Abstract
Background
The related proteins Boi1 and Boi2, which appear to promote polarized growth in S. cerevisiae, both contain a PH (pleckstrin homology) and an SH3 (src homology 3) domain. Previously, we gained evidence that a PH domain-bearing segment of Boi1, which we call Boi1-PH, is sufficient and necessary for function. In the current study, we investigate the binding of Boi1's PH domain to the acidic phospholipids PIP2 (phosphatidylinositol-4,5-bisphosphate) and PS (phosphatidylserine).
Results
Boi1-PH co-sediments with PS vesicles. It does so more readily when these vesicles contain a small amount of PIP2. Boi1-PH is degraded in yeast extracts in a manner that is stimulated by PIP2. Amino-acid substitutions that diminish binding to PIP2 and PS impair Boi1 function. Fusion to a myristoyl group-accepting sequence improves to different degrees the ability of these different mutant versions of Boi1-PH to function. Boi1 and Boi2 are localized to the periphery of buds during much of the budding cycle and to necks late in the cell cycle. Amino-acid substitutions that diminish binding to PIP2 and PS impair localization of Boi1 to the bud, but do not affect the localization of Boi1 to the neck. Conversely, a mutation in the SH3 domain prevents the localization of Boi1 to the neck, but does not impair localization to the bud.
Conclusions
Boi1's PH domain binds to acidic phospholipids, and this binding appears to be important for Boi1 function. The main role of binding to PS may simply be to promote the association of the PH domain with membrane. The higher-affinity binding to PIP2, which apparently promotes a conformational change in the PH domain, may play an important additional role. Boi1 and Boi2 are localized to sites of polarized growth. Whereas the SH3 domain is needed for localization of Boi1 to the neck, the phospholipid-binding portion of the PH domain is important for localization to the bud.
Background
The Bem1-interacting proteins Boi1 and Boi2 were identified from screens for proteins that display two-hybrid interaction with the bud-emergence protein Bem1 [1,2], a protein that is important for the initiation of budding and that contains two SH3 domains. Boi1 and Boi2 are similar in sequence to one another. Each contains an SH3 domain; a SAM (sterile alpha motif) domain; a proline-rich region, which mediates binding to the second SH3 domain of Bem1; and a PH domain [1-3]. Figure 1 shows the relative positions of these sequence features in Boi1.
Figure 1.
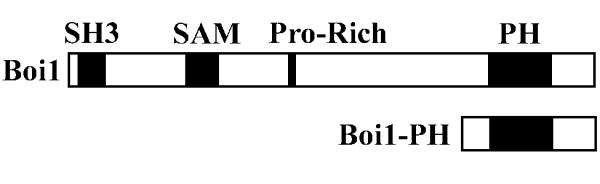
Positions of sequence features in Boi1 and Boi1-PH. Shown are the relative positions of the SH3 domain, SAM domain, proline-rich region, and PH domain. Full-length Boi1 is 980 amino acids long. Boi1-PH is the segment of Boi1 from amino acid position 734 through 980.
Deletion of either BOI1 or BOI2 alone has little effect on growth. However, deletion of both genes together is lethal (at least in some strains), suggesting that Boi1 and Boi2 perform at least one important, shared function [1,2].
The functions of Boi1/Boi2 appear to be linked to those of Cdc42 and Rho3, which are Rho-type GTPases that are important for polarized cell growth [4-9]. Boi1 and Boi2 display two-hybrid interactions with both wild-type and mutationally activated Cdc42 but not with a mutant version of Cdc42 that is predicted to be impaired in the ability to bind nucleotide, suggesting that Boi1 and Boi2 may associate with the GTP-bound ("active") form of Cdc42 [1]. Further evidence that some function of Boi1 is linked to that of Cdc42 is that overexpression of Boi1 inhibits bud emergence and that this inhibition can be suppressed by overexpression of Cdc42 [1,2]. RHO3 can serve as a multicopy suppressor of the lethality caused by deletion of BOI1 and BOI2 [1,2]. These findings are consistent with the possibilities that Boi1 and Boi2 are targets of Cdc42 that promote cell growth in a manner that is regulated by Rho3.
Support for the view that Boi1 and Boi2 are involved in polarized growth comes from the analysis of fission yeast Pob1, which is a homolog of Boi1 and Boi2. In particular, in S. pombe cells, Pob1 is localized to sites of polarized growth, loss of Pob1 function leads to a loss of polarized growth, and overexpression of Pob1 causes cell growth to become depolarized [10].
The PH domain appears to be a critical feature of Boi1: mutations in this domain destroy Boi1 function, and Boi1-PH (see Fig. 1) can substitute in function for Boi1 and Boi2 [1]. In addition, Boi1-PH contains the region of Boi1 that displays the two-hybrid interaction with Cdc42, raising the possibility that the PH domain, itself, mediates or regulates the association with Cdc42 [1].
A generally shared feature of PH domains is the ability to bind acidic phospholipids, usually one or more derived from PI (phosphatidylinositol) and, in some cases, also PS [11,12]. In some proteins, the main role of the binding of PH domains to phospholipids may simply be to promote association with membrane (i.e., to serve as a membrane-localization tag) [13,14]. In other proteins, the binding of PH domains to phospholipids appears to be important for allosteric or other types of regulation [15,16].
Given that 1) Boi1 and Boi2 are important proteins whose functions appear to be linked to those of Cdc42 and Rho3, 2) the PH domain appears to be of particular importance for Boi1 function, and 3) a general role of PH domains may be to bind acidic phospholipids, we wanted to know whether the PH domain of Boi1 binds acidic phospholipids and, if it does, whether this binding is important for the function and proper localization of Boi1. In the current study, we investigate these issues as a first step toward eludicating roles of Boi1's PH domain.
Results
Binding of Boi1-PH to phospholipids
In all binding analyses reported in this study, we used Boi1-PH (see Fig. 1) rather than full-length Boi1, because we wanted to focus on interactions mediated by the PH domain. Also, we needed to have a soluble pool of protein for vesicle co-sedimentation studies, and while most of the Boi1-PH was soluble (at least when overexpressed), virtually none of the full-length Boi1 was soluble (whether or not overexpressed).
Fortuitously, we discovered that Boi1-PH can be proteolyzed in yeast extracts in a manner that is stimulated by PIP2. Figure 2A shows an example of this phenomenon; whereas most of the anti-Boi1 immunoreactivity was present in a single band when Boi1-PH was incubated with control PS vesicles that lacked PIP2, almost all of the anti-Boi1 immunoreactivity was present in faster-migrating bands when Boi1-PH was incubated with PS vesicles that contained PIP2 (lanes 1 and 5). (In this and all subsequent experiments that use PIP2/PS mixed vesicles, the mass ratio of PIP2:PS was 1:20.) Less total anti-Boi1 immunoreactivity was recovered when Boi1-PH was incubated with PIP2/PS mixed vesicles than when incubated with vesicles containing only PS (lanes 1 and 5), suggesting that the faster-migrating species represent degradation products of Boi1-PH. Consistent with this view, although Boi1-PH remained stable when incubated for longer intervals with vesicles that contained only PS, the anti-Boi1 immunoreactive bands became fainter and then disappeared altogether when incubated for longer intervals with PIP2/PS mixed vesicles (data not shown).
Figure 2.
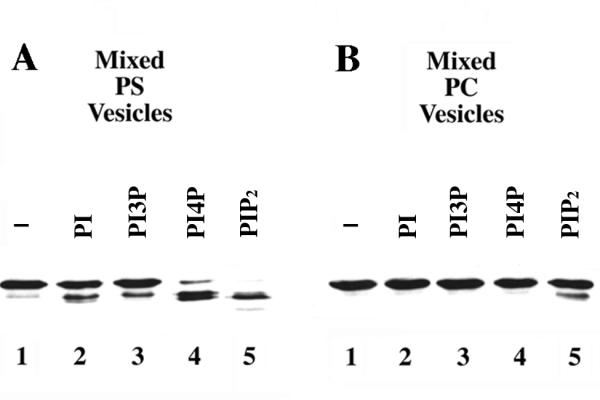
Effects of different phospholipids on the proteolysis of Boi1-PH. Shown are immunoblots, probed with anti-Boi1 antibodies, of aliquots from a Boi1-PH-bearing yeast extract that was incubated, in the presence of 2 mM CaCl2 (to allow proteolysis), with vesicles containing PS (part A) or PC (part B). The vesicles contained either no additional lipid (lanes 1) or a 1:20 (mass:mass) ratio (with respect to PS or PC) of PI (lanes 2), PI3P (lanes 3), PI4P (lanes 4), or PIP2 (lanes 5).
The finding that PIP2 can stimulate degradation of Boi1-PH suggests that PIP2 may bind to Boi1-PH in a manner that results in the exposure of a protease-sensitive site. We do not know the identity of the relevant protease. However, we found that the PIP2-stimulated proteolysis of Boi1-PH is strongly enhanced by Ca++ and is inhibited by EGTA (data not shown), suggesting that this protease may require Ca++ for activity.
Next, we asked whether proteolysis of Boi1-PH could also be stimulated by other inositol-based phospholipids. PI and PI3P (phosphatidylinositol-3-phosphate), presented in PS-based vesicles, appeared to stimulate slightly the proteolysis of Boi1-PH, but did so to a much lesser extent than did PIP2 (Fig. 2A, lanes 1–3 and 5). In contrast, PI4P (phosphatidylinositol-4-phosphate)/PS vesicles stimulated substantial proteolysis of Boi1-PH, although to a lesser extent than did PIP2/PS vesicles (Fig. 2A, lanes 4 and 5). These findings indicate that different phosphatidylinositides can stimulate proteolysis of Boi1-PH but differ greatly in their abilities to do so.
To investigate whether the nature of the bulk lipid in the vesicles affects the ability of phosphatidylinositides to stimulate proteolysis of Boi1-PH, we repeated this analysis using the neutral phospholipid PC (phosphatidylcholine) instead of PS. Using reaction conditions that were otherwise the same as those used when PS was the bulk lipid, no proteolysis of Boi1-PH was detected in the presence of any of the phosphatidylinositides except PIP2, which stimulated only a small amount of proteolysis (Fig. 2B). Thus, PS is much more effective than PC at facilitating the ability of phosphatidylinositides to stimulate proteolysis of Boi1-PH.
To investigate further whether Boi1-PH binds PIP2, we used a vesicle co-sedimentation assay. In this analysis, we asked whether an otherwise soluble fraction of Boi1-PH, after being incubated with PIP2-bearing vesicles, would now sediment in conditions (436,000 × g for 1 hr) that cause vesicles to pellet. (To inhibit the proteolysis of Boi1-PH, we performed this analysis in buffer that lacks Ca++ but that contains EGTA and two other protease inhibitors that were not present in the proteolysis assay.) First, we asked whether Boi1-PH could pellet with vesicles that lacked PIP2. We did not detect any pelleting of Boi1-PH with PC vesicles (data not shown), but found that Boi1-PH could pellet with vesicles composed of PS (Fig. 3, top panel), suggesting that Boi1-PH can bind PS.
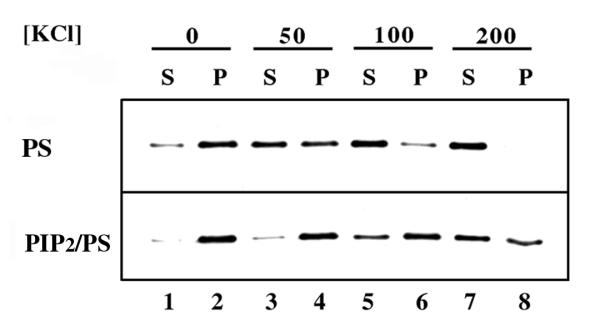
Figure 3.
Co-sedimentation of Boi1-PH with vesicles. The two panels show immunoblots, probed with anti-Boi1 antibodies, of supernatant (S) and pellet (P) fractions resulting from centrifugation of a mixture of Boi1-PH and vesicles composed solely of PS (top panel) or composed of a 1:20 (mass:mass) ratio of PIP2 and PS (bottom panel). For this analysis, prior to centrifugation, aliquots of a cytosolic preparation of yeast extract that contained Boi1-PH were incubated with vesicles in PBS buffer that had the following additional concentrations of KCl: 0 (lanes 1 and 2), 50 (lanes 3 and 4), 100 (lanes 5 and 6), and 200 mM (lanes 7 and 8).
Next, we asked whether the inclusion of PIP2 in the PS vesicles affected the ability of Boi1-PH to pellet. In the starting buffer conditions (phosphate-buffered saline without additional salt), a larger percentage of Boi1-PH pelleted when the vesicles contained PIP2 (Fig. 3, lanes 1 and 2). However, most of the Boi1-PH pelleted whether or not PIP2 was present. So, to try to accentuate the effects of PIP2 on Boi1-PH's co-sedimentation with vesicles, we repeated this analysis using buffer that contained additional concentrations of KCl. In the presence of 50, 100, and 200 mM additional KCl, the ratio of Boi1-PH that pelleted with respect to that which stayed in the supernatant was greater when using vesicles that contained PIP2 (Fig. 3, lanes 3–8), suggesting that Boi1-PH has a higher affinity for PIP2 than for PS.
In similar experiments using mixed PIP2/PC vesicles, all of the Boi1-PH stayed in the soluble fraction (data not shown), supporting the views that the composition of the lipid bilayer affects the ability of Boi1-PH to bind PIP2 and that PS promotes this association.
Mutant versions of Boi1-PH that are impaired in the ability to bind PIP2
To investigate whether binding to PIP2 may be important for Boi1 function, we sought to make mutant versions of Boi1-PH that are defective in the ability to bind PIP2. As a guide for designing such mutants, we used information about one of the PH domains from pleckstrin. Figure 4 shows those positions in the N-terminal PH domain of pleckstrin that have been implicated, based on NMR analysis, to contact PIP2[17]. Boi1's PH domain has identical or similar amino acids at most of the analogous positions (Fig. 4).
Figure 4.

Sequences from a PH domain of pleckstrin and from different versions of Boi1. Asterisks indicate those positions in a segment of pleckstrin's N-terminal PH domain that are implicated in binding to PIP2[17]. Of these positions, those that have the same amino acid at the corresponding position in Boi1 are indicated with dark shading, and those that contain a similar amino acid are indicated with light shading. The sequences of the KK, TK, and KKTK mutant versions of Boi1 are represented by indicating only those amino acids in the mutants that are different from those in the corresponding positions of the wild-type protein.
To obtain mutant versions of Boi1 that might be defective in the ability to bind PIP2, we created three sets of mutations that resulted in amino-acid subsitutions at some of those positions. We call these sets of substitutions "KK" (K785E and K786A), "TK" (T793A and K795E), and "KKTK" (K785E, K786A, T793A, and K795E; a combination of the KK and TK sets) (Fig. 4).
To investigate whether these substitutions affect the ability of Boi1-PH to bind PIP2, we first used the proteolysis assay. We found that, in conditions in which PIP2 stimulated proteolysis of most of the wild-type Boi1-PH, PIP2 did not stimulate the proteolysis of the TK, KK, and KKTK versions of Boi1-PH (Fig. 5A, lanes 3, 6, 9, and 12), suggesting that all three sets of substitutions may disrupt binding to PIP2 and/or PS.
Figure 5.
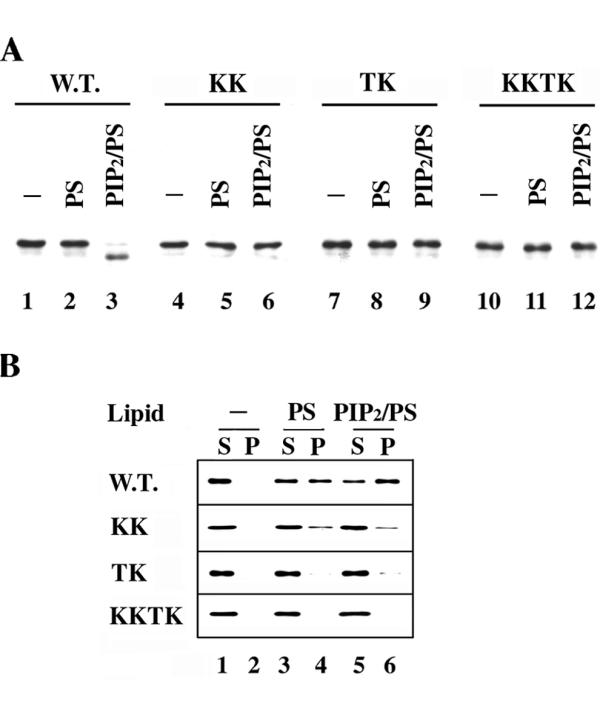
Effects of the KK, TK, and KKTK substitutions on the association of Boi-PH with phospholipids. A). Shown are immunoblots, probed with anti-Boi1 antibodies, of aliquots from yeast cytosolic extracts containing either wild-type or mutant versions of Boi1-PH, after incubation with either no vesicles (lanes 1, 4, 7, and 10), vesicles containing only PS (lanes 2, 5, 8, and 11), or vesicles composed of a 1:20 (mass:mass) mixture of PIP2 and PS (lanes 3, 6, 9, and 12). B). The four panels show immunoblots, probed with anti-Boi1 antibodies, of supernatant (S) and pellet (P) fractions, resulting after centrifugation, of yeast cytosolic extracts containing either wild-type Boi1-PH (top panel), Boi1-PH(KK) (second panel), Boi1-PH(TK) (third panel), or Boi1-PH(KKTK) (bottom panel), with either no vesicles (lanes 1 and 2), vesicles composed only of PS (lanes 3 and 4), or vesicles composed of a 1:20 (mass:mass) mixture of PIP2 and PS (lanes 5 and 6).
To investigate further whether the three sets of substitutions affect the ability of Boi1-PH to bind these lipids, we used the vesicle co-sedimentation assay. In the presence of vesicles composed of only PS, an appreciable amount of Boi1-PH(KK) pelleted (Fig. 5B, second panel, lanes 3 and 4). However, the percentage of Boi1-PH(KK) that pelleted was less than that for wild-type Boi1-PH (Fig. 5B, top and second panels, lanes 3 and 4), suggesting that the KK substitutions reduce somewhat the binding affinity for PS. The TK substitutions had a more obvious effect: no Boi1-PH(TK) was seen to pellet with PS vesicles (Fig. 5B, third panel, lanes 3 and 4), suggesting that the TK substitutions impaired binding to PS to a greater extent than did the KK substitutions. As expected, given that the TK substitutions were sufficient to severely affect pelleting with PS vesicles, none of the Boi1-PH(KKTK) was seen to co-sediment with PS vesicles (Fig. 5B, bottom panel, lanes 3 and 4).
Whereas inclusion of PIP2 in the vesicles increased the amount of wild-type Boi1-PH that co-sedimented, PIP2 did not have an obvious effect on the co-sedimentation behavior of Boi1-PH(KK) (Fig. 5B, top and second panels, lanes 3–6), suggesting that the KK substitutions impair binding to PIP2. In contrast, the TK mutant version of Boi1-PH appeared to retain a slight ability to pellet with PIP2/PS vesicles (Fig. 5B, third panel, lanes 3–6).
None of the Boi1-PH(KKTK) was detected in the pellet fraction even when using vesicles that contained PIP2 (Fig. 5A, bottom panel, lanes 5 and 6), consistent with the view that the KKTK mutant is more severely impaired in the ability to bind PIP2 than are the KK and TK mutants.
Exploring the significance of binding to phospholipids
To investigate whether binding to phospholipid may be important for Boi1 function, we used a red/white, colony-sectoring assay to test whether the KK, TK, and KKTK mutant versions of Boi1 could substitute in function for wild-type BOI1 and BOI2. For this analysis, we used strain PY967, which lacks the genomic copies of BOI1 and BOI2 and which is kept alive by BOI1-bearing plasmid pPB799. pPB799 also contains the ADE3 color marker. Cells that have this plasmid are red, while those that fail to inherit it are white. PY967 cells form thoroughly red colonies (lacking white sectors), because cells that fail to inherit the plasmid lack BOI1 and so do not propagate. However, when transformed with a second plasmid that can substitute in function for BOI1, PY967 cells that fail to inherit pPB799 are still viable and so give rise to white sectors.
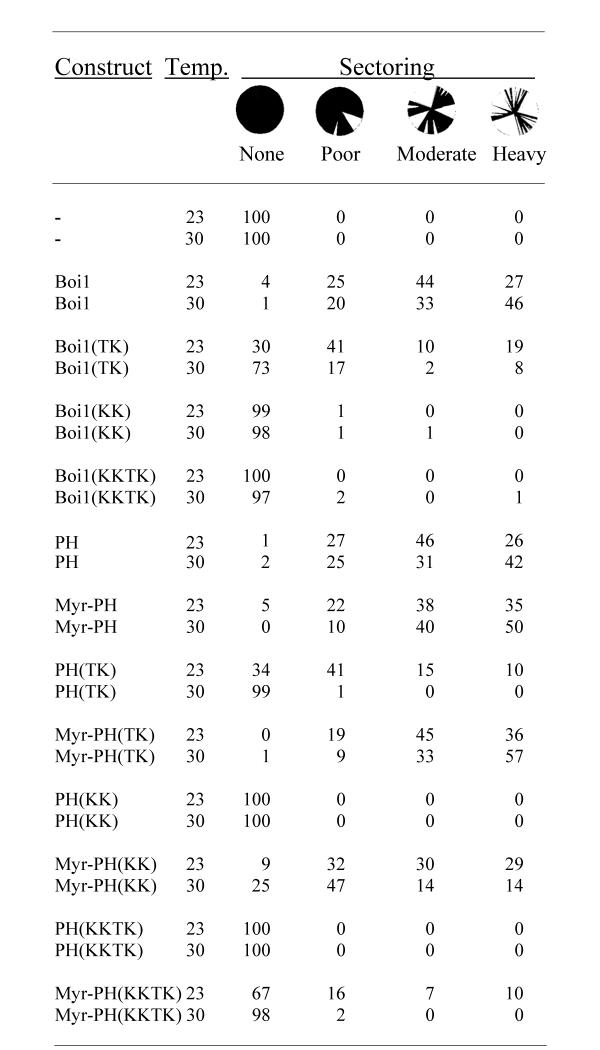
To test the different versions of BOI1 for function, we introduced them on plasmids into strain PY967, at both 23 and 30°C, and then scored the degree of sectoring of the resultant transformant colonies. For each plasmid tested, approximately 200 colonies from each transformation plate were analyzed at each temperature. As negative and positive controls for sectoring, we used an empty plasmid and one that contains wild-type BOI1, respectively. As indicated in Figure 6, the empty plasmid (-) gave no sectoring colonies, indicating that the level of background sectoring in this assay is low. The plasmid that contains wild-type BOI1 gave a range of sectoring abilities, with 71 and 79% of the colonies showing a moderate or heavy degree of sectoring at 23 and 30°C, respectively (Fig. 6).
Figure 6.
Colony-sectoring analysis to test the ability of different mutant versions of Boi1 and Boi1-PH to substitute in function for wild-type Boi1 and Boi2. Values are percentages of colonies that displayed each of the different degrees of sectoring: "none" = no sectoring (i.e., thoroughly red colony), "poor" = one or a few white sectors in an otherwise thoroughly red colony, "moderate" = more than a few white sectors, but where not more than approximately half of the colony contains sectors, and "heavy" = most or all of the colony contains white sectors. The circles illustrate these categories of sectoring, with black representing the red parts of colonies. In this analysis, boi1 boi2 strain PY967, which contains wild-type BOI1 and the color marker ADE3 on one plasmid (pPB799), was transformed with a second plasmid containing the indicated construct that was being tested for function. Transformation plates were incubated at 23 or 30°C, as indicated. The different sets of amino-acid substitutions are indicated in parentheses. "Boi1" = full-length Boi1; "PH" = Boi1-PH; and "Myr-" = fusion to the myristoyl group-accepting sequence MGCTVS. The constructs were encoded by the following plasmids: - (pPB1306), Boi1 (pPB2003), Boi1(TK) (pPB2008), Boi1(KK) (pPB2002), Boi1(KKTK) (pPB1671), PH (pPB1695), Myr-PH (pPB1498), PH(TK) (pPB1669), Myr-PH(TK) (pPB1667), PH(KK) (pPB1697), Myr-PH(KK) (pPB1500), PH(KKTK) (pPB1757), and Myr-PH(KKTK) (pPB1661).
At both temperatures, the TK mutant version of Boi1 allowed a substantial fraction of colonies to show either moderate or heavy sectoring (Fig. 6), suggesting that the TK mutant is able to provide whatever vital function normally is provided by Boi1/Boi2. However, this version of Boi1 appeared to function less efficiently than did wild-type Boi1: at 23°C, the percentage of colonies that showed either moderate or heavy sectoring with Boi1(TK) was only 29%, and, at 30°C, the percentage of colonies that showed moderate or heavy sectoring with Boi1(TK) was only 10%.
At each temperature, almost every one of the colonies of cells that contained either the KK or KKTK mutant versions of Boi1 showed no sectoring (Fig. 6), suggesting that these mutant versions of Boi1 provide little or no function.
To investigate whether the diminished amount of sectoring allowed by the different mutant versions of Boi1 might be due to effects of the amino-acid substitutions on the concentration of Boi1 (e.g., by decreasing the stability of Boi1), we used immunoblotting to compare the concentrations of the mutant proteins to that of wild-type Boi1. This analysis was done using a genomically boi1 BOI2 strain, so that the only version of Boi1 present was the one expressed from a plasmid. As shown in Figure 7A, each of the mutant versions of Boi1 is present at a concentration similar to or higher than that of wild-type Boi1, suggesting that the reduced functions of the mutant versions of Boi1 are not likely to be due to effects on their concentrations.
Figure 7.
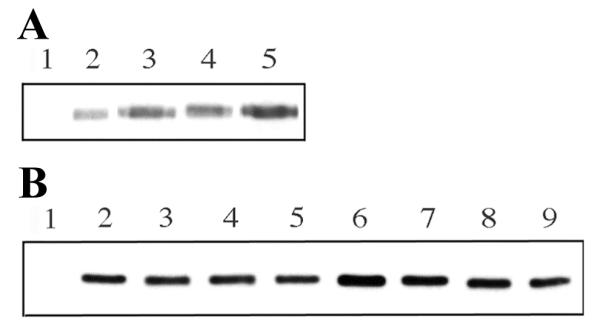
Relative concentrations of different versions of Boi1 and Boi1-PH in yeast extracts. Shown are immunoblots, probed with anti-Boi1 antibodies. A). Extracts were from boi1 strain Y1284 expressing the following versions of Boi1 from the indicated plasmids: lane 1 = no Boi1 (pPB1306); lane 2 = w.t. Boi1 (pPB2003), lane 3 = Boi1(KK) (pPB2002), lane 4 = Boi1(TK) (pPB2008), and lane 5 = Boi1(KKTK) (pPB1671). B). Extracts were from strain PY967 expressing the following versions of Boi1-PH from the indicated plasmids: lane 1 = no Boi1-PH (pPB1306), lane 2 = Boi1-PH (pPB1695), lane 3 = Myr-Boi1-PH (pPB1498), lane 4 = Boi1-PH(KK) (pPB1697), lane 5 = Myr-Boi1-PH(KK) (pPB1500), lane 6 = Boi1-PH(TK) (pPB1669), lane 7 = Myr-Boi1-PH(TK) (pPB1667), lane 8 = Boi1-PH(KKTK) (pPB1757), and lane 9 = Myr-Boi1-PH(KKTK) (pPB1661).
One potential role of the binding of Boi1 to phospholipid is simply to target Boi1 to the plasma membrane. If this were the only role for such binding, then attachment to a membrane-localization tag might be able to restore function to versions of Boi1 that are impaired in the ability to bind phospholipid. One type of membrane-localization tag is the myristoyl group, which is attached to the glycine residue of proteins, such as S. cerevisiae Gpa1 (the α subunit of the pheromone-responsive G protein) [18], that contain at their N-termini the sequence MGXXXS/T (where "X" can be any amino acid) [19]. Thus, we investigated whether fusion to the sequence MGCTVS, which is present at the N-terminus of Gpa1 and which we will henceforth refer to as "Myr", could restore function to the mutant versions of Boi1 that are impaired in the ability to bind phospholipid.
At the time that we initiated this analysis, the only lipid-binding-impaired version of Boi1 that we had generated so far was the KK mutant. In preliminary analyses, we found that Myr did not improve the ability of full-length Boi1(KK) to promote sectoring, but that it did improve the ability of the KK version of Boi1-PH to promote sectoring (data not shown). Therefore, in subsequent tests to ask whether Myr could restore function to mutant versions of Boi1 that are impaired in the ability to bind lipid, we used specifically the Boi1-PH segment of Boi1 rather than full-length Boi1.
First, we checked to see whether Myr had any inhibitory effects on the function of wild-type Boi1-PH. Myr-Boi1(PH) allowed a degree of sectoring similar to that for Boi1-PH without the tag (Fig. 6), suggesting that Myr does not impair the function of Boi1-PH.
Next, we asked whether Myr could improve the ability of the different mutant versions of Boi1-PH to function. At 23°C, 25% of colonies containing Boi1-PH(TK) without the tag showed moderate or heavy sectoring. This value rose to 81% when using the Myr-tagged version of Boi1-PH(TK) (Fig. 6), suggesting that Myr improves the ability of the TK mutant version of Boi1-PH to function. At 30°C, the effect of Myr was even more striking: without the tag, only 1% of the Boi1-PH(TK) colonies showed any degree of sectoring; however, with the tag, 90% of the colonies showed moderate or heavy sectoring (Fig. 6).
Myr also greatly improved the ability of the KK mutant version of Boi1-PH to cause sectoring: Myr-Boi1-PH(KK) allowed 59% moderate or heavy sectoring at 23°C and allowed 28% moderate or heavy sectoring at 30°C, compared to there being no sectoring at either temperature without Myr (Fig. 6). At 23°C, Myr also improved the function of the KKTK (the most severe) mutant version of Boi1-PH: at this temperature, whereas Boi1-PH(KKTK) without Myr did not allow any sectoring, Myr-Boi1-PH(KKTK) allowed 17% of the colonies to show moderate or heavy sectoring (Fig. 6). In constrast, when tested at 30°C, Myr did not affect significantly the amount of sectoring allowed by Boi1-PH(KKTK): 98% of the colonies of cells containing Myr-Boi1-PH(KKTK) were non-sectoring at this temperature (Fig. 6).
It is unlikely that the mechanism by which Myr improves the function of the different mutant versions of Boi1-PH is by increasing their stability, because the presence of Myr did not have an obvious effect on the relative concentrations of the different versions of Boi1-PH (Fig. 7B).
Localization of wild-type and mutant versions of Boi1 and Boi2
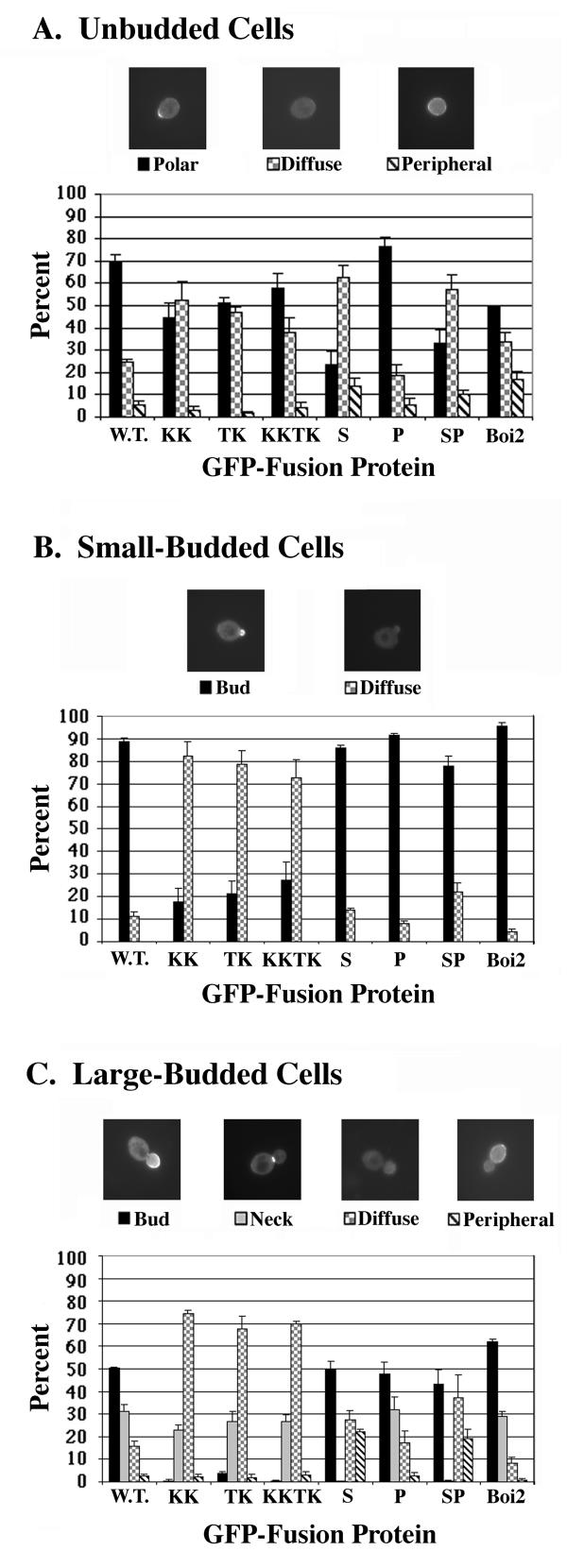
To investigate whether binding to phospholipid may be important for proper localization of Boi1, we used GFP (green fluorescent protein) fusions to identify first the patterns of localization of wild-type Boi1 and Boi2 and then those of the mutant versions of Boi1 that are impaired in the ability to bind phospholipid. The micrographs in Figure 8 show examples of the different patterns of localization observed for wild-type Boi1-GFP. In unbudded cells, Boi1-GFP was seen to be concentrated in a single spot at the cell cortex ("polar" pattern) in 70% of the cells (Fig. 8A). In small-budded cells, Boi1-GFP was concentrated in the bud in 89% of the cells (Fig. 8B). In cells containing large buds, Boi1-GFP was concentrated in the bud in 50% of the cells and was concentrated at the neck in 31% of the cells (Fig. 8C). Boi2-GFP showed distributions of localization patterns generally similar to those for Boi1-GFP (Fig. 8A,8B,8C). Thus, Boi1-GFP and Boi2-GFP both appear to be localized to sites of polarized growth throughout the cell cycle.
Figure 8.
Localization patterns of GFP fusions to different versions of Boi1 and to wild-type Boi2. Parts A-C show analyses for unbudded, small-budded, and large-budded cells, respectively. Micrographs are examples of different patterns of localization of Boi1-GFP. Bars indicate percentages of cells that displayed each of these patterns. In Part C, "peripheral" means localization around the periphery of the mother part of the cell. GFP fusions to the different versions of Boi1 are encoded by the following plasmids: W.T. (wild-type) (pPB1307), KK (pPB1347), TK (pPB2007), KKTK (pPB1689), S (pPB1392), P (pPB1394), and SP (pPB1396). Boi2-GFP ("Boi2") was encoded by plasmid pPB1303.
The KK, TK, and KKTK versions of Boi1-GFP all showed a polar pattern of localization in approximately half of the unbudded cells (Fig. 8A), suggesting that binding to phospholipid is not critical for this pattern of localization. In small-budded cells, the KK, TK, and KKTK mutant proteins showed localization to the bud in only 18, 21, and 27% of the cells, respectively (compared to in 89% of the cells for wild-type Boi1-GFP) (Fig. 8B). Thus, although the KK, TK, and KKTK mutant proteins all were able to localize to the bud in small-budded cells, they were localized and/or retained there less efficiently than was wild-type Boi1-GFP.
A more striking effect of the KK, TK, and KKTK substitutions on the localization of Boi1-GFP was seen in cells that contained large buds: Boi1-TK-GFP was concentrated in the bud in only 4% of large-budded cells, and Boi1-KK-GFP and Boi1-KKTK-GFP were concentrated in the bud in less than 1% of such cells (Fig. 8C). None of the sets of substitutions had an obvious effect on localization to the neck, however (Fig. 8C). These findings suggest that, in large-budded cells, binding to phospholipid may be important for the localization of Boi1 to the bud but not for localization to the neck.
Although our studies focus on roles of Boi1's PH domain, we were also curious to know which other portions of Boi1 contribute to its proper patterns of localization. In particular, we wished to know whether either the SH3 domain or the proline-rich (Bem1-binding) region of Boi1 is important for any pattern of localization. To address this issue, we used the following mutant versions of Boi1: the "S" mutant, which contains a Lys residue in place of a Trp residue at a highly conserved position in the SH3 domain; the "P" mutant, in which seven of the nine Pro residues in the Pro-rich region are replaced with Ala residues (a version of Boi1 that does not bind Bem1); and the "SP" mutant, which contains both the "S" and "P" changes [1].
The most striking effect of the SH3-domain mutation was on the localization of Boi1-GFP to necks: we never saw Boi1-S-GFP localized to necks (Fig. 8C), suggesting that the SH3 domain is required for such localization. In constrast, the SH3-domain mutation had no obvious effect on the localization of Boi1-GFP to the bud in either small- or large-budded cells (Figs. 8B and 8C). However, this mutation increased the frequency of localization to the periphery of large-budded cells, and it did so to a degree that approximated that to which it caused a decrease in the frequency of localization to the neck: in large-budded cells, whereas wild-type Boi1-GFP was localized to the periphery of the mother part of the cell in only 3% of the cells and to the neck in 31% of the cells, Boi1-S-GFP was localized to the periphery of the mother in 22% of the cells (and was never localized to the neck). These findings raise the possibility that failure to localize to the neck increases the likelihood that Boi1 will adopt or retain localization at the periphery of the mother part of the cell.
Another apparent effect of the SH3-domain mutation on the localization of Boi1-GFP was seen in unbudded cells, in which the percentage of cells that showed a polar pattern of localization for Boi1-S-GFP (24%) was lower than for wild-type Boi1-GFP (70%), and in which Boi1-S-GFP gave a higher percentage of cells that showed the diffuse pattern (62%) compared to that given by wild-type Boi1-GFP (24%) (Fig. 8A). These findings raise the possibility that one way in which Boi1-GFP acquires a polar distribution in unbudded cells involves prior localization to the neck, via the SH3 domain.
In all classes of cells, the distributions of localization patterns for Boi1-P-GFP were not notably different from those for wild-type Boi1-GFP, and the distributions of localization patterns for Boi1-S-P-GFP were not notably different from those for Boi1-S-GFP (Figs. 8A,8B,8C), suggesting that the proline-rich region is not important for any of the observed patterns of localization of Boi1-GFP.
Discussion
Binding to phospholipids
Boi1-PH co-sediments with PS vesicles, suggesting that Boi1's PH domain can bind PS. Boi1-PH co-sediments more readily when such vesicles contain a small percentage (5%) of PIP2, suggesting that Boi1-PH also binds PIP2 and that it does so with higher affinity than it binds PS. Given that PIP2 and PS are negatively charged (with PIP2 being more negatively charged than PS), a model to account for these binding behaviors is that Boi1's PH domain binds in a non-specific manner to negatively charged surfaces. The ability to bind different acidic phospholipids appears to be a fairly common attribute of PH domains [11,12]. For example, five out of ten PH domains in one study were judged to bind to PS with affinity similar to that for PIP2 and other inositol-based phospholipids, suggesting that many PH domains bind promiscuously to negatively charged phospholipids [11].
Findings from protease-sensitivity analyses suggest that Boi1-PH binds in a different manner to PIP2 than to PS, however. Specifically, proteolysis of Boi1-PH was stimulated by PIP2/PS mixed vesicles but not by vesicles that contained only PS. For the following reasons, we think that the inability of PS, by itself, to stimulate the proteolysis of PIP2 is not due to an inability of PS to bind to Boi1-PH. First, the ability of PIP2 to stimulate proteolysis of Boi1-PH was greatly enhanced when using PS (rather than PC) as the bulk lipid in the vesicles, suggesting that PS binds to Boi1-PH in this analysis. Second, the proteolysis analysis was conducted in conditions that were very similar to those used in the vesicle co-sedimentation analysis, in which Boi1-PH was found to bind readily to PS. These findings suggest that, in conditions in which both PS and PIP2 bind to Boi1-PH, only PIP2 binds in a manner that triggers the exposure of a protease-sensitive site.
PIP2 was not the only phosphatidylinositide that could stimulate proteolysis of Boi1-PH; PI4P also did so, although less effectively than did PIP2. In contrast, PI3P and PI were comparatively ineffective at stimulating proteolysis. The large difference in the ability of PI4P versus PI3P to stimulate proteolysis supports the view that Boi1's PH domain can discriminate between different phosphorylated versions of PI in some manner that involves more than simply recognizing net charge.
Residues implicated in binding to phospholipid
The KK, TK, and KKTK mutant versions of Boi1-PH contain substitutions at positions in the PH domain that were predicted to be involved in binding PIP2. PIP2/PS vesicles did not stimulate the proteolysis of any of these mutant versions of Boi1-PH, suggesting that each mutant is indeed impaired in the ability to associate with PIP2 and/or PS.
Vesicle co-sedimentation analyses also suggest that each mutant version of Boi1-PH is impaired in the ability to bind PIP2/PS vesicles. However, these analyses suggest that the KK substitutions may impair binding in a different manner than do the TK substitutions. In particular, the co-sedimentation analyses suggest that the TK mutant may be severely impaired in the ability to bind acidic phospholipids generally. In contrast, the KK mutant appears to be impaired more in the ability to recognize specifically PIP2 than in the ability to bind non-specifically to acidic phospholipids.
Significance of binding to phospholipids
Based on colony-sectoring analyses, the KK, TK, and KKTK mutant versions of Boi1 are all impaired in function. The KK (as well as the KKTK) version of Boi1 appeared to be completely non-functional, pointing to the possibility that binding specifically to PIP2 (as opposed to binding to acidic phospholipids generally) may be critical for Boi1 function.
Fusion to a myristoyl group-accepting sequence appeared to completely restore function to Boi1-PH(TK), consistent with the possibility that the TK substitutions may affect primarily association with membrane. The myristoyl group-accepting sequence did not completely restore function to the KK and KKTK versions of Boi1-PH, however, raising the possibility that, rather than simply promoting general association with membrane, one role of binding to PIP2 may be to regulate some other behavior of Boi1 (e.g., the binding of Boi1 to some other protein at the plasma membrane).
If specific binding to PIP2 (and/or to some other phosphatidylinositide, such as PI4P) is critical for Boi1 function, then how could fusion to a myristoyl group-accepting sequence (i.e., to a membrane-localization tag) improve the function of mutant versions of Boi1-PH that are impaired in the ability to bind PIP2? One possibility is that each of these mutant versions of Boi1 retains at least a slight affinity for PIP2 and that the myristoyl group causes these mutant PH domains to be apposed close enough to the lipid bilayer to enable the residual PIP2-binding activity to now be sufficient for binding to PIP2. We speculate that, just as binding to PS may serve to position the PH domain of wild-type Boi1 near the plasma membrane in a manner that facilitates binding to PIP2, the myristoyl group may help to situate the PH domain of the mutant versions of Boi1-PH at the plasma membrane in a manner that facilitates binding to PIP2.
Although the myristoyl group-accepting sequence improved the ability of the KK mutant version of Boi1-PH to function, it failed to improve the function of the corresponding mutant version (KK) of full-length Boi1. This finding is consistent with the possibility that the ability of the myristoyl group to improve function to a version of Boi1 that is impaired in the ability to bind phospholipids may depend on the myristoyl group being near enough to the PH domain to be able to promote a close association with membrane of specifically the PH domain portion of the protein.
Localization of Boi1
A behavior of Boi1 that is affected by mutations that impair binding to phospholipids is the localization of Boi1 to buds. These mutant versions of Boi1 still localize to mother/bud necks, however. In contrast, a mutation in the SH3 domain prevents Boi1 from localizing to necks, but it does not diminish the ability of Boi1 to localize to buds. These findings suggest that binding to phospholipids may promote the localization of Boi1 to (and/or the retention of Boi1 at) the bud, but that this binding is not important for the localization of Boi1 to the neck.
How might binding to phospholipids promote localization of Boi1 to buds? One possibility is that the relevant phospholipid(s) (e.g., PIP2) is itself concentrated in buds. Another possibility is that the binding to phospholipid promotes the binding of Boi1 to some other protein that is itself localized to buds. Other than the unidentified protease that acts on Boi1 in yeast extracts, the only proteins that we have evidence for binding to Boi1 are Bem1 and Cdc42, both of which show localization patterns similar to those for Boi1 ([20,21], and our unpublished data). Amino-acid substitutions in the proline-rich stretch of Boi1 that destroy binding to Bem1 do not affect the localization of Boi1, suggesting that binding to Bem1 is not necessary for any pattern of localization of Boi1.
The situation with respect to Cdc42 is less clear. We have not yet generated mutations in BOI1 that would allow us to probe the role of Boi1's interaction specifically with Cdc42. However, we have observed that the two-hybrid interaction between Boi1-PH and Cdc42 is not reduced by substitutions (i.e., KK, TK, and KKTK) that impair binding to phospholipids. These findings suggest that binding to phospholipid is not necessary for the interaction between Boi1 and Cdc42, and so place in doubt the possibility that the mechanism by which binding to phospholipid promotes localization of Boi1 to buds is by positively regulating the binding of Boi1 to Cdc42. We note, however, that these two-hybrid analyses may be quite limited in terms of what they can reveal about interactions between Boi1 and Cdc42 that might normally occur at the plasma membrane (or at other membranes), because 1) the two-hybrid assay monitors only those interactions that are occurring in the nucleus and 2) in these two-hybrid analyses, we used only versions of Cdc42 that lack a site of isoprenylation (because we were unable to detect two-hybrid interactions between Boi1 and versions of Cdc42 that retain that site). Thus, it is still possible that at the plasma membrane, binding to one or more types of phospholipid influences Boi1's ability to associate with Cdc42. Alternatively, phospholipids might regulate the binding of Boi1 to some other, as-yet unidentified binding partner of Boi1. To us, an important breakthrough toward elucidating the role of the binding of any phospholipid to Boi1 would be the identification of a protein whose binding to Boi1 is regulated by that phospholipid.
Conclusions
From vesicle co-sedimentation and proteolysis-stimulation analyses, we gained evidence that the PH domain of Boi1 binds with higher affinity to PIP2 than to PS and that binding to PIP2 is facilitated by PS and promotes a conformational change in Boi1. Amino-acid substitutions that diminish binding to PS and PIP2 impair Boi1 function, and fusion to a myristoyl group-accepting sequence improves the ability of these mutant versions of Boi1-PH to function, suggesting that binding to phospholipids is important for Boi1 action. Based on the differing extents to which the myristoyl group-accepting sequence improved the ability of the different mutant versions of Boi1-PH to function, we propose that the main role of binding to PS is to promote association with membrane and that binding to PIP2 plays some additional important role. Boi1 and Boi2 are localized to sites of polarized growth, consistent with the view that they are involved in polarized growth. Whereas the SH3 domain is needed for localization of Boi1 to the neck, the phospholipid-binding portion of the PH domain is important for localization to the bud.
Materials and Methods
Materials
The plasmids used in this study are described in Additional file 1: Plasmids and phage . The yeast strains used are Y312 (MATα ura3 leu2 his3 Gal+), Y1284 (MATaboi1::LEU2 ura3 leu2 his3 trp1 met14 Gal+), Y1300 (MATa/MATα ura3/ura3 leu2/leu2 his3/his3 trp1/trp1 met14/met14 Gal+), and PY967 (MATa/Matα boi1::LEU2/boi1::LEU2 boi2::LEU2/boi2::LEU2 ura3/ura3 leu2/leu2 trp1/trp1 ade2/ade2 ade3/ade3 bearing plasmid pPB799).
SC medium is 1.7 g/l yeast nitrogen base without amino acids and ammonium sulfate, 5 g/l ammonium sulfate, 20 g/l glucose, 20 mg/l uracil, 20 mg/l adenine, 80 mg/l L-leucine, 20 mg/l L-histidine, 40 mg/l L-tryptophan, 20 mg/l L-methionine, and 30 mg/l L-lysine. SC-Leu, SC-Ura, and SC-Ura-Leu are SC without leucine, uracil, and both leucine and uracil, respectively. Sgal-Leu is SC-Leu with 20 g/l galactose plus 20 g/l raffinose instead of glucose.
PBS (phosphate-buffered saline, pH 7.0) is 1.42 g/l Na2PO4, 245 mg/l KH2PO4, 8 g/l NaCl, and 0.2 g/l KCl. Protease inhibitors are 40 μg/ml bestatin, 0.7 μg/ml pepstatin A, 1 mM PMSF, 50 μM leupeptin, 1 mM benzamidine, and 1 mM EGTA. 2× SDS sample buffer is 2% sodium dodecyl sulfate, 50 mM Tris (pH6.8), 6 M urea, and bromophenol blue.
Phosphatidylcholine, phosphatidylinositol, and phosphatidylserine were from Avanti Polar Lipids (Alabaster, AL); phosphatidylinositol-4,5-bisphosphate and phosphatidylinositol-4-phosphate were from Calbiochem (San Diego, CA); and phosphatidylinositol-3-phosphate was from Matreya, Inc. (Pleasant Gap, PA).
AP-PH is affinity-purified anti-Boi1 antibody [1]. AP-101 is affinity-purified anti-Boi1 antibody that was generated in the same manner as AP-PH, except that it came from the serum of a different rabbit. Both types of antibody were affinity purified [22] using a 6×His fusion to the C-terminal 194 amino acids of Boi1 (the same fusion that was used as immunogen) [1]. Immobilon P membrane was from Millipore, Inc. (Bedford, MA). ECL reagents were from Amersham Pharmacia Biotech, Inc. (Piscataway, NJ).
Generation of lipid vesicles
PC and PS were stored dissolved in chloroform at concentrations of 20 and 10 mg/ml, respectively. Inositol-based phospholipids were stored at 1 mg/ml dissolved in a 1:1 mixture of chloroform:methanol. Lipids (pure or mixtures) were dried down from these solutions under a stream of nitrogen gas. Bath-type sonication was then used to generate suspensions of vesicles of these lipids in PBS.
Proteolysis assay
The wild-type, KK, TK, and KKTK versions of Boi1-PH were expressed in yeast strain Y312 under the control of the GAL10 promoter from plasmids pPB900, pPB1308, pPB1699, and pPB1755, respectively. Cultures were grown to an O.D.600 of 0.8 in 250 mls of SC-Leu, pelleted, suspended in 250 mls of Sgal-Leu, and incubated at 30°C for 16 hours. The following procedures were then performed at 4°C. Cells were pelleted, and the pellets were rinsed with PBS containing all of the protease inhibitors except leupeptin, benzamidine, and EGTA. Cells were suspended in PBS containing the same protease inhibitors and were then lysed using a French press. Large pieces of cellular debris were removed by centrifugation at 12,000 × g for 10 min in a Sorval SS34 rotor. Soluble fractions were then generated by centrifugation of lysates at 541,000 × g for 1 hr in a TLA 100.4 rotor in an Optima TLX ultracentrifuge (Beckman, Palo Alto, CA). To the cytosolic fractions (supernatants), glycerol was added to a final concentration of 40%. These fractions were stored at -20°C and used within one month. 15 μl of 1 mg/ml lipid vesicle/PBS suspension was mixed with 15 μl of cytosol. 5 mM DTT was included to inactivate any residual PMSF, and then 2 mM CaCl2 was added to allow proteolysis of Boi1-PH. Reaction mixtures were then transferred immediately to a 30°C water bath, and proteolysis was inhibited 5 min later by addition of an equal volume of 2× SDS sample buffer containing 2 mM EGTA. Proteins were separated by SDS-PAGE through a 12% gel, and then proteins were transferred electrophoretically to Immobilon P membrane. Blots were probed with anti-Boi1 antibody AP-PH. Antibody-bound protein bands were detected on X-ray film using ECL. Each of the proteolysis experiments was performed at least three times, with similar results.
Vesicle co-sedimentation assay
Yeast cytosol (from 541,000 × g spins) containing wild-type and mutant versions of Boi1-PH were prepared and stored as described for the proteolysis assay, except that the protease inhibitors leupeptin, benzamidine, and EGTA were included. 15 μl of 1 mg/ml lipid vesicle/PBS suspensions were mixed with 15 μl of cytosol. 30 μl of PBS, with or without additional KCl, was then added to each mixture to reduce the concentration of glycerol to approximately 10%. For the experiment shown in Fig. 3, the concentrations of additional KCl in the final reaction mixture are indicated. For the experiment shown in Fig. 5B, no additional KCl was present. Mixtures were then centrifuged immediately at 436,000 × g for 1 hr in a TLA100 rotor. Supernatants were combined 1:1 with 2× SDS sample buffer, and pellets were dissolved in 2× SDS sample buffer. Equal proportions of each fraction were subjected to SDS-PAGE through a 12% gel, proteins were transblotted Immobilon P membrane, blots were probed with antibody AP-PH, and bound antibodies were detected by ECL. Each of the vesicle co-sedimentation experiments was performed at least three times, with similar results.
Colony-sectoring assay
Plasmids were introduced into strain PY967 using the lithium thiocyanate procedure [23], and transformants were selected on SC-Ura. Concentrations of plasmid DNAs were adjusted so that they would give approximately 400 colonies per petri plate. Transformation plates were incubated at either 23 or 30°C for 4–5 days and then stored at 4°C for one week to allow further development of the red color. To assay sectoring, colonies were viewed using a low-power dissecting microscope. A minimum of 200 colonies were analyzed for each plasmid tested at each temperature.
Analysis of relative protein concentrations
To analyze versions of full-length Boi1, boi1 strain Y1284 was used, selecting for plasmids in SC-Ura. To analyze versions of Boi1-PH, strain PY967 was used, selecting for plasmids in SC-Ura-Leu. Cultures were grown to an O.D.600 of 0.8 at 23°C and then diluted 1:1 with 0.4 M NaOH + 1.7% 2-mercaptoethanol. The resulting lysed-cell suspensions were then incubated on ice for 10 min, and protein was made to precipitate using 10% trichloroacetic acid. Pellets were rinsed first with 70% ethanol and then with acetone, and they were then suspended in 1% SDS + 3 M urea. Protein concentrations were determined using the BCA Protein Assay (Pierce, Rockford, IL). 10 μg of total protein from each sample was subjected to SDS-PAGE through a 12% gel. Proteins were transblotted to Immobilon P membrane, blots were probed with antibody AP-101, and bound antibodies were detected by ECL. Each of the analyses of protein concentration was performed three times, with similar results.
Localization studies
Yeast strain Y1300 was used for localization studies. Cells were grown in SC-Ura to select for plasmids that encode the GFP fusions. Aliquots of cells from exponentially growing cultures were placed directly (without centrifugation) onto glass slides for analysis using a Zeiss Axioplan epifluorescence microscope. No slide sample was analyzed for longer than 5 min, to avoid stressing the cells. Quantification of the different localization patterns was done separately for each morphological category (i.e., analyzing first only unbudded cells, then only small-budded cells, and then only large-budded cells). Cells in which the diameter of the bud was less than 1/4 of the diameter of the mother part of the cell were classified as small-budded cells. For each experiment, we analyzed at least 100 cells in each morphological category, scoring only cells that gave detectable green fluorescence. Figure 8 shows the average values from the results of three such experiments. In some large-budded cells, but in no case more than in 3% of such cells, green fluorescence was seen both at the neck and at the mother periphery. Such cells were placed in the "neck" rather than in the "peripheral" category. Images were captured using a MicroMAX digital camera system and WinView/32 software (Princeton Instruments Inc., Trenton, N.J.). Images were processed using Photoshop 5 software (Adobe Systems Inc., San Jose, CA).
Authors' contributions
M.A.H. performed the lipid-binding analyses, assisted in the localization and colony-sectoring studies, and prepared the figures. H.S.L. conducted localization and colony-sectoring analyses and constructed most of the plasmids and phage. A.B. constructed some of the plasmids and phage and assisted in the colony-sectoring analyses.
All authors read and approved the final manuscript.
Supplementary Material
This table describes the plasmids and phage used in this study
Acknowledgments
Acknowledgements
We thank B. Dennehey, C. Garvin, S. Ems-McClung, D. Daleke, W. Saxton, E. Raff, and J. Bonner for helpful discussions. This work was supported by USPHS grant GM46271 and CTR grant 4620.
Contributor Information
Mark A Hallett, Email: hallettm@iupui.edu.
H Shuen Lo, Email: slo@bio.indiana.edu.
Alan Bender, Email: abender@bio.indiana.edu.
References
- Bender L, Lo HS, Lee H, Kokojan V, Peterson J, Bender A. Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in yeast. J Cell Biol. 1996;133:879–894. doi: 10.1083/jcb.133.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Matsui R, Akada R, Toh-e A. Yeast src homology region 3 domain-binding proteins involved in bud formation. J Cell Biol. 1996;133:865–878. doi: 10.1083/jcb.133.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Ponting CP, Hofmann K, Bork P. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 1997;6:249–253. doi: 10.1002/pro.5560060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo JE, Rossi G, Brennwald P. The Rho GTPase Rho3 has a direct role in exocytosis that is distinct from its role in actin polarity. Mol Biol Cell. 1999;10:4121–4133. doi: 10.1091/mbc.10.12.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AEM, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai J, Toh-e A, Matsui Y. Genetic analysis of the Saccharomyces cerevisiae RHO3 gene, encoding a Rho-type small GTPase, provides evidence for a role in bud formation. Genetics. 1996;142:359–369. doi: 10.1093/genetics/142.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI, Pringle JR. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Toh-e A. Yeast RHO3 and RHO4 ras superfamily genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and BEM1. Mol Cell Biol. 1992;12:5690–5699. doi: 10.1128/mcb.12.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NGG, Guo L, Imai J, Toh-e A, Matsui Y, Tamonoi F. Rho3 of Saccharomyces cerevisiae, which regulates the actin cytoskeleton and exocytosis, is a GTPase which interacts with Myo2 and Exo70. Mol Cell Biol. 1999;19:3580–3587. doi: 10.1128/mcb.19.5.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toya M, Iino Y, Yamamoto M. Fission yeast Pob1p, which is homologous to budding yeast Boi proteins and exhibits subcellular localization close to actin patches, is essential for cell elongation and separation. Mol Biol Cell. 1999;10:2745–2757. doi: 10.1091/mbc.10.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J Biol Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- Rameh LE, Arvidsson A, Carraway KL, III, Couvillon AD, Rathbun G, Crompton A, VanRenterghem B, Czech MP, Ravichandran KS, Burakoff SJ, Wang D, Chen C, Cantley LC. A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J Biol Chem. 1997;272:22059–22066. doi: 10.1074/jbc.272.35.22059. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Falasca M, Ferguson KM, Schlessinger J. Regulatory recruitment of signalling molecules to the cell membrane by pleckstrin-homology domains. Trends Cell Biol. 1997;7:237–242. doi: 10.1016/S0962-8924(97)01065-9. [DOI] [PubMed] [Google Scholar]
- Paris S, Beraud-Dufour S, Robineau S, Bigay J, Antonny B, Chabre M, Chardin P. Role of protein-phospholipid interactions in the activation of ARF by the guanine nucleotide exchange factor Arno. J Biol Chem. 1997;272:22221–22226. doi: 10.1074/jbc.272.35.22221. [DOI] [PubMed] [Google Scholar]
- Kam JL, Miura K, Jackson TR, Gruschus J, Roller P, Stauffer S, Clark J, Aneja R, Randazzo PA. Phosphoinositide-dependent activation of the ADP-ribosylation factor GTPase-activating protein ASAP1. J Biol Chem. 2000;275:9653–9663. doi: 10.1074/jbc.275.13.9653. [DOI] [PubMed] [Google Scholar]
- Qian X, Vass WC, Papageorge AG, Anborgh PH, Lowy DR. N Terminus of Sos1 Ras exchange factor: critical roles for the Dbl and Pleckstrin homology domains. Mol Cell Biol. 1998;18:771–778. doi: 10.1128/mcb.18.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan JE, Hajduk PJ, Yoon HS, Fesik SW. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature. 1994;371:168–170. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- Stone DE, Cole GM, De Barros Lopes M, Goebl M, Reed SI. N-myristoylation is required for function of the pheromone-responsive Gα protein of yeast: conditional activation of the pheromone response by a temperature-sensitive N-myristoyl transferase. Genes & Dev. 1991;5:1969–1981. doi: 10.1101/gad.5.11.1969. [DOI] [PubMed] [Google Scholar]
- Resh MD. Myristylation and palmitylation of Src family members: The fats of the matter. Cell. 1994;76:411–413. doi: 10.1016/0092-8674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Bi E, Harkins HA, Zahner JE, De Virgilio C, Chant J, Corrado K, Fares H. Establishment of cell polarity in yeast. C S H Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- Ziman M, Preuss D, Mulholland J, O'Brien JM, Botstein D, Johnson DI. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol Biol Cell. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted JB. Analysis of cytoskeletal structures using blot-purified monospecific antibodies. Meth Enzymol. 1986;134:467–472. doi: 10.1016/0076-6879(86)34112-0. [DOI] [PubMed] [Google Scholar]
- Keszenman-Pereyra D, Hieda K. A colony procedure for transformation of Saccharomyces cerevisiae. Curr Genet. 1986;13:21–23. doi: 10.1007/BF00365751. [DOI] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Bender A, Sprague GF., Jr Pheromones and pheromone receptors are the primary determinants of mating specificity in the yeast Saccharomyces cerevisiae. Genetics. 1989;121:463–476. doi: 10.1093/genetics/121.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C, DeMarini DJ, Pringle JR. SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiae that is expressed specifically in sporulating cells. Microbiology. 1996;142:2897–2905. doi: 10.1099/13500872-142-10-2897. [DOI] [PubMed] [Google Scholar]
- Botstein D, Falco SC, Stewart SE, Brennan M, Scherer S, Stinchcomb DT, Struhl K, Davis RW. Sterile host yeasts (SHY): A eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979;8:17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Rose AB, Broach JR. Propagation and expression of cloned genes in yeast: 2-μm circle-based vectors. Meth Enzymol. 1990;185:234–279. doi: 10.1016/0076-6879(90)85024-i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table describes the plasmids and phage used in this study