electrophoretic patterns and the presence of reproductive barriers between C. moschata and the Sororia Group taxa (e.g., hybridization is possible only with C. argyrosperma as the female parent) (3, 34).
We analyzed 16 individuals of C. moschata from a wide distributional range extending from Mexico to Bolivia (Table 1). The genetic variation found among these specimens may be a result of landrace divergence from a common ancestor, although more population-level work is needed on this question. Northern South America, in particular Colombia, have been identified as areas of high diversity for C. moschata, where landraces exhibiting primitive characteristics (e.g., very small and dark seeds, highly lignified and warty rinds, sometimes bitter fruits) are common (1, 35). An origin for this squash near the southern limit of C. sororia (Fig. 1B) would also make sense in view of the fact that this is the wild species most closely related to C. moschata. Present data, therefore, point to a single origin for C. moschata somewhere in northern lowland South America from a wild ancestor that has yet to be identified.
Conclusion
This research has demonstrated the utility of a mitochondrial gene to study evolutionary relationships among crop plants and closely related wild relatives. The mtDNA nad1 gene provided the clearest and most detailed phylogeny of the genus Cucurbita to date, and it appears to be the first mtDNA region to provide intraspecific-level variation in any Angiosperm. mtDNA has been widely used in molecular systematic studies of animals, and our findings suggest that a similarly rich source of information is potentially available from some economically important plants.
Our data support other lines of evidence in suggesting that six separate domestications occurred in the genus Cucurbita, and they add important new evidence bearing on the question of more than one domestication of C. pepo. Future studies using additional genetic markers should clarify the relations between some of the free-living and domesticated species considered here, and draw more restricted geographic boundaries around potential regions of crop domestication.
Supplementary Material
Acknowledgments
We thank Adam Eyre-Walker for his assistance with the linkage disequilibrium analysis. Michelle Waycott, Harilaos Lessios, Ken Olsen, Michael Nee, Patty Jo Watson, Anthony J. Ranere, and two reviewers provided many useful comments on the manuscript. They, however, are not responsible for its content. This research was supported by the Smithsonian Tropical Research Institute (STRI) and a STRI postdoctoral fellowship to O.S.
Abbreviation
- ML
maximum likelihood
Footnotes
References
- 1.Nee M. Econ Bot. 1990;44:56–68. [Google Scholar]
- 2.Andres T C. In: Biology and Utilization of the Cucurbitaceae. Bates DM, Robinson RW, Jeffrey C, editors. Ithaca, NY: Cornell Univ. Press; 1990. pp. 102–119. [Google Scholar]
- 3.Merrick L C. In: Evolution of Crop Plants. Smartt J, Simmonds N W, editors. Essex, U.K.: Longman; 1995. pp. 97–105. [Google Scholar]
- 4.Sauer, C. O. (1950) in Handbook of South American Indians, ed. Steward, J. (Bureau of American Ethnology Bulletin No. 143, U.S. Govt. Printing Office, Washington, DC), pp. 487–543.
- 5.Smith B D. Science. 1997;276:932–934. [Google Scholar]
- 6.Doebley J. Econ Bot. 1990;44:6–27. [Google Scholar]
- 7.Buckler E S, IV, Holtsford T P. Mol Biol Evol. 1996;13:612–622. doi: 10.1093/oxfordjournals.molbev.a025621. [DOI] [PubMed] [Google Scholar]
- 8.Olsen K M, Schaal B A. Proc Natl Acad Sci USA. 1999;96:5586–5591. doi: 10.1073/pnas.96.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsen K M, Schaal B A. Am J Bot. 2001;88:131–142. [PubMed] [Google Scholar]
- 10.Wilson H D, Doebley J, Duvall M. Theor Appl Genet. 1992;84:859–865. doi: 10.1007/BF00227397. [DOI] [PubMed] [Google Scholar]
- 11.Decker-Walters D S. Econ Bot. 1988;42:4–15. [Google Scholar]
- 12.Decker-Walters D S, Walters T W, Cowan C W, Smith B D. J Ethnobiol. 1993;13:55–72. [Google Scholar]
- 13.Jobst J, King K, Hemleben V. Mol Phylogenet Evol. 1998;9:204–219. doi: 10.1006/mpev.1997.0465. [DOI] [PubMed] [Google Scholar]
- 14.Katzir N, Tadmor, Tzuri G, Leshzeshen E, Mozes-Daube N, Danin-Poleg Y, Paris HS. Acta Hortic. 2000;510:433–439. [Google Scholar]
- 15.King K, Jobst J, Hemleben V. J Mol Evol. 1995;41:996–1005. doi: 10.1007/BF00173181. [DOI] [PubMed] [Google Scholar]
- 16.Demesure B, Doszi N, Petit R J. Mol Ecol. 1995;4:129–131. doi: 10.1111/j.1365-294x.1995.tb00201.x. [DOI] [PubMed] [Google Scholar]
- 17.Schneider, S. Roessli, D. & Excoffier, L. (2000) ARLEQUIN Ver. 2000. A Software Population Genetics Data Analysis (Genetics and Biometry Laboratory, University of Geneva, Geneva).
- 18.Awadalla P, Eyre-Walker A, Smith M J. Science. 1999;286:2524–2525. doi: 10.1126/science.286.5449.2524. [DOI] [PubMed] [Google Scholar]
- 19.Posada D, Crandall K A. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 21.Swofford D L. paup* Ver. 4.0b8. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 22.Wolfe K H, Li W-H, Sharp P M. Proc Natl Acad Sci USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer J D. In: Molecular Systematics of Plants. Soltis P S, Soltis D E, Doule J, editors. New York: Chapman & Hall; 1992. pp. 36–49. [Google Scholar]
- 24.Gugerli F, Senn J, Anzidei M, Madaghiele A, Büchler U, Sperinsen C, Vendramin G G. Mol Ecol. 2001;10:1489–1497. doi: 10.1046/j.1365-294x.2001.01285.x. [DOI] [PubMed] [Google Scholar]
- 25.Mitton J B, Kreiser B R, Latta R G. Mol Ecol. 2001;9:91–97. doi: 10.1046/j.1365-294x.2000.00840.x. [DOI] [PubMed] [Google Scholar]
- 26.Sauer J D. Historical Geography of Crop Plants: A Select Roster. London: CRC; 1993. [Google Scholar]
- 27.Piperno D R, Pearsall D M. The Origins of Agriculture in the Lowland Neotropics. San Diego: Academic; 1998. [Google Scholar]
- 28.Piperno D R, Andres T C, Stothert K E. J Arch Sci. 2000;27:193–208. [Google Scholar]
- 29.King F B. In: Prehistoric Food Production in Eastern North America. Ford R I, editor. University of Michigan, Ann Arbor, MI: Museum of Anthropology; 1985. pp. 73–97. [Google Scholar]
- 30.Fritz G. Am Antiquity. 1999;64:417–429. [Google Scholar]
- 31.Smith B D. Lat Am Ant. 1997;8:342–383. [Google Scholar]
- 32.Andres T C. Cucur Gen Coop Rep. 1987;10:69–71. [Google Scholar]
- 33.Merrick L C. In: Biology and Utilization of the Cucurbitaceae. Bates D M, Robinson R W, Jeffrey C, editors. Ithaca, NY: Cornell Univ. Press; 1990. pp. 77–95. [Google Scholar]
- 34.Wessel-Beaver L. Cucur Gen Coop Rep. 2000;23:62–63. [Google Scholar]
- 35.Wessel-Beaver L. Cucur Gen Coop Rep. 2000;23:54–55. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.