Abstract
We previously reported the identification of a novel nuclear compartment detectable in heat-shocked HeLa cells that we termed stress-induced Src-activated during mitosis nuclear body (SNB). This structure is the recruitment center for heat shock factor 1 and for a number of RNA processing factors, among a subset of Serine-Arginine splicing factors. In this article, we show that stress-induced SNBs are detectable in human but not in hamster cells. By means of hamster>human cell hybrids, we have identified three human chromosomes (9, 12, and 15) that are individually able to direct the formation of stress bodies in hamster cells. Similarly to stress-induced SNB, these bodies are sites of accumulation of hnRNP A1-interacting protein and heat shock factor 1, are usually associated to nucleoli, and consist of clusters of perichromatin granules. We show that the p13-q13 region of human chromosome 9 is sufficient to direct the formation of stress bodies in hamster>human cell hybrids. Fluorescence in situ hybridization experiments demonstrate that the pericentromeric heterochromatic q12 band of chromosome 9 and the centromeric regions of chromosomes 12 and 15 colocalize with stress-induced SNBs in human cells. Our data indicate that human chromosomes 9, 12, and 15 contain the nucleation sites of stress bodies in heat-shocked HeLa cells.
INTRODUCTION
Stress treatments trigger a complex modification of the cell metabolism, including arrest of DNA replication, transcription, RNA processing, and translation. At the same time, they activate the synthesis of a small number of ubiquitous heat shock proteins that are necessary for cell survival (Lindquist, 1986; Morimoto, 1993). At the morphological level, this profound alteration of the cell metabolism is accompanied by drastic rearrangements of the nuclear structure (O'Keefe et al., 1994; Tani et al., 1996).
Among the rearrangements occurring in heat-shocked cells, we and others have previously reported the formation of novel nuclear compartments termed either stress bodies or hnRNP A1-interacting protein (HAP) bodies (Jolly et al., 1997; Chiodi et al., 2000). HAP bodies usually lie in proximity of the nucleoli and do not coincide with other nuclear compartments such as nucleoli, Cajal bodies, kinetochores, promyelocytic leukemia bodies, or the speckles enriched in splicing factors (Jolly et al., 1997, 1999; Chiodi et al., 2000). Although the composition of these nuclear compartments is still largely undefined, it is known that they contain heat shock factor 1 (HSF1) and numerous RNA binding proteins, including heterogeneous nuclear ribonucleoprotein (hnRNP) HAP, hnRNP M, Src-activated during mitosis (Sam68), and a subset of SR (Serine–Arginine) splicing factors (Jolly et al., 1999; Weighardt et al., 1999; Denegri et al., 2001). To account for the close relationship with Sam68 nuclear bodies (SNBs) detectable in unstressed cells, we have recently proposed to rename HAP bodies “stress-induced SNBs” (Denegri et al., 2001). Although the appearance of stress-induced SNBs temporally coincides with the expression of heat shock genes, they are not sites of transcription (Jolly et al., 1997, 1999; Chiodi et al., 2000). RNA synthesis, however, is required for the formation of these structures and, in fact, the recruitment of hnRNP HAP is efficiently prevented by RNA polymerase inhibitors such as actinomycin D and 5,6-dichlorobenzimidazole riboside (Weighardt et al., 1999). The role of RNA is further indicated by the fact that stress-induced SNBs correspond to cluster of perichromatin granules, namely a highly packed form of ribonucleoprotein complexes (Fakan, 1994) that are recruited from the whole nucleoplasm (Chiodi et al., 2000). On the other hand, not all the components of these bodies are recruited in a transcription-dependent manner. This is the case of HSF1, which has been shown to form stress bodies even in transcriptionally inactive mitotic cells (Jolly et al., 1999). Contrary to interphase cells in which a substantial cell-to-cell variation in the number of HSF1 bodies is observed, mitotic cells always display four HSF1 bodies that range in size from 0.3 to 1.4 μm. These bodies are associated with mitotic chromosomes, suggesting that specific, even although still unidentified, chromosomal loci could serve as recruitment centers for HSF1 (Jolly et al., 1999).
It is known that many nuclear subdomains are dynamically associated with specific genetic loci. This is exemplified by the association between the nucleolus and the chromosomal domains that contain the ribosomal genes. In humans, ribosomal genes are distributed on five chromosomal loci called nucleolar organizer regions or NORs, which are located on the short arm of acrocentric Homo sapiens (HSA) chromosomes (HSA13, 14, 15, 21, and 22) (Henderson et al., 1972). On exit from mitosis, mini-nucleoli form around individual NORs and then fuse into large nucleoli, incorporating both active and silent NORs (Ochs et al., 1985; Sullivan et al., 2001). Cajal bodies provide another example of an association between nuclear bodies and specific gene loci. Indeed, Cajal bodies have been recently shown to bind to the histone gene clusters (Frey and Matera, 1995) and to the clusters of genes encoding the U1, U2, and U3 snRNA (Matera, 1998). In all cases, the association is mediated by nascent transcripts (Frey et al., 1999). Colocalization with specific chromosomal domains has also been reported in the case of some transcription factors. Indeed, transcription factor X-linked a-thalassaemia/mental retardation colocalizes with pericentromeric heterochromatin and with the short arms of acrocentric chromosomes (McDowell et al., 1999), whereas PSE-binding transcription factor and Oct1 transcription factors preferentially associate with a small region on chromosome 6 and with chromosome 7 in a cell-cycle–dependent manner (Pombo et al., 1998). It has been suggested that, similar to the nucleoli, these domains may bring particular genes to a region where the appropriate transcription and processing factors are concentrated, thereby facilitating gene expression.
In this article, we investigated the relationship between stress-induced SNBs and chromatin domains, and we identified three human chromosomes that are involved in the formation of these nuclear bodies. Moreover, we show that stress-induced SNBs colocalize with the heterochromatic regions on human chromosomes 9, 12, and 15.
MATERIALS AND METHODS
Cell Lines
HeLa cells were grown in DMEM (Sigma, St. Louis, MO), 10% fetal bovine serum, 50 μg/ml gentamicin, and 2 mM l-glutamine. B14-150, Chinese hamster ovary cells, and hamster>human somatic cell hybrids were grown in RPMI-1640 (Sigma), 10% fetal bovine serum, 50 μg/ml gentamicin, and 2 mM l-glutamine. Hamster>human somatic cell hybrids are listed in Table 1 (Rocchi et al., 1986). Characterization of the hybrids was refined by reverse painting (Antonacci et al., 1995). Hamster cells containing a single copy of a human supernumerary chromosome monochromosomic hybrid (MCH cells) were previously described (Raimondi et al., 1991). We verified by fluorescence in situ hybridization (FISH) the presence of the correct human chromosomes in monochromosomal hamster>human cell hybrids (our unpublished results). Heat shock (1 h at 42°C followed by 1 h at 37°C) was performed in complete medium supplemented with 40 mM HEPES buffer (Sigma).
Table 1.
Formation of stress-induced SNBs in hamster>human cell hybrids
| Hybrid | Chromosomes
|
Number of
bodies
|
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | X | Y | 0 | 1 | >1 | |
| B14-150 | |||||||||||||||||||||||||||
| HY-166T4 | X | X | X | X | X | X | 19 | 54 | 27 | ||||||||||||||||||
| HY-8F6 | X | X | X | X | X | X | X | X | X | X | X | X | 49 | 23 | 28 | ||||||||||||
| YXY-95S | o | o | o | o | o | ||||||||||||||||||||||
| HY-3124E | X | X | X | X | X | X | X | 14 | 40 | 46 | |||||||||||||||||
| HY-1916 | X | X | X | X | X | X | X | X | 23 | 17 | 60 | ||||||||||||||||
| HY-75E1 | X | X | X | X | X | X | 15 | 26 | 59 | ||||||||||||||||||
| GM-10501 | X | X | X | X | X | X | X | X | X | 25 | 40 | 35 | |||||||||||||||
| GM-10662A | o | o | o | o | o | o | o | ||||||||||||||||||||
| GM-10156C | o | ||||||||||||||||||||||||||
| GM-10479A | o | ||||||||||||||||||||||||||
| GM-11010A | o | ||||||||||||||||||||||||||
| GM-13140 | o | ||||||||||||||||||||||||||
| GM-10611A | X | 35 | 65 | ||||||||||||||||||||||||
| Y-E210TC | X | 17 | 83 | ||||||||||||||||||||||||
| GM-11418 | X | 25 | 75 | ||||||||||||||||||||||||
The human chromosome arrays in eight different multi-chromosomal and seven monochromosomal hamster>human cell hybrids are indicated. Chromosomes are scored either by X or by o, depending on the ability of the hybrid clone to form stress bodies as determined by indirect immunofluorescence with anti-HAP antibodies. For each positive cell hybrid clone, the percentage of cells without or with one or more than one stress bodies is indicated on the right side of the Table. These figures were calculated on an average of three independent experiments counting at least 500 cells.
Indirect Immunofluorescence
Cells grown on coverslips were washed once with phosphate-buffered saline (PBS), fixed for 7 min in 4% formaldehyde, and subsequently permeabilized in 0.5% Triton X-100 for 7 min on ice. Primary antibodies were diluted at working concentration in PBS containing 5% skimmed milk (Difco, Detroit, MI) and were then added to the coverslips. Primary antibodies used were affinity-purified rabbit anti-HAP polyclonal antibody (Weighardt et al., 1999), and rat anti-HSF1 monoclonal antibody (mAb) 10H8 (NeoMarkers, Fremont, CA). After 1 h at 37°C in a humid chamber, coverslips were washed three times with PBS. Secondary antibodies used were rhodamine-conjugated goat anti-rabbit immunoglobulin (Ig)G antibody (Jackson ImmunoResearch Laboratories, West grove, PA) and fluorescein isothiocyanate (FITC)-conjugated goat anti-rat IgG antibody (Sigma). Secondary antibodies were diluted at the final concentration recommended by the supplier in PBS-made 5% skimmed milk and were added to coverslips. After 1 h at 37°C in a humid chamber, coverslips were washed three times with PBS, rinsed, and mounted in 90% glycerol in PBS. Confocal microscopy was performed with a TCS-NT digital scanning confocal microscope (Leica, Deerfield, IL) equipped with a 63X/NA = 1,32 oil immersion objective. We used the 488 nm laser line for excitation of FITC (detected at 500 nm < λFITC < 540 nm) and the 543 nm laser line for the rhodamine fluorescence (detected at >590 nm). The pinhole diameter was kept at 1 μm. Images were exported to Adobe Photoshop (Adobe Systems, Mountain View, CA).
Electron Microscopy Analysis
HeLa cells and monochromosomal GM-10611A hamster>human cell hybrids containing HSA9 were harvested by trypsinization, immediately fixed in 4% formaldehyde (2 h at 4°C) in the culture medium, and were then incubated in 2% OsO4 for 1 h at room temperature. Cell pellets were embedded in agar (2% in H2O), rinsed several times with S-rensen buffer (pH 7.2), and dehydrated in ethanol. Finally, the cells were embedded in LR White resin and polymerized at 60°C for 24 h. Thin sections from formaldehyde-fixed cells were collected on nickel grids covered with a Formvar-carbon film and stained with the EDTA technique (Bernhard, 1969). Specimens were observed with a EM900 electron microscope (Zeiss, Jena, Germany) equipped with a 30-μm objective aperture and operating at 80 kV.
Western Blot Analysis
Western blot analysis was performed on total extracts prepared from human HeLa and from hamster B14-150 and HY-1916 cells as previously described (Montecucco et al., 2001) using affinity-purified anti-HAP rabbit antibodies (Weighardt et al., 1999) and GTU-88 mAb to γ-tubulin (Sigma). Primary antibodies were revealed with horseradish peroxidase-conjugated goat anti-rabbit antibodies and enhanced chemiluminescence system (Amersham, Buckinghamshire, UK). BenchMark prestained protein ladder (Life Technologies, Milano, Italy) was used as molecular weight markers. To better appreciate differences in molecular size, proteins were resolved onto a 7.5% SDS-PAGE. Competition experiments were performed as described by Sambrook et al. (1989) using bacteria expressing the glutathione S-transferase-23 recombinant antigen used for the production of anti-HAP antibodies (Weighardt et al., 1999).
FISH
The following probes were used: pHuR98 specific for a satellite III DNA subfamily on HSA9 (Rocchi et al., 1991); pDMX1, pBR12, and pMC15 hybridizing to α-satellite sequences on X-chromosome; HSA12; and HSA15 (Baldini et al., 1990; Archidiacono et al., 1995). Probes were labeled with biotinylated-16-dUTP (Roche Molecular Biochemicals, Indianapolis, IN) using a nick-translation system (Life Technologies) according to the protocol provided by the supplier. The labeled probes were resuspended in hybridization buffer (50% formamide, 10% Dextran sulfate, 1× Denhardt's solution, 0.1% SDS, 40 mM Na2HPO4, pH 6.8, and 2× SSC) at a final concentration of 5 μg/ml and they were denatured at 70°C for 10 min.
FISH on metaphase spreads was carried out essentially as previously reported (Raimondi et al., 1996). For FISH analysis on interphase heat-shocked HeLa cells, cells were rinsed with PBS and fixed in 4% formaldehyde for 15 min at room temperature. After washing three times for 15 min each at room temperature in PBS, cells were permeabilized with 0.5% Triton X-100 for 7 min at 4°C. Slides were rinsed twice in PBS, air-dried, and the cells were denatured in 70% formamide/2× SSC. Hybridization was performed overnight at 42°C in 20 μl of hybridization buffer containing 5 μg/ml denatured probe. Stringent washings were performed in 50% formamide/2× SSC at 42°C, and 0.1× SSC at 60°C. After in situ hybridization, cells were washed three times in PBS and incubated with anti-HAP polyclonal antibody and with FITC-conjugated avidin DCS (Vector Laboratories, Peterborough, UK) for 1 h at room temperature in PBS-rendered 5% skimmed milk. Cells were then rinsed in PBS and then incubated with rhodamine-conjugated anti-rabbit IgG goat antibodies (Jackson ImmunoResearch Laboratories) with biotin-conjugated antiavidin D antibody (Vector Laboratories) and finally with FITC-conjugated avidin. Slides were counterstained with 4,6-diamidino-2-phenylindole (0.01 μg/ml) and mounted in 90% glycerol in PBS containing 2% DABCO antifade [1,4 diazobicyclo-(2.2.2) octane; Sigma]. Confocal microscopy was performed with a TCS-NT digital scanning confocal microscope (Leica).
RESULTS
Human Chromosomes Confer to Hamster Cells the Ability to Form Stress-Induced Nuclear Bodies
Stress-induced SNBs are exclusively detectable in human cells. Indeed, we failed to observe similar structures in a number of non-human cell lines, including green monkey Cos7 and mouse NIH-3T3 cells (our unpublished results), indicating that specific human DNA sequences or genes could be involved in the formation of these bodies. We decided to exploit this species specificity to identify, by means of hamster>human somatic cell hybrids, human chromosomes able to direct the assembly of stress bodies in hamster cells. For the sake of clarity, hereafter we will use “stress-induced SNBs” and “stress bodies” to refer to the structures detectable, respectively, in human cells and in hamster>human cell hybrids.
We initially tested the rabbit antibodies against human HAP in Western blot analysis of extracts of hamster B14-150 cells. As shown in Figure 1, these antibodies recognized a single protein band with an apparent molecular mass of ∼110 kDa, significantly smaller than human HAP (∼150 kDa, 917 amino acids) (Weighardt et al., 1999). The recognition was specific because it was competed by the recombinant glutathione S-transferase-HAP antigen (Figure 1). Although we presently do not know the reason of this difference in size, the fact that a minor band with the same electrophoretic mobility of the hamster protein is detectable also in HeLa cells raises the possibility that it could originate from alternative splicing. Similar to human HAP, the hamster protein (HAP*) had a punctuated distribution in the cell nucleus with exclusion of nucleoli (Figure 2). However, in B14-150 hamster cells, neither a moderate heat shock (1 h at 42°C followed by 1 h recovery at 37°C, Figure 2) nor a more drastic treatment (1 h at 45°C, our unpublished results) caused the recruitment of HAP* to stress bodies.
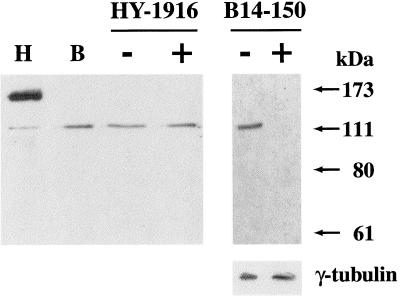
Figure 1.
Western blotting analysis of hamster B14-150 cells with anti-HAP antibodies. (Left panel) Total extracts of human HeLa cells (H) hamster B14-150 cells (B) and hamster>human somatic cell hybrid HY-1916 were fractionated onto a 7.5% SDS-PAGE and analyzed in Western blotting with an immunopurified polyclonal antibody directed against the recombinant human HAP protein (Weighardt et al., 1999). HY-1916 cells were analyzed both before (−) and after (+) heat shock. (Right panel) To prove the antibody specificity, Western blotting analysis of B14-150 cells was also performed in the absence (−) or in the presence (+) of purified recombinant antigen (see “Materials and Methods”). The same filter was hybridized with the G.T.U-88 mAb to γ-tubulin as a control for protein loading. The antibody–antigen complex was revealed with a peroxidase-conjugated anti-rabbit antibody and the enhanced chemiluminescence system. Molecular mass markers are indicated.
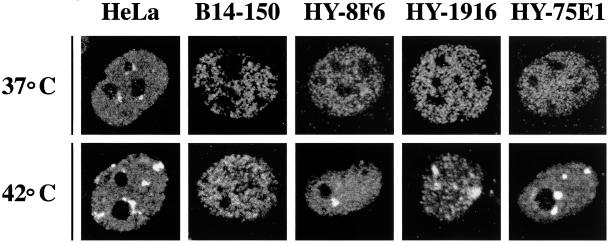
Figure 2.
The heat shock-induced redistribution of HAP in hamster cells requires specific human chromosomes. Human HeLa cells, hamster B14-150 recipient cells, and HY-8F6, HY-1916, and HY-75E1 multi-chromosomal hamster>human cell hybrids were fixed with formaldehyde and analyzed by indirect immunofluorescence with anti-HAP rabbit polyclonal antibodies. The antigen–antibody complex was revealed with a rhodamine-conjugated sheep anti-rabbit secondary antibody. HAP distribution was revealed by confocal laser microscopy both in unstressed (37°C) and in heat-shocked (42°C) cells.
We next analyzed whether stress bodies could be detectable with anti-HAP antibodies in hamster>human somatic cell hybrids. We initially tested a panel of eight multi-chromosomal cell hybrids altogether accounting for the whole set of human chromosomes (Table 1). Nuclear bodies, similar to those observed in heat-shocked HeLa cells, were detected in six cell hybrids incubated 1 h at 42°C and allowed to recover 1 h at 37°C. We concluded that the human chromosomes present in the remaining two hybrids YXY-95S and GM-10662A, namely HSA 2, 3, 6, 7, 8, 13, 14, 16, 21, and X, were unable to direct the formation of these structures (Table 1). Differences, however, existed between the six positive hybrids concerning the number and size of stress bodies, suggesting that some chromosomal sets were more compatible with the formation of these structures. In particular, the recruitment of HAP* was very efficient in two hybrid clones, HY-1916 and HY-75E1, in which multiple bodies were detectable in most of the cells (Figure 2). It is worth noticing that none of these two clones contains HSA19 on which the single human HAP gene has been mapped (DuPont et al., 1997), ruling out the possibility that the formation of stress bodies is due to the expression of the human protein in hamster cells. In fact, Western blot analysis showed that the electrophoretic mobility of HAP* was identical in B14-150 and in YH-1916 and was not affected by stress (Figure 1).
Individual Human Chromosomes Direct the Formation of Stress Bodies in Hamster Cells
The analysis in the previous section demonstrated that certain sets of human chromosomes could confer to hamster cells the ability to form stress bodies in response to heat shock. We next investigated whether the same result could be obtained with single chromosomes.
We observed that HSA12 was shared by all the hybrids displaying stress bodies. To assess whether this chromosome was responsible for the assembly of these structures, we analyzed Y-E210TC cells, which contained a single copy of HSA12 as the only human chromosome. As shown in Table 1, stress bodies were detectable in 83% of heat-shocked Y-E210TC cells. However, contrary to what was observed with multi-chromosomal hamster>human cell hybrids, a single body was detectable in Y-E210TC cells, suggesting that additional human chromosomes could be involved in the redistribution of HAP*. To verify this hypothesis, we analyzed the distribution of HAP* in a number of monochromosomal cell hybrids (Table 1). Stress bodies were not observed in GM-10156C and GM-10479A cells containing, respectively, only HSA7 and HSA14, namely two chromosomes excluded on the basis of the analysis in the previous section. Also, no redistribution of HAP* was observed in hybrids containing HSA18 and HSA20 (GM-11010A and GM-13140), which were instead present in multi-chromosomal hamster>human somatic cell hybrids that scored positive (Table 1). On the contrary, stress bodies occurred in most of the cells containing a single copy of HSA15 or HSA9 (GM-11418 and GM-10611A in Table 1), two of the chromosomes present in the hamster>human hybrids with multiple bodies. Although the different number of stress bodies in GM10501 and HY1916 cells (Table 1), which contain both HSA12 and 15, seems to suggest the involvement of additional chromosomes, our analysis demonstrates that at least three human chromosomes (HSA9, 12, and 15) can individually direct the recruitment of HAP* to stress bodies after heat shock.
Stress Bodies in Hamster>Human Cell Hybrids Are Similar to Stress-Induced SNBs
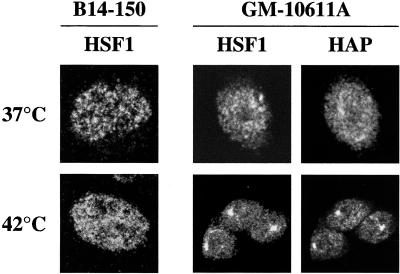
To understand whether the stress bodies in hamster>human hybrids were comparable with stress-induced SNBs observed in human cells, we investigated in more detail the nature of these structures. We initially determined the distribution of HSF1, another protein recruited to stress-induced SNBs (Jolly et al., 1997; Weighardt et al., 1999). As shown in Figure 3, HSF1 was distributed throughout the nuclear volume of parental B14-150 hamster cells both before and after heat shock. Because HSF1 is recruited to stress-induced SNBs before HAP (Jolly et al., 1999; Weighardt et al., 1999), this result indicates that hamster cells are defective for the formation of the whole structure and not simply for the recruitment of HAP*. The presence of HSA9 in hamster>human hybrids (GM-10611A in Figure 4) was sufficient to direct the recruitment of both HSF1 and HAP* to stress bodies. An identical result was obtained with hybrid cells containing HSA12 or HSA15 (our unpublished results), indicating that each of these three chromosomes provided all the elements necessary for the formation of these structures.
Figure 3.
Human chromosome 9 directs the recruitment of HAP and HSF1 to stress-induced SNBs in hamster cells. Unstressed (37°C) and heat-shocked (42°C) recipient B14-150 hamster cells and monochromosomal GM-10611A hamster>human cell hybrids containing HSA 9 were analyzed by indirect immunofluorescence with the anti-HSF1 10H8 mAb and the anti-HAP polyclonal antibody. The distribution of HSF1 and HAP proteins was revealed with FITC-conjugated goat anti-rat IgG antibody and rhodamine-conjugated goat anti-rabbit IgG antibody, respectively. Confocal laser images of the same GM-10611A cells stained with anti-HSF1 and anti-HAP antibodies are shown.
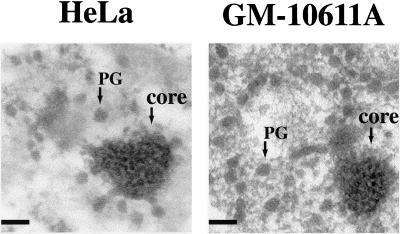
Figure 4.
Ultrastructural analysis of stress bodies in human cells and hamster>human cell hybrids. HeLa cells and GM-10611A hamster>human hybrid were heat shocked at 42°C for 1 h and after 1 h of recovery at 37°C, were fixed with formaldehyde, stained with the EDTA technique, and analyzed in electron microscopy. The dense “core” of the stress bodies and the clustering of perichromatin granules are detectable in both cell lines. Scale bars, 0.1 μm.
We next analyzed the ultrastructure of the bodies present in GM-10611A cells containing HSA9. As shown in Figure 4, similar to stress-induced SNBs in HeLa cells (Chiodi et al., 2000), they consisted of clusters of perichromatin granules. These structures were also detectable in cell hybrids containing HSA12 or HSA15, but not in parental B14-150 hamster cells (our unpublished results).
Thus, our analysis indicates that at least three different human chromosomes can, individually, confer to hamster cells the ability to form nuclear stress bodies with the same protein composition and ultrastructure of stress-induced SNBs described in human cells.
Stress-Induced SNBs Colocalize with the Pericentromeric Heterochromatin of HSA9
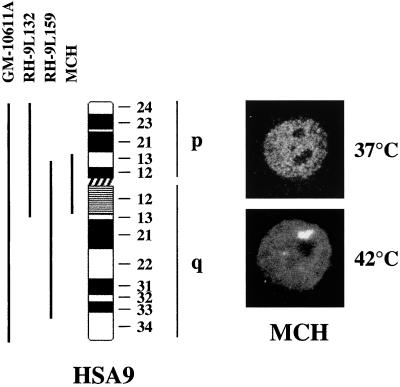
To define more precisely the chromosomal domain involved in the formation of stress bodies, we analyzed two cell hybrids containing, respectively, most of the long arm of HSA9 along with the p13-cen region (RH-9L159 in Figure 5) or the entire short arm and the q13-cen region (RH-9L132 in Figure 5) of the same chromosome. After heat shock, stress bodies were detectable in both cell hybrids, thus mapping the region involved in this process to the p13 to q13 of HSA9 shared by the two chromosomal fragments (our unpublished results). To confirm this conclusion, we took advantage of the fact that a human supernumerary mini-chromosome spanning the 9p13-q13 region had been previously characterized in our laboratory (Raimondi et al., 1991). MCH cells containing a single mini-chromosome as the only human chromosome (Raimondi et al., 1996) were therefore challenged for the ability to form stress bodies after heat shock. As shown in Figure 5, indirect immunofluorescence analysis showed that stress bodies were, indeed, detectable in MCH cells.
Figure 5.
Ideogram of HSA9. The different chromosomal fragments present in GM-10611A, RH-9L132, RH-9L159, and in MCH hamster>human cell hybrids are shown. On the right part of the figure are shown confocal laser microscopy images of MCH cells, either unstressed (37°C) or heat shocked (42°C), stained with anti-HAP antibodies and rhodamine-conjugated goat anti-rabbit IgG antibodies.
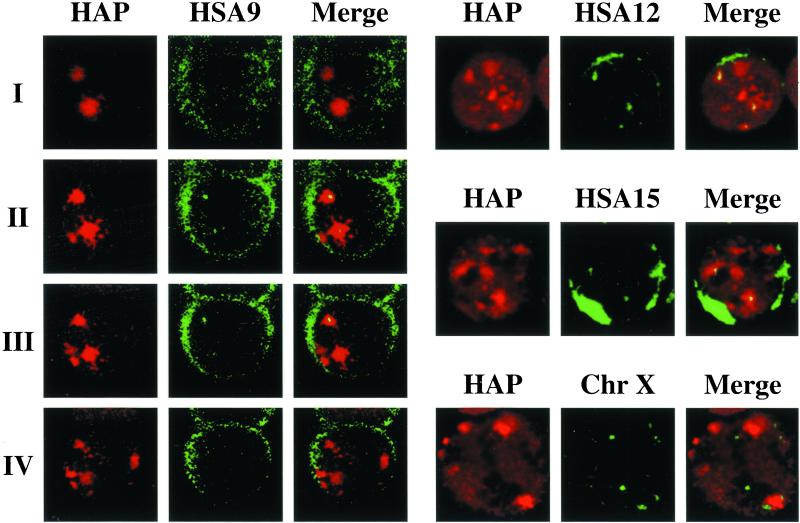
A distinguishing feature of this portion of HSA9 is the presence of an extended heterochromatic region that forms the pericentromeric q12 band and is composed of arrays of different types of satellite DNA (Finelli et al., 1996). Because satellite DNAs are among the most divergent sequences in evolution, we hypothesized that they could correspond to the missing genetic elements required for the formation of stress bodies in hamster cells. Moreover, taking into account the fact that heterochromatic regions are thought transcriptionally silent and that stress-induced SNBs do not correspond to sites of transcription (Chiodi et al., 2000), we reasoned that this pericentromeric heterochromatic region could act as a scaffold for the assembly of these structures. To verify this hypothesis, we studied the distribution of the q12 band (7–8 Mb) of HSA9 relative to that of stress-induced SNBs. HeLa cells were heat shocked and hybridized to the biotinylated pHuR98 probe directed to a subfamily of satellite III DNA specific for the heterochromatic region of HSA9 (Rocchi et al., 1991). On mitotic chromosomal spreads, this probe specifically decorated, as expected, the pericentromeric region of HSA9 (Rocchi et al., 1991), and most of the cells (87%) contained two or three copies of this chromosome (not shown). Cells were costained with anti-HAP antibodies and were analyzed by confocal laser microscopy (see “Materials and Methods”). In most of the cells (scored as positive in Table 2), at least one hybridization signal colocalized with a stress body, and the colocalization extended through successive optical sections (Figure 6). In the remaining cells, the hybridization signals were neither embedded nor adjacent to the stress bodies, and these cells were therefore scored as negative (Table 2). The chi-square test indicated that the association between HSA9 and stress-induced SNBs was statistically significant under the null hypothesis of a 50% association. As a control, we studied the spatial relationship between HAP bodies and the centromeric region of X chromosome that, on the basis of the data in Table 1, did not appear to be involved in the formation of these bodies. As exemplified in Figure 6, most the hybridization signals obtained with the pDMX1 probe, directed against the X chromosome specific α-satellite (Archidiacono et al., 1995), did not colocalize with stress-induced SNBs. The significant chi-square test of the data in Table 2, under the same null hypothesis as above, confirmed the absence of colocalization between the X chromosome and stress bodies.
Table 2.
Frequency of association of specific chromosomal bands with stress-induced SNBs
| Chromosome | Probe | Association with
stress-induced SNBs (percentage of cells)
|
||
|---|---|---|---|---|
| + | − | n | ||
| 9 | Sat III | 75a | 25 | 279 |
| 12 | Sat α | 70b | 30 | 64 |
| 15 | Sat α | 80c | 20 | 58 |
| X | Sat α | 24d | 76 | 96 |
Stress-induced SNBs and interphase territories were labeled, optical sections through whole nuclei were collected at 0.5 μm intervals, and overlap between the stress-induced SNBs and signals obtained with satellite specific probes was scored as (+) or (−). The assignment to either (+) or (−) classes was independently done by two individuals. The number of cells (n) analysed as indicated.
Probability of a specific association between HSA9 and stress- induced SNBs: χ21 = 69.25; P << 0.0001.
Probability of a specific association between HSA12 and stress-induced SNBs: χ21 = 12.65; P << 0.0001.
Probability of a specific association between HSA15 and stress-induced SNBs: χ21 = 30.37; P << 0.0001.
Probability of a random association between X chromosome and stress-induced SNBs: χ21 = 24.04; P << 0.0001.
Figure 6.
Stress-induced SNBs colocalize with pericentromeric heterochromatin of HSA9, and centromeric regions of HSA12 and 15. HeLa cells were heat shocked at 42°C for 1 h and were allowed to recover 1 h at 37°C. After fixation in formaldehyde, cells were hybridized to biotinylated probes specific for satellite III DNA of HSA9, and α-satellite sequences on chromosome X, HSA12, and HSA15 (see “Materials and Methods”). Cells were costained with anti-HAP polyclonal antibodies to detect stress-induced SNBs. The distribution of HAP and of the biotinylated probes was revealed by indirect immunofluorescence with a rhodamine-conjugated goat anti-rabbit antibody and with FITC-conjugated avidin (see “Materials and Methods” for details). Confocal laser microscopy images of hybridized cells were taken to reveal the distribution of HAP (in red) and of the biotinylated probe (green). Images were merged to detect colocalization between satellite DNA sequences and stress bodies. On the left, confocal laser microscopy images of four consecutive optical sections (I–IV, 0.5 μm in depth) of the same cell hybridized to a probe (pHuR98) that recognizes the pericentromeric heterochromatin of HSA9. On the right, images of cells hybridized to probes specific for α-satellite of HSA12, HSA15, and chromosome X.
Therefore, considering the results obtained with the X chromosome as a reference value for a random association between a chromosomal domain and stress-induced SNBs, a chi-square test based on the comparison of the data obtained with HSA9 and X chromosomes reinforced the conclusion of a specific association between HSA9 and stress-induced bodies (one-tailed chi-square[1] = 78.597; P ≪ 0.0001).
Stress-Induced SNBs Colocalize with the Centromeric Regions of HSA12 and HSA15
The results in the previous section demonstrate that although statistically significant, the association between the pericentromeric region of HSA9 and stress-induced SNBs is incomplete. Indeed, HSA9 failed to colocalize with stress-induced SNBs in 25% of the cells (see Table 2), as if this chromosome were not always involved in the formation of these structures. Two additional evidences supported this interpretation. First, even in cells scored as positive, we rarely observed colocalization of all the hybridization signals with stress bodies. Second, stress bodies usually outnumbered the HSA9 homologs. Indeed, most of the cells contained two to three copies of HSA9, whereas stress bodies ranged in number between one and seven, and cells with two, three, four, and five bodies had approximately the same frequency (∼17%). We wondered whether the additional chromosomes able to direct the formation of stress bodies in hamster cells, namely HSA12 and 15, could colocalize with stress-induced SNBs in HeLa cells. On the basis of the results obtained with HSA9, we hypothesized that the centromeric heterochromatin of these two chromosomes could contain recruiting centers for the assembly of these structures and could colocalize with stress bodies. HeLa cells were, therefore, hybridized to either pBR12 or pMC15 containing, respectively, α-satellite DNA sequences exclusively present on HSA12 and 15 (Baldini et al., 1990; Archidiacono et al., 1995). Analysis of mitotic spreads showed that most of the cells had two to three copies of HSA12 and 15 (78 and 83%, respectively, our unpublished results). Exponentially growing HeLa cells were heat shocked, hybridized to either one of the two probes, and then stained with anti-HAP antibodies. As shown in Figure 6, both centromeric regions colocalized with stress-induced SNBs, and the association was statistically significant (Table 2). However, similar to what was observed with HSA9, each chromosome colocalized with a subset of stress-induced SNBs (see Figure 6). This phenomenon was particularly evident in cells containing more than three bodies.
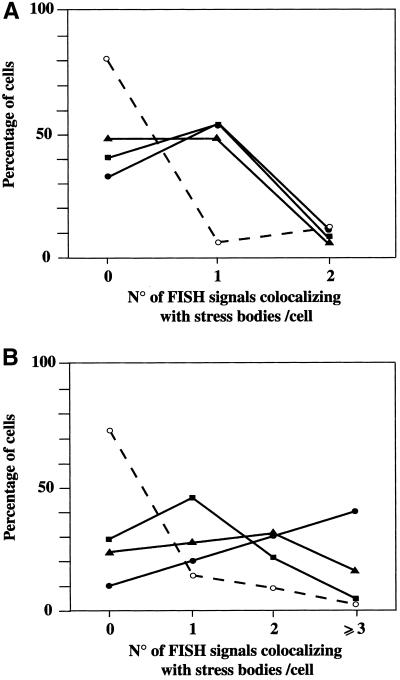
To make our analysis more quantitative, we counted for each chromosome (HAS9, 12, 15, and X) the number of hybridization signals colocalizing with stress-induced SNBs. We considered two groups of cells: those containing one or two stress-induced SNBs (Figure 7A) and those in which more than two bodies were detectable (Figure 7B). As shown in Figure 7, most of the signals obtained with the probe specific for chromosome X did not colocalize with stress-induced SNBs, regardless of their number in the cell. A completely different behavior was observed with HSA 9, 12, and 15, which instead associated with stress-induced SNBs. However, as shown in Figure 7A, in cells containing at most two stress-induced SNBs, usually only one homolog of each chromosome colocalized with stress bodies, implying that the recruiting centers for the two bodies were located on different chromosomes (for instance, HSA9 and HSA15). Although this asymmetric behavior of the two chromosomal homologs was less marked in cells with more than two stress bodies (Figure 7B), it was still evident in the case of HSA9. This result suggests a difference in the chromatin structure the two homologs, at least for what concerns the nucleation site of stress-induced SNBs. In conclusion, this analysis indicates that a subset of human chromosomes, including HSA9, 12, and 15, contains recruiting centers for the formation of stress-induced SNBs.
Figure 7.
Quantitative analysis of the colocalization between specific chromosomes and stress-induces SNBs. Heat-shocked HeLa cells were independently hybridized to biotinylated probes the for heterochromatic region of HSA9 (pHuR98), HSA12 (pBR12), HSA15 (pMC15), and chromosome X (pDMX1). Cells were costained with anti-HAP polyclonal antibodies to detect stress-induced SNBs. The distribution of HAP and of the biotinylated probe was revealed by indirect immunofluorescence with a rhodamine-conjugated goat anti-rabbit antibody and with FITC-conjugated avidin. Confocal laser microscopy images were taken, and for each chromosome, we counted the number hybridization signals colocalizing with stress bodies. (A) Colocalizing signals in cells with one or two stress-induced SNBs. We analyzed 68 cells (25 of which had a single body) stained for HSA9, 17 cells (8 with one body) stained for HSA12, 15 cells (7 with one body) stained for HSA15, and 16 cells (7 with one body) stained for chromosome X. (B) Cells with more than two stress-induced SNBs were analyzed as in A. We analyzed 211 cells for HSA9, 47 cell for HSA12, 43 cells for HSA15, and 80 cells for chromosome X. ▪, HSA9. ▴, HSA12. ●, HSA15. ○ and dashed line, chromosome X.
DISCUSSION
In this paper, we have exploited the species specificity of stress-induced SNBs to identify the human chromosomes that direct the formation of stress bodies in heat-shocked hamster cells. By means of panels of multi- and monochromosomal hamster>human somatic cell hybrids, we have identified three human chromosomes, HSA9, 12, and 15, individually conferring to recipient hamster cells the ability to form stress bodies. Our analysis warrants the conclusion that these bodies have the same features of stress-induced SNBs described in human cells (Weighardt et al., 1999; Chiodi et al., 2000; Denegri et al., 2001). Indeed, similar to stress-induced SNBs, they are sites of accumulation of hnRNP HAP and of the HSF1 factor. Moreover, they are usually associated to the nucleoli and consist of clusters of highly packed forms of ribonucleoprotein complexes, the perichromatin granules.
Specific Chromosomal Domains Are Involved in the Formation of Stress-Induced SNBs
Two alternative hypotheses can be raised to account for the fact that at least three human chromosomes can individually assist the formation of stress bodies in hamster>human cell hybrids. According to one hypothesis, rodent cells would fail to form stress bodies because they lack specific trans-acting factors that are instead coded by genes on different human chromosomes. Several considerations argue against this possibility. The most convincing argument is that both in human and in hamster>human cell hybrids, the number of stress bodies is linked to the number of chromosomes (Jolly et al., 1997, and this paper). Moreover, in light of the high level of evolutionary conservation of the coding genome among mammals, it seems unlikely that a gene present on at least three different human chromosomes and involved in the redistribution of several RNA processing factor has no counterparts in hamster cells. Because of these considerations, we favor the alternative possibility whereby specific domains of HSA9, 12, and 15 would provide a sort of nucleation site for the formation of stress-induced SNBs. This possibility is consistent with the fact that, as shown in this paper, the pericentromeric heterochromatic q12 band of HSA9 and the centromeric regions of HSA12 and 15 colocalize with stress-induced SNBs in HeLa cells (Figure 6). In addition, in the case of HSA9, we have shown that the pericentromeric heterochromatic q12 band (7–10 Mb) is part of the shortest portion of the chromosome (p13-q13; 20–25 Mb) able to direct the formation of stress bodies in hamster cells (Figure 5).
The existence of multiple recruiting centers distributed on different chromosomes can account for the observation that in HeLa cells, the number of stress-induced SNBs usually exceeds the number of homologs of a single chromosome. Our analysis suggests that the number and identity of the chromosomal domains activated as recruiting centers differ among cells. Although the parameters that direct this choice are still to be discovered, the moment during cell cycle when cells are stressed, the duration, and the intensity of the stress treatment are likely to have a role.
The q12 heterochromatic band of HSA9 is one of the largest unsequenced portions of the human genome (Consortium, 2001). It is known, however, that it consists of arrays of tandemly repeated satellite DNA, including α, β, and satellite III (Finelli et al., 1996). Similar heterochromatic regions are present on acrocentric chromosomes (such as HSA15), proximal to the NORs, on HSA 1, and on the Y chromosome (Finelli et al., 1996), all of which have been shown to be associated with the nucleoli (Manuelidis and Borden, 1988; Sullivan et al., 2001). Not all of the chromosomes associated to the nucleoli are able to direct the formation of stress-induced SNBs, and in fact, similar structures are undetectable in a hamster>human monochromosomal cell hybrid containing the acrocentric HSA14. However, the behavior of HSA9 suggests a relationship between the activity of the recruiting center and the nuclear position of the chromosome. Indeed, contrary to what occurs with HSA15 and 12, only one HSA9 homolog preferentially associates with the stress-induced SNBs (Figure 7). Intriguingly, we have observed that the homolog associated with a stress body usually lies adjacent to the nucleolus, whereas the other one has a more variable position. This different distribution of the two HSA9 homologs, one always compartmentalized on the nucleolus and the second either adjacent to the nucleolus or near to the nuclear membrane, has already been reported (Manuelidis and Borden, 1988). It is tempting to speculate that the two HSA9 homologs exist in different chromatin conformations, one of which preferentially associates with stress-induced SNBs.
Constitutive centromeric heterochromatin is characterized by a distinctive higher-order chromatin structure (Gilbert and Allan, 2001) and by the presence of long tracts of tandemly repeated highly methylated satellite DNA (Bird, 1992; Festenstein et al., 1999). Recent investigations have identified a number of proteins recruited to heterochromatic domains of the cell nucleus. Among them, the best characterized is the heterochromatin protein 1 (HP1), originally identified in Drosophila and conserved in evolution from fission yeast to humans (Jones et al., 2000). HP1 is viewed as a structural adapter participating in the assembly of macromolecular complexes in chromatin. It has been suggested that HP1 has a role in targeting pericentromeric heterochromatin to specific nuclear compartments. Indeed, in response to treatments with inhibitors of histone deacetylalase, pericentromeric heterochromatic regions lose their association with HP1 proteins and relocate specifically toward the nuclear periphery (Taddei et al., 2001). Histone deacetylation and methylation are important for the association of HP1 with heterochromatin. Interestingly, acetylated histone H4 is found transiently enriched in heterochromatin regions only at the time of DNA replication in midS phase (Taddei et al., 1999) and is then deacetylated from an activity recruited to the chromatin by methyl-binding proteins (Rountree et al., 2000). The presence of acetylated histones in midS phase can be relevant for the formation of stress-induced SNBs and can account for our observation that the interval required for the assembly of these structures is shorter in midS phase than in other moments of the cell cycle (Weighardt et al., 1999). We speculate that DNA replication, by perturbing the chromatin structure, could provide the first step toward the establishment of the proper configuration for the ensuing assembly of stress-induced SNBs.
It is known that heat shock and other stress treatments drastically affect the nuclear structure and the distribution of proteins, such as nucleolin (Daniely and Borowiec, 2000; Wang et al., 2001), normally present in specific nuclear compartments. It is conceivable, therefore, that stress treatments could also trigger a complex rearrangement of the heterochromatic fiber, producing the displacement of some factors and binding of others, including HSF1. HSF1 is probably one of the first proteins recruited to stress bodies. However, its presence in stress bodies does not seem to be sufficient for the recruitment of HAP and probably other RNA processing factors in these structures, as indicated by the fact that osmotic stress can induce the recruitment of HSF1 (Jolly et al., 1999) but not of HAP (our unpublished results). We think that some distinctive, still unknown feature of the heterochromatic regions of HSA9, 12, and 15 is required for recruitment of RNA processing factors to stress-induced SNBs. Indeed, a role for specific heterochromatic regions as recruitment center for a subset of nuclear factors has been already proposed. For instance, in a number of human cell lines, the Polycomb group (PcG) complex forms unique discrete structure termed PcG bodies. These bodies are tightly associated with large pericentromeric heterochromatin regions (1q12) on chromosome 1 (Saurin et al., 1998) and are likely to represent storage domains where surplus PcG proteins are stored until required by the cell. As recently suggested, similar to PcG bodies, stress-induced SNBs would be depots for HSF1 and a number of RNA processing factors (Jolly et al., 1999; Chiodi et al., 2000).
On the other hand, stress-induced SNBs seem to originate from preexisting SNBs that probably correspond to sites through which transcripts pass along their path toward the nuclear envelope (Chen et al., 1999). Heat shock, by altering the movement of transcripts, would induce the appearance of stress-induced SNBs. Although the functional clarification of both SNBs and stress-induced SNBs still deserves further investigation, it is conceivable that the recruitment of ribonucleoprotein complexes to these nuclear domains can be important for the correct processing of transcripts. In this perspective, our finding that specific heterochromatic regions can play a role in the organization of stress-induced SNBs reveals a previously underestimated role of heterochromatin in the organization of nuclear compartments and in the underlying functions.
ACKNOWLEDGMENTS
We thank Centro Grandi Strumenti of the University of Pavia for the confocal microscopy facility. This work was supported by a grant from Associaziona Italiana per la Ricerca sul Cancro (to G.B.), by a grant from Ministero dell'Università e della Ricerca Scientifica e Tecnologica-Consiglio Nazionale delle Ricerche “Biomolecole per la salute umana” L. 95/95, and by a grant from Progetto Strategico Tecnologie di base della postgenomica (to F.C.). M.D. was supported by a fellowship of Consiglio Nazionale delle Ricerche. The authors thank A. Lisa and G. Zei for the statistical analysis and D. Arena for technical assistance.
Footnotes
DOI: 10.1091/mbc.01–12–0569.
REFERENCES
- Antonacci R, Marzella R, Finelli P, Lonoce A, Forabosco A, Archidiacono N, Rocchi M. A panel of subchromosomal painting libraries representing over 300 regions of the human genome. Cytogenet Cell Genet. 1995;68:25–32. doi: 10.1159/000133882. [DOI] [PubMed] [Google Scholar]
- Archidiacono N, Antonacci R, Marzella R, Finelli P, Lonoce A, Rocchi M. Comparative mapping of human alphoid sequences in great apes using fluorescence in situ hybridization. Genomics. 1995;25:477–484. doi: 10.1016/0888-7543(95)80048-q. [DOI] [PubMed] [Google Scholar]
- Baldini A, Rocchi M, Archidiacono N, Miller OJ, Miller DA. A human alpha satellite DNA subset specific for chromosome 12. Am J Hum Genet. 1990;46:784–788. [PMC free article] [PubMed] [Google Scholar]
- Bernhard W. A new staining procedure for electron microscopical cytology. J Ultrastruct Res. 1969;27:250–265. doi: 10.1016/s0022-5320(69)80016-x. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Chen T, Boisvert FM, Bazett-Jones DP, Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Biol Cell. 1999;10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodi I, Biggiogera M, Denegri M, Corioni M, Weighardt F, Cobianchi F, Riva S, Biamonti G. Structure and dynamics of hnRNP-labeled nuclear bodies induced by stress treatments. J Cell Sci. 2000;113:4043–4053. doi: 10.1242/jcs.113.22.4043. [DOI] [PubMed] [Google Scholar]
- Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:806–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Daniely Y, Borowiec JA. Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J Cell Biol. 2000;149:799–810. doi: 10.1083/jcb.149.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denegri M, Chiodi I, Corioni M, Cobianchi F, Riva S, Biamonti G. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol Biol Cell. 2001;12:3502–3514. doi: 10.1091/mbc.12.11.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPont BR, Garcia DK, Sullivan TM, Naylor SL, Oesterreich S. Assignment of SAFB encoding Hsp27 ERE-TATA binding protein (HET)/scaffold attachment factor B (SAF-B) to human chromosome 19 band p13. Cytogenet Cell Genet. 1997;79:284–285. doi: 10.1159/000134744. [DOI] [PubMed] [Google Scholar]
- Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Festenstein R, Sharghi-Namini S, Fox M, Roderick K, Tolaini M, Norton T, Saveliev A, Kioussis D, Singh P. Heterochromatin protein 1 modifies mammalian PEV in a dose- and chromosomal-context-dependent manner. Nat Genet. 1999;23:457–461. doi: 10.1038/70579. [DOI] [PubMed] [Google Scholar]
- Finelli P, Antonacci R, Marzella R, Lonoce A, Archidiacono N, Rocchi M. Structural organization of multiple alphoid subsets coexisting on human chromosomes 1, 4, 5, 7, 9, 15, 18, and 19. Genomics. 1996;38:325–330. doi: 10.1006/geno.1996.0635. [DOI] [PubMed] [Google Scholar]
- Frey MR, Bailey AD, Weiner AM, Matera AG. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol. 1999;9:126–135. doi: 10.1016/s0960-9822(99)80066-9. [DOI] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Allan J. Distinctive higher-order chromatin structure at mammalian centromeres. Proc Natl Acad Sci USA. 2001;98:11949–11954. doi: 10.1073/pnas.211322798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson AS, Warburton D, Atwood KC. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci USA. 1972;69:3394–3398. doi: 10.1073/pnas.69.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Morimoto R, Robert-Nicoud M, Vourc'h C. HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J Cell Sci. 1997;110:2935–2941. doi: 10.1242/jcs.110.23.2935. [DOI] [PubMed] [Google Scholar]
- Jolly C, Usson Y, Morimoto RI. Rapid and reversible relocalization of heat shock factor 1 within seconds to nuclear stress granules. Proc Natl Acad Sci USA. 1999;96:6769–6774. doi: 10.1073/pnas.96.12.6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DO, Cowell IG, Singh PB. Mammalian chromodomain proteins: their role in genome organization and expression. Bioessays. 2000;22:124–137. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Manuelidis L, Borden J. Reproducible compartmentalization of individual chromosome domains in human CNS cells revealed by in situ hybridization and three-dimensional reconstruction. Chromosoma. 1988;96:397–410. doi: 10.1007/BF00303033. [DOI] [PubMed] [Google Scholar]
- Matera AG. Of coiled bodies, gems, and salmon. J Cell Biochem. 1998;70:181–192. [PubMed] [Google Scholar]
- McDowell TL, Gibbons RJ, Sutherland H, O'Rourke DM, Bickmore WA, Pombo A, Turley H, Gatter K, Picketts DJ, Buckle VJ, Chapman L, Rhodes D, Higgs DR. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci USA. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A, Rossi R, Ferrari G, Scovassi AI, Prosperi E, Biamonti G. Etoposide induces the dispersal of DNA ligase I from replication factories. Mol Biol Cell. 2001;12:2109–2118. doi: 10.1091/mbc.12.7.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Cells in stress: transcriptional activation of heat shock genes. Science. 1993;259:1409–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- Ochs RL, Lischwe MA, Shen E, Carroll RE, Busch H. Nucleologenesis: composition and fate of prenucleolar bodies. Chromosoma. 1985;92:330–336. doi: 10.1007/BF00327463. [DOI] [PubMed] [Google Scholar]
- O'Keefe RT, Mayeda A, Sadowski CL, Krainer AR, Spector DL. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J Cell Biol. 1994;124:249–260. doi: 10.1083/jcb.124.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Cuello P, Schul W, Yoon JB, Roeder RG, Cook PR, Murphy S. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 1998;17:1768–1778. doi: 10.1093/emboj/17.6.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi E, Balzaretti M, Moralli D, Vagnarelli P, Tredici F, Bensi M, De Carli L. Gene targeting to the centromeric DNA of a human minichromosome. Hum Gene Ther. 1996;7:1103–1109. doi: 10.1089/hum.1996.7.9-1103. [DOI] [PubMed] [Google Scholar]
- Raimondi E, Ferretti L, Young BD, Sgaramella V, De Carli L. The origin of a morphologically unidentifiable human supernumerary minichromosome traced through sorting, molecular cloning, and in situ hybridization. J Med Genet. 1991;28:92–96. doi: 10.1136/jmg.28.2.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi M, Archidiacono N, Ward DC, Baldini A. A human chromosome 9-specific alphoid DNA repeat spatially resolvable from satellite 3 DNA by fluorescent in situ hybridization. Genomics. 1991;9:517–523. doi: 10.1016/0888-7543(91)90419-f. [DOI] [PubMed] [Google Scholar]
- Rocchi M, Roncuzzi L, Santamaria R, Archidiacono N, Dente L, Romeo G. Mapping through somatic cell hybrids and cDNA probes of protein C to chromosome 2, factor X to chromosome 13, and α1-acid glycoprotein to chromosome 9. Hum Genet. 1986;74:30–33. doi: 10.1007/BF00278781. [DOI] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saurin AJ, Shiels C, Williamson J, Satijn DP, Otte AP, Sheer D, Freemont PS. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan GJ, Bridger JM, Cuthbert AP, Newbold RF, Bickmore WA, McStay B. Human acrocentric chromosomes with transcriptionally silent nucleolar organizer regions associate with nucleoli. EMBO J. 2001;20:2867–2877. doi: 10.1093/emboj/20.11.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Maison C, Roche D, Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol. 2001;3:114–120. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- Taddei A, Roche D, Sibarita JB, Turner BM, Almouzni G. Duplication and maintenance of heterochromatin domains. J Cell Biol. 1999;147:1153–1166. doi: 10.1083/jcb.147.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani T, Derby RJ, Hiraoka Y, Spector DL. Nucleolar accumulation of poly (A)+ RNA in heat-shocked yeast cells: implication of nucleolar involvement in mRNA transport. Mol Biol Cell. 1996;7:173–192. doi: 10.1091/mbc.7.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Guan J, Wang H, Leeper D, Iliakis G. Regulation of DNA replication after heat shock by replication protein a-nucleolin interactions. J Biol Chem. 2001;276:20579–20588. doi: 10.1074/jbc.M100874200. [DOI] [PubMed] [Google Scholar]
- Weighardt F, Cobianchi F, Cartegni L, Chiodi I, Villa A, Riva S, Biamonti G. A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J Cell Sci. 1999;112:1465–1476. doi: 10.1242/jcs.112.10.1465. [DOI] [PubMed] [Google Scholar]