Abstract
Myotonic dystrophy type 1 (DM1) is an autosomal dominant neuromuscular disorder associated with a (CUG)n expansion in the 3′-untranslated region of the DMPK (DM1 protein kinase) gene. Mutant DMPK mRNAs containing the trinucleotide expansion are retained in the nucleus of DM1 cells and form discrete foci. The nuclear sequestration of RNA binding proteins and associated factors binding to the CUG expansions is believed to be responsible for several of the splicing defects observed in DM1 patients and could ultimately be linked to DM1 muscular pathogenesis. Several RNA binding proteins capable of co-localizing with the nuclear-retained mutant DMPK mRNAs have already been identified but none can account for the nuclear retention of the mutant transcripts. Here, we have employed a modified UV crosslinking assay to isolate proteins bound to mutant DMPK-derived RNA and have identified hnRNP H as an abundant candidate. The specific binding of hnRNP H requires not only a CUG repeat expansion but also a splicing branch point distal to the repeats. Suppression of hnRNP H expression by RNAi rescued nuclear retention of RNA with CUG repeat expansions. The identification of hnRNP H as a factor capable of binding and possibly modulating nuclear retention of mutant DMPK mRNA may prove to be an important link in our understanding of the molecular mechanisms that lead to DM1 pathogenesis.
INTRODUCTION
Myotonic dystrophy type 1 (DM1) is an autosomal, dominantly inherited neuromuscular disorder with a global incidence of 1 per 8000 (1). Adult onset DM1 is primarily characterized by myotonia, muscle wasting and weakness, but also affects a number of organs and results in cataracts, cardiac conduction abnormalities, testicular atrophy, male baldness and insulin resistance (1). The mutation responsible for the disease is a (CUG)n repeat expansion in the 3′-untranslated region (3′-UTR) of the DM protein kinase (DMPK) gene (2–4). This repeat ranges in size from 5 to 37 in the normal population to between 50 and 1000 in adult onset cases (1).
Amongst several proposed molecular mechanisms, the RNA dominant mutational model proposes that triplet repeat expansion causes a gain-of-function at the RNA level (5,6), possibly by sequestering essential cellular RNA binding proteins (7–10). Targeting and destruction of mutant DMPK mRNA releases these factors thus allowing restoration of several of the normal myotube functions (11,12). In support of the gain-of-function model, transgenic mice containing CUG repeats in an unrelated mRNA display myotonia and a myopathy phenotype (13). Mice transgenic for the human DMPK region with expanded CTG repeats display muscular and brain abnormalities (14). Several features of DM1 pathogenesis can be explained by aberrant alternative-splicing defects (15). Misregulation of insulin receptor (IR) (16), muscle-specific chloride channel (CLC-1) (17,18) and cardiac troponin T (cTNT) (19) splicing is linked with common symptoms of DM1 such as insulin resistance, skeletal muscle membrane hyperexcitability characteristic of myotonia and cardiac conduction defects (16,20).
Several CUG repeat binding proteins have been identified to date (9,21–26). CUG-BP1 is one of the first CUG binding proteins identified. While this protein does not co-localize with the nuclear foci formed by mutant DMPK transcripts, it has been shown that expression levels of CUG-BP1 are increased in DM1 (23,27). Functional analyses indicate that increased expression of CUG-BP1 could be implicated for the aberrant regulation of cTNT, IR and CIC-1 by binding to U/G-rich motifs in introns adjacent to the regulated splice site (28–30). Muscleblind (MBNL) protein family members in humans have also been shown to bind to CUG repeats and can also co-localize with the nuclear foci (10,31–34). Recently, a muscleblind (MBNL1) knock-out mouse was produced that displayed muscle, eye and RNA splicing abnormalities that are characteristic of DM1 disease (33). Although MBNL1 protein depletion in mice helps explain some of the molecular mechanism involved in DM1, it is reasonable to hypothesize that there are additional CUG binding factors which work coordinately with these aforementioned CUG binding proteins.
To address this possibility, we utilized a modified RNA/protein crosslinking assay to search for proteins that bind DM1-derived CUG repeat containing transcripts. This assay identified the heterogeneous nuclear ribonucleprotein H (hnRNP H) as a novel protein capable of binding RNA with CUG repeats when a branch point sequence is located downstream. HnRNP H is best known for its role as an alternative splicing factor and pre-mRNA cleavage and polyadenylation (35–39). Surprisingly, we show that knock-down of endogenous hnRNP H expression by siRNAs in cells expressing an EGFP gene fused to CUG repeats leads to release of nuclear sequestrated transcripts and restoration of EGFP expression. These results could provide insight into the mechanisms implicated in the nuclear sequestration of mutant DMPK transcripts in DM1.
MATERIALS AND METHODS
DNA clones
The DMPK clone containing 100 CUG repeats (pRMK-100) was digested with SacI and EcoRI restriction endonucleases and cloned into the pGEM vector and further modified by deletion of a SacI/SacII fragment (40). Among several subclones containing a variety of CUG repeats, the clones containing 5, 46 and 85 CUG repeats were selected following sequence confirmation. (CUG)85′ which is the (CUG)85 clone with mutated 3′ branch site was created by PCR using two primers (5′-GAACGGGGCTCGAAGCTTCCTT-3′ and 5′-CTAGACTGGAATTCGGCTTATGGTCACTGATC-3′) and cloned into the pBluescript II SK vector. RNAs were transcribed in vitro using T7 RNA polymerase on linearized DNA plasmid templates in a 20 μl reaction. For the generation of biotinylated RNA, 10% of the UTP in the transcription reaction was replaced with biotinylated UTP (Roche) in a 100 μl reaction. RNA produced from the transcription reaction was mixed with four volumes of sterile water and further purified using a MicroSpin™-G50 column (Amersham Biosciences, Piscataway, NJ). To create the hnRNP H-EGFP fusion gene, hnRNP H was amplified by PCR using two primers (5′-CAGCCATATGCTCGAGTGATG-3′ and 5′-CTTTGTTAGCAGCCGGATCC-3′) and the product cloned into the XhoI/BamHI sites of the pLEGFP-C1 vector (Clonetech).
Extract preparation and hnRNH H purification
The total cell extracts of HeLa and DM1 cells were prepared as follows. Cells (1 × 108) were harvested and washed with buffer D, sequentially mixed with 100 μl of buffer D, and sonicated for 15 s at 4°C. After 5 min of micro-centrifugation at 4°C, the supernatants were collected and used as total cell extracts. The HeLa nuclear extract was used for the purification of CUG binding proteins. The extract was precipitated with 30% ammonium sulfate, the supernatant was harvested following a 10 min centrifugation at 4°C: 300 μl of extract was incubated with 300 μl of pre-washed Streptavidin M-280 Dynabeads for 30 min at room temperature. The extract was recovered by microcentrifugation and used for purification of CUG binding proteins. The pre-treated extract was incubated with 30 μg of E.coli tRNA for 20 min at room temperature, and 30 μg of biotinylated CUG RNA was mixed and incubated for an additional 30 min. The buffer D pre-washed beads were incubated for 30 min and washed three times again with 1 ml of buffer D. The bound proteins were eluted by buffer D containing 200 mM KCl. The eluted proteins were separated in 15% SDS–PAGE gel. The 50 kDa fragment was gel-purified and sequenced by mass spectrometry in the protein microsequencing facility of the City of Hope. Recombinant hnRNP H protein was purified as described by others (41).
Immunodepletion of hnRNP H
Anti-hnRNP H antiserum (100 μl) was incubated with 200 μl of Protein A conjugated Dynabeads at room temperature for 1 h. The beads were washed with 1 ml of PBS 5 times and incubated with 50 μl of the nuclear extract at 4°C for 1 h. The mixture was spun for 5 min, and the supernatant was used as the hnRNP H immuno-depleted extract. The depletion of hnRNP H was confirmed by western blotting with anti-hnRNP H antisera. For the reconstituted extract, the 20 μl of the hnRNP H-depleted extract was mixed with 20 ng of recombinant hnRNP H protein, and incubated at room temperature for 10 min.
In vitro and in vivo crosslinking assay
A total of 10 μl of extract was mixed with E.coli tRNA (Sigma) at a final concentration of 2 mg/ml and incubated at room temperature for 10 min. G50 column-purified labeled RNA (1 μl) was mixed and incubated at room temperature for 20 min. The reaction mixture was pipetted into a Petri dish maintained at 4°C on ice water. UV crosslinking was performed using a UV Stratalinker 2400 (Stratagene, San Diego, CA) 5 cm from the light source for 10 min. The irradiated samples were digested with 10 μg of RNAse A for 10 min at 37°C and resolved in a 10 or 15% SDS–PAGE gel. For UV crosslinking in the presence of the anti-hnRNP H antibody, 2 μl of the antisera was mixed with the extract for 10 min prior to the crosslinking treatment.
For in vivo UV crosslinking, DM1 cells were placed on Petri plates and irradiated by UV as described above. Extracts were prepared following sonication and incubated in the presence of antibody-conjugated Protein A conjugated Dynabeads as described in the section for the immunodepletion assay.
Native gel assays
Non-denaturing composite gel electrophoresis was performed as described previously (42). The reaction was mixed with an equal volume of non-denaturing loading dye containing bromophenol blue and 50% glycerol.
RNAi assays
For synthesis of the siRNA targeting hnRNP H, two sets of oligos were designed (AS1, 5′-AAGGTGGAGAGGGATTCGTGGCCTGTCTC-3′; S1, 5′-AACCACGAATCCCTCTCCACCCCTGTCTC-3′). siRNA was synthesized using the Silencer siRNA construction kit from Ambion (Austin, TX). For siRNA assays, 293T or DM1 cells that were 30% confluent were transfected with 100 ng of the EGFP-(CUG)85 reporter gene and the anti-hnRNP H siRNA or a scrambled siRNA (IDT, Coralville, IA) in a final concentration of 10 nM using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The suppression of EGFP expression was tested 24 h later. For suppression of hnRNP H, the cells were harvested after 72 h followed by total RNA isolation. For knock-down of hnRNP F, a Dicer substrate dsRNA (43) (sense, 5′-GUUAGGAACAUUUUGAGUUACUUGAA-3′; and antisense, 5′-UUCAAGUAACUCAAAUGUUCCUAACAA-3′) was transfected into HEK293T cells at a final concentration of 20 nM as described above. Three days later, cells were harvested and divided into two aliquots. One aliquot was used to prepare total RNA for northern blot assays and the other for preparation of a total cell extract. For northern blot analyses, a total of 20 μg of RNA was loaded in each well of a 1% agarose gel and electrophoresed and blotted onto a nylon membrane (Hybond). For hybridization, a hnRNP F-specific oligonucleotide (5′-AAGTAACTCAAATGTTCCTAACAA-3′) was used.
Primary human muscle cell cultures
DM1, CDM1 and normal control myoblasts were obtained from the quadriceps of 15-week-old aborted fetuses. The DM1 fetus had ∼750 CUG repeats (verified by Southern blot analysis). Skeletal muscle biopsies were approved by Laval University and the CHUL's ethical committees. Myoblasts were grown in MCDB-120 supplemented with 15% heat-inactivated fetal bovine serum, 5 μg/ml insulin, 0.5 mg/ml BSA, 10 ng/ml human hrEGF, 0.39 μg/ml dexamethasone, 50 μg/ml streptomycin and 50 μg/ml penicillin. Differentiation was carried out in DMEM supplemented with 10 μg/ml insulin, 10 μg/ml apo-transferrin, 50 μg/ml streptomycin and 50 μg/ml penicillin.
In situ hybridization
Detection of foci was performed using 10 ng of a Cy3-labeled (CAG)10 oligo (IDT, Coralville, IA) as described (44). Detection by immunofluorescence involved using two antibodies (first, anti-hnRNP H and second, anti-rabbit antibody conjugated with FITC) as described previously (45).
RESULTS
UV crosslinking of a mutant DMPK 3′-UTR binding protein
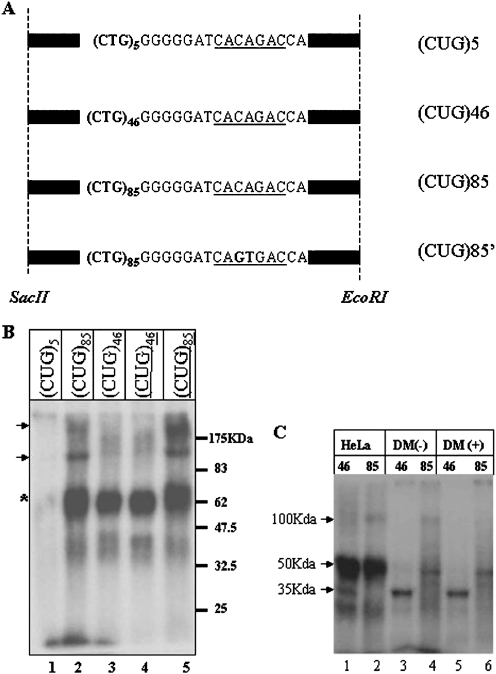
The probes used in the in vitro crosslinking assays consisted of a partial sequence of the 3′-UTR region of the DMPK gene and contained either (CUG)5, (CUG)46 or (CUG)85 repeats and a 3′ splicing branch site (Figure 1A). The DNA was linearized and used as template for in vitro transcription. The radioactively labeled RNA was incubated in a HeLa cell extract followed by UV crosslinking. We were unable to detect specific crosslinking products when the DMPK RNA with five CUG repeats (CUG)5 was used for the assays, despite a previous report that several proteins bind to the CUG elements (Figure 1B, Lane 1) (46). In contrast to (CUG)5 RNA, when the (CUG)46 RNA was used as a probe, we obtained a crosslink to a 50 kDa protein indicated by ‘*’ (lane 3). Interestingly, the amount of crosslinking to this protein was increased using the (CUG)85 RNA probe (lane 2). Additional products were also observed, with approximate molecular weights of 100 and 200 kDa (indicated by the arrowhead in Figure 1B). To aid in the purification of the crosslinked protein(s), we tested crosslinking to RNAs containing biotinylated UTP. The binding patterns of unmodified and biotinylated RNAs were identical, demonstrating that the biotinylated RNA could be used for purification of the bound protein(s) (lanes 4 and 5).
Figure 1.
UV crosslinking of CUG repeat RNAs in HeLa nuclear extracts. (A) RNA clones used. The fragments of the DMPK gene with 5, 46 or 85 CTG repeats were cloned and transcribed in vitro with T7 RNA polymerase. Black bars represent the vector sequence common to all three clones. The 3′ branch site is underlined. (CUG)85′ is the clone with 85 repeats of CUG and a mutated branch site. (B) UV crosslinking using HeLa nuclear extracts. Lane 1, (CUG)5; lane 2, (CUG)85; lane 3, (CUG)46; lane 4, biotinylated (CUG)46 (underlined); lane 5, with biotinylated (CUG)85 (underlined). (C) UV crosslinking in DM extracts. Lanes 1 and 2, HeLa total cell extracts; lanes 3 and 4, total DM1 cell extracts before cell differentiation; lanes 5 and 6, DM1 extracts after differentiation.
To further investigate of the potential biological relevance of the 50 kDa protein in DM1, we performed the UV crosslinking assay using extracts prepared from fetal DM1 myoblasts containing 750 repeats (DM1) (Figure 1C). Extracts were adjusted to similar concentrations prior to the crosslinking assays. The 50 kDa protein crosslinked products were observed with the (CUG)85 probe but not with the (CUG)46 probe when the DM1 cell extract was used (Figure 1C, lanes 3 and 4). We did however observe binding of a 35 kDa protein in both HeLa and DM1 extracts using the (CUG)46 probe. Because the 35 kDa protein did not bind to the longer CUG repeats, it was not investigated further.
HnRNP H can bind and dimerize in the presence of expanded CUG repeats
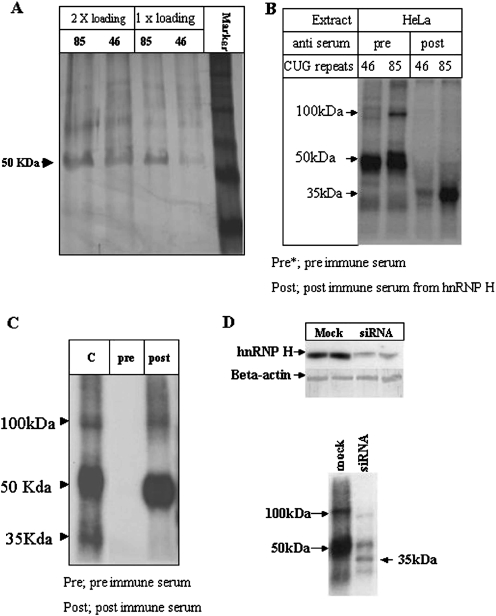
To identify the 50 kDa protein, we prepared a nuclear HeLa cell extract, and the interacting protein was further purified from the extract using affinity purification with biotinylated (CUG)85 RNA. The RNA–protein complex was purified using streptavidin-conjugated beads and the bound proteins were washed and eluted in high salt: the 50 kDa protein eluted in 200 mM salt (Figure 2A). The relative differences in eluted 50 kDa protein from the (CUG)46 versus (CUG)85 RNAs may reflect differences in the binding efficiencies to these two different substrates. The proteins eluted from the biotinylated CUG substrates were excised from the SDS–PAGE gel, eluted and micro-sequenced. The sequences we obtained from three peptides were each derived from hnRNP H (Table 1, for 50 kDa).
Figure 2.
Purification and identification of the CUG repeat binding protein. (A) Purification of the binding protein. The eluted proteins from the RNA affinity column using the (CUG)46 or (CUG)85 RNAs were separated in a SDS–PAGE gel. (B) UV crosslinking assays in extracts treated with pre-immune (pre) or hnRNP H anti-sera (post). (C) The crosslinking products were treated with pre-immune sera (pre, lane 2) or anti-hnRNP H (post, lane 3). (D) The levels of hnRNP H and beta-actin were compared from cells that were mock-transfected or transfected with anti-hnRNP H siRNAs (top panel). The siRNA-treated cell extracts were used in UV crosslinking assays (bottom panel).
Table 1.
Peptides identified by mass spectrometry
| Sequenced protein | Identified hnRNP H peptide sequences |
|---|---|
| 50 kDa | STGEAFVQFASQEIAEK |
| HTGPNSPDTANDGFVR | |
| YGDGGSTFQSTTGHCVHMR | |
| VHIEIGPGR | |
| DLNYCFSGMSDHR | |
| VHIEIGPDGR | |
| 100 kDa | YVEVFK |
| DLNYCFSGMSDHR | |
| VHIEIGPDGR |
Anti-hnRNP H antibodies (a gift from Drs Black and Helfman) were used to confirm the identity of the CUG binding protein (Figure 2B). HeLa cell extracts were treated with either pre-immune or post-immune antisera. UV crosslinking assays performed on the extracts treated with post-immune sera demonstrated loss of the 50 and 100 kDa bands, and revealed enhanced binding of the 35 kDa protein (Figure 2B). Seemingly, there is competitive binding of hnRNP H and the 35 kDa protein for the (CUG)85 RNA, and to a lesser extent to the (CUG)46 RNA. The specific inhibition of 100 kDa complex is suggestive of dimer formation by hnRNP H on this template.
To investigate this hypothesis, we performed immuno-precipitations using hnRNP H antiserum-conjugated beads in HeLa cell extracts UV-crosslinked to the (CUG)85 probe (Figure 2C). Bands migrating at 50 and 100 kDa were generated from this crosslinking whereas the 35 kDa protein was absent, demonstrating the specificity of the antibody. The larger 100 kDa product was then purified and micro-sequenced. As observed previously with the 50 kDa band, all of the sequenced peptides are derived from hnRNP H (Table 1, lower column). To eliminate the possibility of cross-contamination of the 100 kDa protein with the abundant 50 kDa product, the crosslinking was repeated using HeLa cell extracts prepared from cells treated with an anti-hnRNP H siRNA (Figure 2D). If the 100 kDa product is indeed a dimer of hnRNP H, then its expression should also be knocked-down. Northern gel analyses of hnRNP H mRNA in cells treated with the anti-hnRNP H siRNA versus a mock siRNA (scrambled) showed a significant reduction in the amount of hnRNP H mRNA (Figure 2D, top panel). Extracts were prepared from the siRNA-transfected cells and tested in the crosslinking assay (Figure 2D, lower panel). Both the 50 and 100 kDa products were strongly reduced, demonstrating that the 100 kDa crosslinking product requires hnRNP H. Interestingly, binding of the unidentified 35 kDa protein was restored when hnRNP H was depleted from the extracts.
It has been shown that hnRNP H and F interact to form a heterodimer (47). Although our data suggest that hnRNP H itself may dimerize on the longer repeat template (Table 1, Figure 2C and D), it is still possible that the 100 kDa complex is comprised of the two proteins. To test whether or not this is the case, RNAi was used to reduce the level hnRNP F. Although the RNA level of hnRNP F was reduced by 80% (Supplementary figure, left panel), we did not observe an effect on the relative amount of the 100 kDa product in the UV crosslinking assay (Supplementary figure, right panel).
HnRNP H dimerization requires additional cellular factors
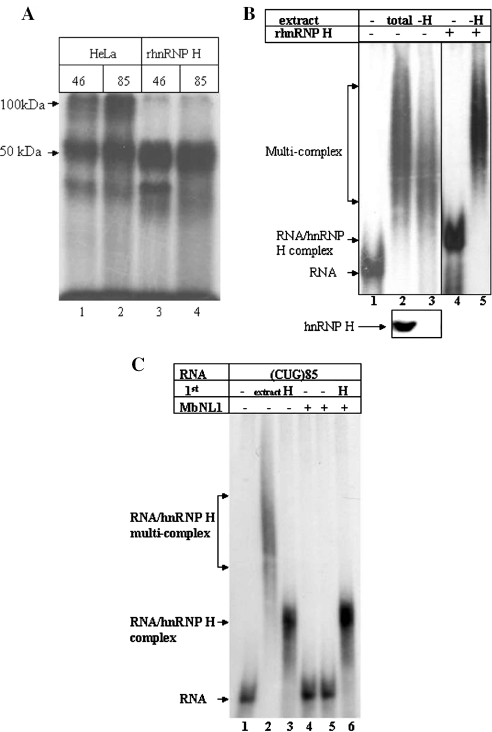
In order to assess the binding requirements of hnRNP H with the DMPK-derived RNA containing CUG repeats, we produced a purified recombinant hnRNP H in bacteria. This protein was capable of forming the 50 kDa complex in our crosslinking assays but did not form the 100 kDa complex (Figure 3A, lanes 3 and 4). These results suggest that the recombinant protein may lack essential post-translational modifications or the dimerization requires one or more nuclear co-factors.
Figure 3.
An additional cellular factor(s) is required for dimer formation of hnRNP H. (A) Recombinant hnRNP H does not dimerize by itself. A UV crosslinking assay was carried out using (CUG)46 or (CUG)85 RNAs incubated in total HeLa cell extracts (lanes 1 and 2) or with recombinant hnRNP H (lanes 3 and 4). (B) A cellular factor is required for hnRNP H dimerization. Lane 1, (CUG)85 RNA alone; lane 2, (CUG)85 RNA incubated with total HeLa cell extract; lane 3, (CUG)85 RNA incubated with a hnRNP H-depleted HeLa cell extract; lane 4, (CUG)85 RNA incubated with 10 ng of recombinant hnRNP H; lane 5, (CUG)85 RNA with hnRNP H immuno-depleted extract to which 10 ng of purified recombinant hnRNP H was added prior to (CUG)85 RNA addition. To confirm immuno-depletion of hnRNP H, the total amount of hnRNP H was compared prior to (lane 2, bottom panel) and following (lane 3, bottom panel) immuno-depletion. (C) Recombinant MBNL1 has no effect on hnRNP H-mediated complex formation. Lane 1, (CUG)85 RNA alone; lane 2, (CUG)85 RNA incubated with total cell extract; lane 3, (CUG)85 RNA incubated with 10 ng of recombinant hnRNP H; lane 4, (CUG)85 RNA incubated with 100 ng of recombinant MBNL1; lane 5, (CUG)85 RNA incubated with 500 ng of MBNL1; lane 6, (CUG)85 RNA incubated with 10 ng of hnRNP H and 500 ng of MBNL1.
We next immuno-depleted a HeLa total cell extract of endogenous hnRNP H. Complex formations were resolved in a native gel assay (Figure 3B). A western blot analysis was performed to assess the depletion of endogenous hnRNP H from the extracts (Figure 3B, bottom panel). When hnRNP H was depleted from the protein extract, there was a reduction in the high molecular weight complex (Figure 3B, lane 3). When the depleted extract was reconstituted with recombinant hnRNP H, formation of the larger complex was restored (Figure 3B, lane 5). These results indicate that the recombinant protein is capable of binding to the substrate RNA, but requires additional cellular factor(s) for the formation of large complexes. MBNL1 and 2 are CUG repeat binding proteins that co-localize with the foci that contain mutant DMPK mRNAs with large CUG repeats (9,10). We next sought to determine if recombinant MBNL1 could facilitate dimerization of hnRNP H. But, as can be seen in Figure 3C, recombinant MBNL1 had no effect on hnRNP H binding or dimerization.
Binding of hnRNP H requires the CUG repeats and a splicing branch point
Based on previous data (Figures 1B, 1C, 2A and 2B), we speculated that the binding of hnRNP H to CUG repeats is proportional to the length of the repeats. The recombinant form of hnRNP H was incubated with different length CUG repeats and complexes were resolved in a native gel (Figure 4A). No binding to the (CUG)5 RNA was observed (lanes 1 and 3). When the (CUG)46 RNA was used in this assay, only a small fraction of the protein was gel-shifted (marked as ‘*’ in lane 6). In contrast, the (CUG)85 RNA was much more gel-shifted (marked as ‘*’ in lane 9). Similar patterns of complex formation were observed for each RNA incubated with total cell extracts (lane 5 versus 8). hnRNP H binds to CUG repeats in a proportional way, which depends on the length of the CUG repeats.
Figure 4.
Binding of hnRNP H to CUG repeats is proportional to the length of the repeats and requires the 3′ splicing branch site of exon 16. (A) lane 1, (CUG)5 RNA only; lane 2, (CUG)5 RNA with total cell extract; lane 3, (CUG)5 with recombinant hnRNP H; lane 4, (CUG)46 RNA only; lane 5, (CUG)46 and total extract; lane 6, (CUG)46 and recombinant hnRNP H; lane 7, (CUG)85 RNA only; lane 8, (CUG)85 and total extract; lane 9, (CUG)85 and recombinant hnRNP H. (B) Binding requires the 3′ branch site. Lane 1, (CUG)85 clone alone; lane 2, (CUG)85 incubated in the total cell extract; lane 3, the RNA was incubated in the presence of 10 ng of recombinant hnRNP H; lane 4, (CUG)85 RNA with the mutated 3′ branch site; lane 5, the mutant RNA with 10 ng of recombinant hnRNP H protein.
The different RNAs used in these assays have CUG repeats as well as a splicing acceptor site derived from a downstream sequence of the DMPK coding region (46). To understand the role of this 3′ branch site in the binding reaction, a mutant (CUG)85 clone containing a mutant form of the branch site was created (marked as (CUG)85′, Figure 1A). Interestingly, the binding of hnRNP H was abolished when the mutant branch site containing probe was used (Figure 4B, lanes 4 and 5). When the mutated RNA was used for the binding assay with the total cell extract, the formation of the large complex was also reduced (data not shown). Our results indicate that CUG repeats and the splicing branch point of Exon 16 are both necessary for hnRNP H binding to the transcripts.
HnRNP H co-localizes to CUG repeat RNAs in vivo
We next wanted to ascertain whether endogenous hnRNP H co-localizes with CUG repeats in patient-derived DM1 cells expressing mutant DMPK transcripts with 750 CUG repeats. An in situ hybridization was performed to reveal both the mutant transcripts (red) and endogenous hnRNP H (green) (Materials and methods) (Figure 5A). Although there was some apparent hnRNP H co-localization with the foci, much of the hnRNP H immunostaining was randomly dispersed throughout the cell nucleus. To determine if binding of hnRNP H to the mutant DMPK transcripts in vivo, a UV crosslinking assay was carried out on DM1 cells (Figure 5B). DM1 myoblasts were UV-irradiated to crosslink protein–RNA interactions and hnRNP H was immuno-purified using the anti-hnRNP H antibody. No hnRNP H was immuno-purified using the pre-immune sera (Figure 5B, lane 1, bottom panel), whereas hnRNP H anti-sera precipitated hnRNP H from both irradiated and non-irradiated cells (Figure 5B, lanes 2 and 3, bottom panel). A phenol extraction was then performed on equal volumes of each immuno-purified sample to isolate bound RNA. The purified RNA was separated in a denaturing gel and hybridized with a radioactively labeled (CAG)10 probe (Figure 5B, top panel). Mutant DMPK mRNA was detected in UV-irradiated samples that were immuno-purified with hnRNP H anti-serum. When the parallel experiment was carried out using normal myoblast cells, no bound RNA was detected in UV-treated immuno-precipitated samples (data not presented). These results are consistent with recent findings by Thornton and colleagues who demonstrated that hnRNP H and F co-localize, to a limited extent, with nuclear foci-containing poly-CUG RNA in DM1 patient brain neurons (48).
Figure 5.
RNA foci of DM1 cells contain hnRNP H. (A) Co-localization assay for hnRNP H and RNA foci in DM1 cell. First column, immuno-staining of endogenous hnRNP H; second column, in situ hybridization with a (CAG)10 probe; third column, superimposed images using a double filter. (B) HnRNP H interacts with CUG repeats in vivo. DM1 myoblast extracts were crosslinked by UV irradiation. HnRNP H in the total extract was immuno-purified using the hnRNP H antibody. HnRNP H-associated RNAs were extracted and resolved in a denaturing gel, blotted to a nylon membrane and probed with a 32P-labeled (CAG)10 DNA (top panel). Lane 1, extract prepared from the UV-irradiated cells was treated with pre-immune sera; lane 2, extract from non-irradiated cells was treated with anti-hnRNP H antibody; lane 3, extract from irradiated cells treated with anti-hnRNP H antisera. To monitor the immuno-purification procedure, an aliquot of the treated samples was analyzed by western blotting (bottom panel).
RNAi-mediated knock-down of hnRNP H expression rescues CUG repeat-containing RNAs from nuclear retention
It has been previously shown that expression of an EGFP gene fused to expanded CUG repeats in myoblasts results in a severe reduction of EGFP expression due to nuclear retention of the transcripts (49). If hnRNP H is in part responsible for nuclear retention, one would expect that knocking-down expression of hnRNP H should restore the EGFP-CUG repeat reporter gene expression. To test this possibility, 293T cells were transfected with the EGFP-CUG reporter gene and several siRNAs (Figure 6A). Transfection of the EGFP-(CTG)5 reporter plasmid and scrambled siRNA in 293T cells resulted in strong EGFP expression indicating that transcripts with only five CUG repeats are readily exported to the cytoplasm and translated (Figure 6A, panel 1). In contrast, the reporter plasmid encoding EGFP-(CTG)85 transfected in the presence of a scramble or anti-EGFP siRNA resulted in only low labels of EGFP expression (Figure 6A, panels 2 and 3). However, EGFP expression was restored when siRNAs directed against hnRNP H were co-transfected with the EGFP-(CTG)85 (Figure 6A, panel 4).
Figure 6.
Suppression of hnRNP H expression can rescue the nuclear retention of RNA with CUG repeats. (A) HEK 293T cells were transfected with either the eGFP-(CUG)5 (Panel 1) or the eGFP-(CUG)85 (Panel 2) reporter genes alone. An irrelevant siRNA (Panel 3) or an anti-hnRNP H siRNA (Panel 4) were co-transfected with the eGFP-(CUG)85 reporter. (B) SiRNA-mediated gene-specific knock-down of hnRNP H (see panels of Figure 6A for lane identities). (C) SiRNA-mediated expression knock-down of hnRNP can restore expression of the eGFP-(CUG)85 reporter gene in primary myoblasts. The left panel shows transfection of myoblasts with an irrelevant siRNA, the right panel shows expression of the reporter in myoblasts transfected with the anti-hnRNP H siRNA.
To insure that the rescue of EGFP expression was a direct effect of the suppression of hnRNP H, aliquots of each cell sample were processed to prepare total RNAs. Northern analysis confirmed downregulation of hnRNP H mRNA by the specific siRNAs (Figure 6B). Parallel experiments were performed using normal myoblasts, but the transfection efficiency was <20% (data not presented). Myoblasts were co-transfected with the EGFP-(CTG)85 reporter gene and each of the siRNAs (Figure 6C). When the myoblasts were transfected with the EGFP-(CTG)85 reporter gene and an irrelevant siRNA, the level of EGFP expression was very low (Figure 6C, left panel). In contrast, cells transfected with the anti-hnRNP H siRNA resulted in rescue of EGFP expression (Figure 6C, right panel). These results strongly support a role for hnRNP H in the nuclear retention of DMPK mutant transcripts.
DISCUSSION
Through this work, we have identified a novel protein involved in binding mutant DMPK mRNA. HnRNP H is not a CUG binding protein per se, since it requires both a CUG expansion containing at least 46 repeats and a distal sequence containing a splicing branch point, which produces a rare splicing isoform harboring Exon 16 of DMPK that does not contain the CUG repeats (46). Under our experimental conditions, we did not detect binding of other known CUG binding proteins, such as the Muscleblind family members or CUG-BP1. Some of the reasons for this could be the use of HeLa cell extracts in conditions that did not favor optimum expression of these proteins, the use of a >10-fold excess of a non-specific RNA competitor or simply our protein–RNA crosslinking method. However, our data show that binding of hnRNP H to mutant DMPK-derived RNAs is specific and enhanced by cellular factors present in cell extracts that have yet to be identified. In the presence of these factors, we observed the formation of a complex migrating ∼100 kDa in the native gel assays. This complex is absent from extracts made from cells transfected with small interfering RNAs directed against hnRNP H or when the extracts have been depleted of endogenous hnRNP H. These results suggest the formation of a complex comprised of the mutant RNA, hnRNP H and unidentified docking molecules present in our protein extracts. The formation of such a large complex in the cell nucleus could be linked to the DMPK mRNA-containing foci present in DM1 cells.
For several years, mouse models of DM1 have been used to elucidate factors involved in DM1 muscle pathogenesis. The first model to show the true involvement of a CUG expansion in DM1 was developed by the Thornton (13) and Gourdon (14,50) laboratories. These mice either expressed the human skeletal actin gene fused to CTG repeats or the complete human DM1 locus. The mice in both these models developed several of the hallmark clinical symptoms of DM1, such as myotonia and myopathy. The typical formation of nuclear foci was also observed in histological sections. What these mice lacked however was the characteristic muscle weakness and wasting of DM1 patients suggesting other factors are involved in the pathogenesis, such as haplo-insufficiency of the DMPK protein, over-expression of CUG-BP1 (30) or reduced expression of the downstream SIX5 gene in humans (51). Recently, a mouse knock-out of the gene that encodes the MBLN1 protein which binds CUG repeats (also CCUG repeats typical of myotonic dystrophy type 2) has been made (33). These mice developed abnormalities in RNA splicing, and eye and muscle defects typical of DM1. These mouse model combined the SIX5 and DMPK knock-out mice provide the great majority of symptoms observed in DM1 ranging from the myotonia to cataract formation and muscle weakness and wasting. However, what is missing from these models is the identification of the factor responsible for mutant DMPK mRNA sequestration in the nucleus of DM1 cells. Some reports have provided evidence that reducing mutant transcript accumulation could restore some molecular features such as the proper alternative-splicing of the insulin receptor in DM1 cells in vitro (11,12).
A compelling set of data that links hnRNP H to DM1 pathogenesis is the RNAi-mediated knock-down of hnRNP H experiments. Experiments with EGFP RNAs containing expanded CUGs previously designed by Amack and Mahadevan (49) showed that these transcripts are retained in the nucleus as measured by reduced EGFP expression. Using a similar approach, we showed that EGFP expression could be restored in cells expressing these mutant RNAs if the level of endogenous hnRNP H is reduced. We performed RNAi experiments in differentiated DM1 myoblasts to assess whether foci number and intensity were reduced in cells depleted of hnRNP H, but poor transfection levels and the inability to identify transfected cells did not allow us to obtain conclusive results with these cells (data not shown).
The mechanisms that underlie DM1 and DM2 pathogenesis are proving to be extremely complex and challenging to comprehend. The pathological CTG and CCTG expansions responsible for causing these diseases create a true gain-of-function that alters cellular metabolism in unpredictable ways, from splicing defects to altering gene expression. In both these diseases, RNA and protein nuclear sequestration seem to be at the root of most these disturbances. Our data suggest that hnRNP H plays a pivotal role in this process.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank the Black lab for the recombinant hnRNP H clone and the anti-hnRNP H antibody, and the Helfman lab for the anti-hnRNP H antibody. D. Kim is a Beckman Fellow. M.-A.L. is the recipient of a doctoral studentship from the Canadian Institutes of Health Research. This work was supported by a grant from the Arnold and Mabel Beckman Foundation, the National Institutes of Health (AI29329 and HL074704), and a grant to J.P. from the Canadian Institutes of Health Research. Funding to pay the Open Access publication charges for this article was provided by NIH funding.
Conflict of interest statement. None declared.
REFERENCES
- 1.Harper P.S. Myotonic Dystrophy. 3rd edn. London: W.B. Saunders; 2001. [Google Scholar]
- 2.Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barcelo J., O'Hoy K., et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 3.Fu Y.H., Pizzuti A., Fenwick R.G., Jr, King J., Rajnarayan S., Dunne P.W., Dubel J., Nasser G.A., Ashizawa T., de Jong P., et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- 4.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T., et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. doi: 10.1016/0092-8674(92)90418-c. [DOI] [PubMed] [Google Scholar]
- 5.Tapscott S.J. Deconstructing myotonic dystrophy. Science. 2000;289:1701–1702. doi: 10.1126/science.289.5485.1701. [DOI] [PubMed] [Google Scholar]
- 6.Filippova G.N., Thienes C.P., Penn B.H., Cho D.H., Hu Y.J., Moore J.M., Klesert T.R., Lobanenkov V.V., Tapscott S.J. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nature Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- 7.Caskey C.T., Swanson M.S., Timchenko L.T. Myotonic dystrophy: discussion of molecular mechanism. Cold Spring Harb. Symp. Quant. Biol. 1996;61:607–614. [PubMed] [Google Scholar]
- 8.Timchenko L.T., Caskey C.T. Trinucleotide repeat disorders in humans: discussions of mechanisms and medical issues. FASEB J. 1996;10:1589–1597. doi: 10.1096/fasebj.10.14.9002550. [DOI] [PubMed] [Google Scholar]
- 9.Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fardaei M., Rogers M.T., Thorpe H.M., Larkin K., Hamshere M.G., Harper P.S., Brook J.D. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 11.Furling D., Doucet G., Langlois M.A., Timchenko L., Belanger E., Cossette L., Puymirat J. Viral vector producing antisense RNA restores myotonic dystrophy myoblast functions. Gene Ther. 2003;10:795–802. doi: 10.1038/sj.gt.3301955. [DOI] [PubMed] [Google Scholar]
- 12.Langlois M.A., Lee N.S., Rossi J.J., Puymirat J. Hammerhead ribozyme-mediated destruction of nuclear foci in myotonic dystrophy myoblasts. Mol Ther. 2003;7:670–680. doi: 10.1016/s1525-0016(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 13.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 14.Seznec H., Agbulut O., Sergeant N., Savouret C., Ghestem A., Tabti N., Willer J.C., Ourth L., Duros C., Brisson E., et al. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum. Mol. Genet. 2001;10:2717–2726. doi: 10.1093/hmg/10.23.2717. [DOI] [PubMed] [Google Scholar]
- 15.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 16.Savkur R.S., Philips A.V., Cooper T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nature. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 17.Charlet B.N., Savkur R.S., Singh G., Philips A.V., Grice E.A., Cooper T.A. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 18.Mankodi A., Takahashi M.P., Jiang H., Beck C.L., Bowers W.J., Moxley R.T., Cannon S.C., Thornton C.A. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 19.Philips A.V., Timchenko L.T., Cooper T.A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 20.Furling D., Marette A., Puymirat J. Insulin-like growth factor I circumvents defective insulin action in human myotonic dystrophy skeletal muscle cells. Endocrinology. 1999;140:4244–4250. doi: 10.1210/endo.140.9.7057. [DOI] [PubMed] [Google Scholar]
- 21.Tian B., White R.J., Xia T., Welle S., Turner D.H., Mathews M.B., Thornton C.A. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA. 2000;6:79–87. doi: 10.1017/s1355838200991544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu X., Timchenko N.A., Timchenko L.T. Cardiac elav-type RNA-binding protein (ETR-3) binds to RNA CUG repeats expanded in myotonic dystrophy. Hum. Mol. Genet. 1999;8:53–60. doi: 10.1093/hmg/8.1.53. [DOI] [PubMed] [Google Scholar]
- 23.Timchenko L.T., Miller J.W., Timchenko N.A., DeVore D.R., Datar K.V., Lin L., Roberts R., Caskey C.T., Swanson M.S. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timchenko N.A., Welm A.L., Lu X., Timchenko L.T. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPbeta mRNA and regulates translation of C/EBPbeta isoforms. Nucleic Acids Res. 1999;27:4517–4525. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhagwati S., Ghatpande A., Leung B. Identification of two nuclear proteins which bind to RNA CUG repeats: significance for myotonic dystrophy. Biochem. Biophys. Res. Commun. 1996;228:55–62. doi: 10.1006/bbrc.1996.1615. [DOI] [PubMed] [Google Scholar]
- 26.Kino Y., Mori D., Oma Y., Takeshita Y., Sasagawa N., Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- 27.Roberts R., Timchenko N.A., Miller J.W., Reddy S., Caskey C.T., Swanson M.S., Timchenko L.T. Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc. Natl Acad. Sci. USA. 1997;94:13221–13226. doi: 10.1073/pnas.94.24.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timchenko N.A., Patel R., Iakova P., Cai Z.J., Quan L., Timchenko L.T. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 29.Timchenko N.A., Iakova P., Cai Z.J., Smith J.R., Timchenko L.T. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol. Cell. Biol. 2001;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timchenko N.A., Cai Z.J., Welm A.L., Reddy S., Ashizawa T., Timchenko L.T. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 31.Ho T.H., Charlet B.N., Poulos M.G., Singh G., Swanson M.S., Cooper T.A. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dansithong W., Paul S., Comai L., Reddy S. MBNL1 is the primary determinant of focus formation and aberrant IR splicing in DM1. J. Biol. Chem. 2004;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 33.Kanadia R.N., Johnstone K.A., Mankodi A., Lungu C., Thornton C.A., Esson D., Timmers A.M., Hauswirth W.W., Swanson M.S. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 34.Fardaei M., Larkin K., Brook J.D., Hamshere M.G. In vivo co-localization of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buratti E., Baralle M., De Conti L., Baralle D., Romano M., Ayala Y.M., Baralle F.E. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHbeta genes. Nucleic Acids Res. 2004;32:4224–4236. doi: 10.1093/nar/gkh752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caputi M., Zahler A.M. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 2002;21:845–855. doi: 10.1093/emboj/21.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C.D., Kobayashi R., Helfman D.M. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arhin G.K., Boots M., Bagga P.S., Milcarek C., Wilusz J. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 2002;30:1842–1850. doi: 10.1093/nar/30.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bagga P.S., Arhin G.K., Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 1998;26:5343–5350. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ordway J.M., Detloff P.J. In vitro synthesis and cloning of long CAG repeats. Biotechniques. 1996;21:609–610. doi: 10.2144/96214bm08. [DOI] [PubMed] [Google Scholar]
- 41.Markovtsov V., Nikolic J.M., Goldman J.A., Turck C.W., Chou M.Y., Black D.L. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D.H., Edwalds-Gilbert G., Ren C., Lin R.J. A mutation in a methionine tRNA gene suppresses the prp2-1 Ts mutation and causes a pre-mRNA splicing defect in Saccharomyces cerevisiae. Genetics. 1999;153:1105–1115. doi: 10.1093/genetics/153.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D.H., Behlke M.A., Rose S.D., Chang M.S., Choi S., Rossi J.J. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 44.Taneja K.L. Localization of trinucleotide repeat sequences in myotonic dystrophy cells using a single fluorochrome-labeled PNA probe. Biotechniques. 1998;24:472–476. doi: 10.2144/98243rr02. [DOI] [PubMed] [Google Scholar]
- 45.O'Brien R.M., Lucas P.C., Yamasaki T., Noisin E.L., Granner D.K. Potential convergence of insulin and cAMP signal transduction systems at the phosphoenolpyruvate carboxykinase (PEPCK) gene promoter through CCAAT/enhancer binding protein (C/EBP) J. Biol. Chem. 1994;269:30419–30428. [PubMed] [Google Scholar]
- 46.Tiscornia G., Mahadevan M.S. Myotonic dystrophy: the role of the CUG triplet repeats in splicing of a novel DMPK exon and altered cytoplasmic DMPK mRNA isoform ratios. Mol. Cell. 2000;5:959–967. doi: 10.1016/s1097-2765(00)80261-0. [DOI] [PubMed] [Google Scholar]
- 47.Chou M.Y., Rooke N., Turck C.W., Black D.L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang H., Mankodi A., Swanson M.S., Moxley R.T., Thornton C.A. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 49.Amack J.D., Mahadevan M.S. The myotonic dystrophy expanded CUG repeat tract is necessary but not sufficient to disrupt C2C12 myoblast differentiation. Hum. Mol. Genet. 2001;10:1879–1887. doi: 10.1093/hmg/10.18.1879. [DOI] [PubMed] [Google Scholar]
- 50.Gourdon G., Radvanyi F., Lia A.S., Duros C., Blanche M., Abitbol M., Junien C., Hofmann-Radvanyi H. Moderate intergenerational and somatic instability of a 55-CTG repeat in transgenic mice. Nature Genet. 1997;15:190–192. doi: 10.1038/ng0297-190. [DOI] [PubMed] [Google Scholar]
- 51.Sarkar P.S., Appukuttan B., Han J., Ito Y., Ai C., Tsai W., Chai Y., Stout J.T., Reddy S. Heterozygous loss of Six5 in mice is sufficient to cause ocular cataracts. Nature Genet. 2000;25:110–114. doi: 10.1038/75500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.