Abstract
Using the serial analysis of gene expression technique, we surveyed transcriptomes of three major tissues (panicles, leaves, and roots) of a super-hybrid rice (Oryza sativa) strain, LYP9, in comparison to its parental cultivars, 93-11 (indica) and PA64s (japonica). We acquired 465,679 tags from the serial analysis of gene expression libraries, which were consolidated into 68,483 unique tags. Focusing our initial functional analyses on a subset of the data that are supported by full-length cDNAs and the tags (genes) differentially expressed in the hybrid at a significant level (P < 0.01), we identified 595 up-regulated (22 tags in panicles, 228 in leaves, and 345 in roots) and 25 down-regulated (seven tags in panicles, 15 in leaves, and three in roots) in LYP9. Most of the tag-identified and up-regulated genes were found related to enhancing carbon- and nitrogen-assimilation, including photosynthesis in leaves, nitrogen uptake in roots, and rapid growth in both roots and panicles. Among the down-regulated genes in LYP9, there is an essential enzyme in photorespiration, alanine:glyoxylate aminotransferase 1. Our study adds a new set of data crucial for the understanding of molecular mechanisms of heterosis and gene regulation networks of the cultivated rice.
Cultivated rice (Oryza sativa) is one of the vital crops for human consumption, providing staple food for more than one-half of the world's population. To meet the demand from population growth, an estimated 50% yield increase in grain production, including that of rice, is expected by the year 2030 (Horton, 2000). Exploiting the potential of heterosis has become a key strategy for increasing productivity of crop plants (Xiao et al., 1995). When genetically distant breeding lines (often inbred) are artificially crossed, the resulting hybrid is characterized by an increased yield compared to its parental lines, taking advantage of a phenomenon termed heterosis or hybrid vigor (Shull, 1952). However, the practice of producing promising hybrid breeds is largely a trial-and-error exercise and detailed molecular mechanisms for increased productivity in a hybrid are yet to be elucidated, especially when molecular markers empowering such research procedures are not entirely available. It is still indispensable to discover more heterosis-associated genes by acquiring data large-scale to compare gene expression profiles between a hybrid and its parental lines.
In this study, we focused our attention on a super-hybrid rice, Liang-You-Pei-Jiu (LYP9), which produces 20% to 30% more grains per hectare than other hybrid or nonhybrid higher-yield rice crops (Lu and Zhou, 2000). Its paternal cultivar, 93-11, is an indica variety (Oryza sativa L. subsp. indica), the major rice subspecies grown in China and many other Asian-Pacific regions. The maternal cultivar, Pei-Ai 64s (PA64s), has a major genetic background of indica and minor gene flows from japonica and javanica, two other major cultivated rice subspecies. PA64s is also a temperature-sensitive genic male-sterile rice cultivar with male-sterility at 24°C.
We used the serial analysis of gene expression (SAGE) technique (Velculescu et al., 1995) to study transcriptome expression profiles among LYP9 and its parental lines. SAGE technique has been widely used to generate transcriptome profiles in animal systems, especially for cancer research, in large scale and at affordable cost (Zhang et al., 1997; Hermeking, 2003). It is useful for large-scale discovery of new transcripts, especially for identifying the relatively rare ones, and for acquisition of quantitative information on copy numbers of expressed transcripts under discrete conditions. For higher plants, several groups have applied this technology to study general expression profiles in selected tissues and growth conditions in Arabidopsis (Arabidopsis thaliana; Ekman et al., 2003; Lee and Lee, 2003; Fizames et al., 2004) and in rice (Matsumura et al., 1999, 2003; Gibbings et al., 2003; Gowda et al., 2004). We now report an attempt in identifying differentially expressed genes in a hybrid rice strain and its parental cultivars, hoping to provide new data and insights into the molecular mechanism of heterosis.
RESULTS
Basic Datasets and Tag-to-Gene Assignment
To build gene expression profiles of a parent-hybrid triad, we constructed nine SAGE libraries from mRNAs harvested in parallel growth stages from three basic rice tissues at distinct growth stages of the hybrid plant and its parental cultivars: (1) leaves at the milky stage of rice grain maturation, (2) panicles at the pollen-maturing stage, and (3) roots at the first tillering stage. From the libraries, we collectively sequenced 26,690 SAGE clones that yielded 465,679 individual tags and 68,483 different (or unique species of) SAGE tags (Table I).
Table I.
Data from rice SAGE libraries
| Librarya | No. of Total Tags | No. of Unique Tags |
|---|---|---|
| P1 | 47,086 | 11,878 |
| P2 | 46,820 | 13,927 |
| P3 | 67,793 | 19,602 |
| N1 | 69,550 | 22,890 |
| N2 | 52,515 | 15,398 |
| N3 | 48,238 | 18,084 |
| L1 | 68,549 | 23,178 |
| L2 | 36,226 | 9,873 |
| L3 | 28,902 | 10,871 |
| Total | 465,679 | 68,483 |
P, N, and L stand for PA64s, 93-11, and LYP9, respectively. Numbers 1, 2, and 3 denote libraries made from materials of panicles, leaves, and roots, respectively. Panicles were collected from the top one-third portion of panicles at the pollen-maturing stage. Leaves were harvested from the first leaves at the milky stage of grain ripening phase. Roots were collected at the first tillering stage.
To do comparative analyses on these SAGE tags, we prepared four essential datasets. The first dataset contained 841,788 virtual tags that included all 10-mer sequences downstream from (or 3′ to) all CATG sites in an indica genome sequence assembly (Yu et al., 2002) in both forward and reverse directions. The second dataset covered tags that were exclusively confirmed with full-length cDNAs (FL-cDNAs) of a japonica rice variety, Nipponbare (a nonredundant set of 20,259 FL-cDNAs; nonredundant Knowledge-based Oryza Molecular Biological Encyclopedia (nr-KOME)-cDNAs; Kikuchi et al., 2003). A total of 11,941 tags were matched to one or more FL-cDNA sequences (17.4% of total tags), of which 96% (11,458 tags) matched to a single and unique cDNA. In comparison, when the alignment was not limited to those annotated by FL-cDNAs but all SAGE tags, 31.2% of the total tags were assigned to a single location on the rice chromosomes. In this study, we annotated all tags (genes) based on the FL-cDNA dataset and did not use computer-predicted genes. We also largely ignored the sequence variations between indica and japonica rice and a small fraction of the tags were disqualified due to sequence variations between the two subspecies. The third and fourth datasets were collections of expressed sequence tags (ESTs) and proteins, respectively, brought together from our own and the public databases.
Distribution of SAGE Tags in Rice Genome
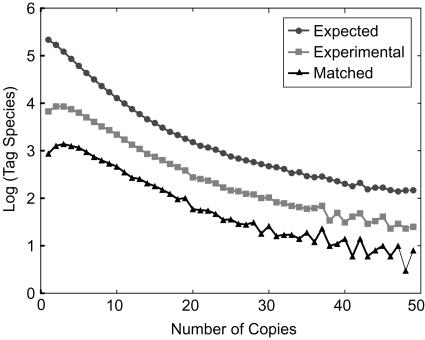
To evaluate sampling bias, redundancy, and data quality, we did several standard analyses and benchmarked our expression analysis only on FL-cDNA confirmed tags (the entire dataset is also publicly available). To evaluate sampling biases, we first plotted SAGE tags as a function of their redundancy (copy numbers) from three datasets: the experimentally acquired SAGE tags, a subset of them that were confirmed by FL-cDNAs, and predicted tags based on rice genome sequences (Fig. 1). Nearly identical distributions were observed for all three datasets. The number of tags decreased from more than 10,000 to 100 when copy numbers increased from 1 to 50. A slight difference between the predicted and real sites was observed in the low-copy fraction (1–5 copies), where a decreased number of tags were seen in the experimental data and even in the subset supported by the FL-cDNA dataset. One straightforward reason for this disparity is that a minor sampling bias may exist for rare transcripts among the methods employed in different data acquisition protocols of SAGE and cDNA cloning.
Figure 1.
Total numbers of tag species as function of their redundancy. Experimental results (black squares) were compared to the expected distribution (black circles). Tags that match to known FL-cDNA (black triangles) were also plotted. The experimental results have a similar trend as the theoretical prediction.
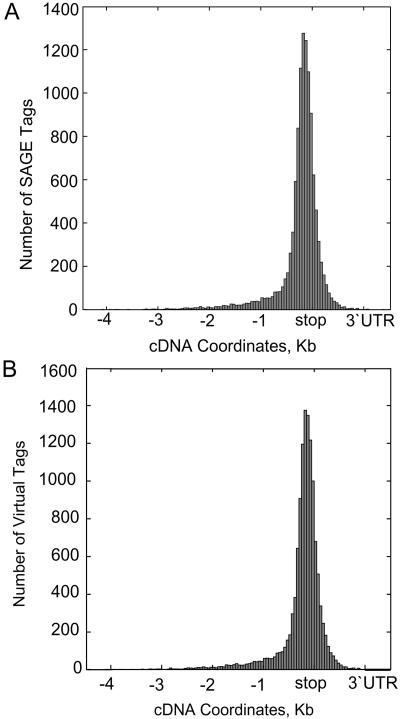
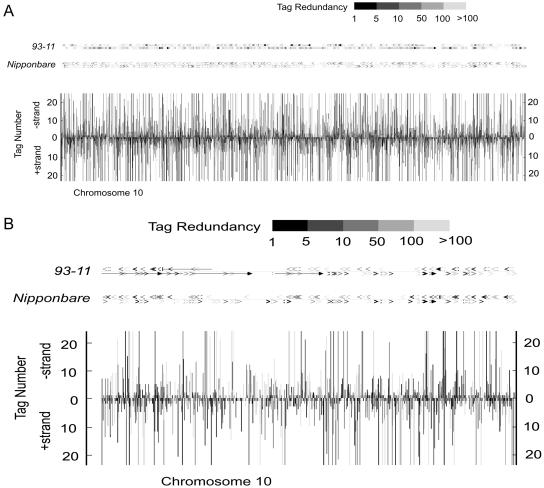
We next evaluated relative positioning of SAGE tags to the 3′-untranslated region (UTR) of genes, where they were targeted (Fig. 2; Chen et al., 2000). In this exercise, we first took the FL-cDNA dataset, aligned it to the genome sequence, and extracted a dataset composed of cDNA-verified virtual SAGE tags. We then established the position of the virtual tags in two plots: one containing the virtual tags that matched to our experimental tags (Fig. 2A) and the remaining tags that did not match to our experimental data (Fig. 2B). The two distributions are rather narrow and nearly identical, peaking at 100 nucleotides upstream of a stop codon and suggesting a parity of the two data sets. We also positioned SAGE tags over predicted genes to assess how SAGE tags distributed over rice chromosome length, taking advantage of their high density. Figure 3 depicts such an alignment on rice chromosome 10. Two sets of predicted genes assembled and annotated by Beijing Genomics Institute (BGI) were used, one from the 93-11 (indica) genome and the other from the Nipponbare (japonica) genome (Goff et al., 2002; Yu et al., 2002, 2005). The experimentally acquired SAGE tags were specifically mapped to chromosomal locations where genes reside. They appeared distributed quite evenly over gene-rich regions and had an overall low density in gene-poor regions. We also went one step further and investigated several multigene families in details, for which we had both SAGE tags and corresponding genes annotated, including those of small GTP binding protein, α-amylase, Suc transporter, ammonium transporter, nitrate transporter, and chitinase (Table II). The result was very encouraging; approximately 90% of our SAGE tags were specific enough in distinguishing members of these gene families, albeit most of them were seen as more than one copy among different libraries.
Figure 2.
Distributions of SAGE tags and their positions near 3′-UTR centered on a stop codon. The distribution of experimentally confirmed tags is shown in A. The distribution of virtual tags based on FL-cDNA data but not confirmed by experimental data is shown in B. Note that the distributions are nearly identical.
Figure 3.
Distribution of rice SAGE tags on chromosome 10. Tags were anchored directionally on both forward (vertical bars, bottom) and reverse (vertical bars, top) strands. Vertical bars indicate the redundancy of tags. Annotations of 93-11 and Nipponbare genomes are shown above, where exons and genes are depicted with arrowheads and arrows, respectively. One or more copies of the tags were mapped in each site. The entire chromosome 10 (A) and a highlighted q-arm portion (B) are shown here.
Table II.
Tag distribution of selected gene families
| Gene | Tag | Copies per Tagb | |
|---|---|---|---|
| Small GTP-Binding Protein | |||
| AK071126 | OsRac1 | GTTGAATCAA | 3 |
| AK069345 | OsRac3 | ATGCAATGTT | 13 |
| AK100585 | OsRacB | GAGAAATCTG | 49 |
| AK067796 | OsRacD | CTCATCTCCTa | 7 |
| AK061099 | ORRab2 | TGTAACTAAA | 20 |
| AK061116 | OsRab5A | TTCGTTCTGT | 34 |
| AK072731 | OsRan | GGCAGTGTGC | 3 |
| AK061714 | OsRan | AATTTGTTGG | 251 |
| α-Amylase | |||
| AK059671 | OsAmyc2 | ATTAGTGACA | 1 |
| AK073487 | OsAmy | CTCAAACCAG | 11 |
| Suc Transporter | |||
| AK065430 | OsSUT4 | TATAGTACTG | 7 |
| AK100027 | OsSUT | CAGATAATAT | 26 |
| AK073105 | OsSUT5 | TGAGAGTGTT | 1 |
| Ammonium Transporter | |||
| AK073718 | OsAMT1p | CGTACGTGTC | 64 |
| AK065288 | OsAMT2;1 | CCTATTAAAT | 8 |
| AK102106 | OsAMT2 | AGAATTATCT | 1 |
| Nitrate Transporter | |||
| AK101480 | NRT1;2 | ATCAGGTTAC | 16 |
| AK068409 | NRT | ACATACGCAA | 3 |
| Chitinase | |||
| AK061042 | Chia1a | TTGGGCGTCA | 5 |
| AK071196 | Chia1d | AATAAAGTAG | 2,100 |
| AK100973 | OsChib3a | GGAGTGTATT | 17 |
| AK067962 | OsChib3H-b | TGTGATCGATa | 14 |
| AK073267 | OsChib3H-d | TGTGATCGAC | 2 |
| AK071453 | Rcb4 | CTTATGTTGT | 14 |
| AK059767 | Chitinase 1 | GTCAGCAGCT | 2 |
| AK060033 | Chitinase 2 | GTGTTCTAAT | 10 |
| AK071013 | chia4a | TTGCAAGTGA | 3 |
| AK070067 | Chitinase | AAGTATTAAAa | 2 |
| AK106178 | Chitinase | TTACATCTAA | 3 |
| AK063939 | OsChib3H-a | CTCTGGGACA | 1 |
| AK111415 | Chitinase | TGTTGATTGC | 3 |
| AK099355 | Chitinase | TTATTGTGAA | 7 |
Tags match more than one gene.
Numbers are the sum of tags found in all nine SAGE libraries.
Finally, we compared our SAGE data to that of 144,083 tags from Arabidopsis root libraries (Fizames et al., 2004). The result revealed a similar distribution of genes between the two studies in various abundance classes, with minor variation largely due to sampling depth (Table III). In most of the SAGE studies, over 80% of unique tags were found present at low copy numbers (≤5 tags), and around 50% of them were detected only once in a genome (Chrast et al., 2000; Lee et al., 2001; Fizames et al., 2004).
Table III.
Distribution of tags from SAGE libraries of rice and Arabidopsis
| Copy Nos.a
|
Rice
|
Arabidopsisb
|
||
|---|---|---|---|---|
| Unique | Percent in All | Unique | Percent in All | |
| ≥100 | ||||
| Tag | 500 | 0.7 | 80 | 0.2 |
| Matched | 306 | 2.6 | ||
| 21–99 | ||||
| Tag | 3,015 | 4.4 | 670 | 1.3 |
| Matched | 1,677 | 14.0 | ||
| 6–20 | ||||
| Tag | 8,543 | 12.5 | 3,023 | 5.8 |
| Matched | 3,635 | 30.4 | ||
| 2–5 | ||||
| Tag | 18,550 | 27.1 | 12,191 | 23.4 |
| Matched | 3,929 | 32.9 | ||
| 1 | ||||
| Tag | 37,875 | 55.3 | 36,114 | 69.3 |
| Matched | 2,394 | 20.0 | ||
| Total | ||||
| Tag | 68,483 | 100 | 52,078 | 100 |
| Matched | 11,941 | 100 | ||
Tags are categorized according to their copy numbers: ≥100, 21 to 99, 6 to 20, 2 to 5, and single copies. Matched indicates the number of unique tags that matched to FL-cDNAs.
Published SAGE data from Arabidopsis roots (Fizames et al., 2004).
Comparisons of Our SAGE Results with Other Gene Expression Data
Various EST sequencing and proteomics approaches have been widely used in studying gene expression, especially in identifying tissue-specific genes, even though exactly matched datasets for comparison are somewhat sparse in the public data depositaries. We nonetheless compared our data to those acquired from a cDNA library (L499) constructed by Takuji Sasaki and coworkers from mRNAs of rice panicles at the flowering stage. From 6,502 available EST sequences, we found 26 panicle-specific genes annotated in the database, which were only detected in libraries generated from panicles but not in those from other tissues (Table IV). Panicle specificity of all listed genes was found within our dataset, with one or more confirmations. We noticed a lower confirmation rate (percentage of EST-identified genes confirmed in our SAGE study) between the two studies in the panicle of PA64s (five out of 26 genes were detected); the result was consistent with the fact that PA64s is male-sterile. In addition, the expression levels of these genes were overall higher in panicles of 93-11 and LYP9 at a similar magnitude in both cultivars. Although some of the genes were not strictly speaking panicle specific, detectable in leaves and roots, they were expressed at a higher level in panicles than in the other two tissues. In rare cases (such as AK070187), we also observed variable expression levels between our SAGE and the EST data, but it was indistinguishable if they were due to differences of source materials or methods used for data acquisition.
Table IV.
Comparison of expression profiles of panicle-specific genes between SAGE and EST
| Gene
|
Tag
|
UniGene Cluster
|
EST Counta
|
SAGEb
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P2 | P3 | N1 | N2 | N3 | L1 | L2 | L3 | Total | ||||
| AK070187 | AAGCTGTCGT | Os.12697 | 64 | 0 | 0 | 0 | 43 | 2 | 0 | 47 | 0 | 0 | 92 |
| AK071196 | AATAAAGTAG | Os.4976 | 27 | 11 | 0 | 0 | 494 | 238 | 0 | 835 | 0 | 6 | 1,584 |
| AK071325 | CAGGCTTTTT | Os.37927 | 21 | 0 | 0 | 0 | 71 | 7 | 0 | 43 | 0 | 0 | 121 |
| AK101023 | TTCTGCGGTG | Os.49687 | 19 | 0 | 0 | 0 | 38 | 0 | 0 | 29 | 0 | 0 | 67 |
| AK100805 | GGCAATGCAC | Os.4041 | 19 | 0 | 0 | 0 | 20 | 0 | 0 | 23 | 0 | 0 | 43 |
| AK072792 | CACATATATA | Os.2402 | 15 | 1 | 0 | 1 | 10 | 0 | 0 | 14 | 0 | 0 | 26 |
| AK109576 | TGTGAGATCG | Os.11067 | 12 | 4 | 0 | 0 | 142 | 10 | 0 | 157 | 0 | 0 | 313 |
| AK107558 | CAATATGATGc | Os.12275 | 12 | 0 | 1 | 0 | 73 | 7 | 0 | 102 | 0 | 3 | 186 |
| AK071282 | ATTAACACGA | Os.12275 | 12 | 0 | 0 | 0 | 6 | 0 | 0 | 13 | 0 | 0 | 19 |
| AK070631 | ACGGTGGACG | Os.8140 | 6 | 0 | 0 | 0 | 12 | 3 | 0 | 13 | 0 | 0 | 28 |
| AK072672 | ACCGGTGATA | Os.5819 | 6 | 0 | 0 | 0 | 18 | 0 | 0 | 17 | 0 | 0 | 35 |
| AK106951 | GCATCGCCTT | Os.10688 | 5 | 0 | 0 | 0 | 74 | 4 | 0 | 51 | 0 | 0 | 129 |
| AK072232 | TGGCCCCAAA | Os.4957 | 5 | 0 | 0 | 0 | 12 | 0 | 0 | 22 | 0 | 0 | 34 |
| AK070646 | TTTTTTTCCG | Os.9384 | 5 | 0 | 0 | 0 | 33 | 0 | 0 | 32 | 0 | 0 | 65 |
| AK069853 | ATCGAGTCTT | Os.2405 | 4 | 0 | 0 | 0 | 23 | 0 | 0 | 18 | 0 | 0 | 41 |
| AK070815 | CTGCATTTGT | Os.8166 | 4 | 1 | 1 | 0 | 16 | 0 | 0 | 26 | 0 | 0 | 44 |
| AK069283 | TTCAACTCGC | Os.7026 | 3 | 0 | 0 | 0 | 7 | 0 | 0 | 9 | 0 | 1 | 17 |
| AK107837 | AATAACTCTC | Os.10395 | 2 | 0 | 0 | 0 | 28 | 2 | 0 | 46 | 0 | 6 | 82 |
| AK109762 | CGTGTTCGGT | Os.20322 | 2 | 0 | 0 | 0 | 7 | 0 | 0 | 17 | 0 | 0 | 24 |
| AK111098 | TGTCCTTCCA | Os.9665 | 2 | 0 | 0 | 0 | 163 | 19 | 0 | 176 | 0 | 1 | 359 |
| AK071058 | AGCTTAAGAG | Os.4956 | 2 | 0 | 0 | 0 | 7 | 3 | 0 | 14 | 0 | 0 | 24 |
| AK060272 | GTTATAGTCC | Os.12736 | 2 | 0 | 1 | 0 | 5 | 2 | 3 | 13 | 0 | 3 | 27 |
| AK070171 | GTTGAGGAGG | Os.52968 | 1 | 0 | 0 | 0 | 6 | 0 | 0 | 9 | 0 | 0 | 15 |
| AK107744 | CTGCTGATGA | Os.55257 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 9 | 0 | 0 | 10 |
| AK070921 | CCTACTCCTA | Os.25495 | 1 | 2 | 0 | 0 | 133 | 58 | 0 | 110 | 0 | 3 | 306 |
| AK071236 | CCCGATCGAA | Os.5821 | 1 | 0 | 0 | 0 | 14 | 5 | 0 | 21 | 0 | 0 | 40 |
EST data are from public data that were acquired from a cDNA library made from panicle material of Nipponbare at flowering stage (http://www.ncbi.nlm.nih.gov/UniGene/library.cgi?ORG=Os&LID=499).
The tag numbers are listed as individual SAGE libraries.
Tag matches more than one gene.
We further compared genes that were identified in LYP9 with a set of EST data published recently (Zhou et al., 2003). In Zhou's study, three shoot (young leaves) cDNA libraries were constructed from LYP9 and PA64s at the trefoil stage, but mRNAs from 93-11 were extracted from root tissues at the tillering stage. Approximately 10,000 EST sequences were obtained from each library. The EST sequences were annotated according to nr-KOME-cDNAs by using BLASTN with threshold values of E-value < 1E-30 and identities >95% over a length of 100 bp (Wu et al., 2002). A P-value of 0.05 or less was considered as significant when evaluated for differential expressions. We found 121 leaf-associated genes that were both matched to our SAGE tags and differentially expressed in LYP9 (118 up-regulated and three down-regulated). Among them, 21 genes (17%) showed consistent results between SAGE and EST studies. Five and three of the consistently up-regulated genes were involved in photosynthetic pathways and antioxidative enzymes, respectively. One obvious possibility for the relatively low confirmation rate (percentage of EST-identified genes confirmed in our SAGE study) between the two studies is that the leaf materials were collected at different developmental stages, where they were harvested from leaves at the trefoil or tillering stage (rapid growing stage) for the EST study and at the (grain) milky stage for the SAGE experiment. Another possibility has to do with different cloning efficiencies between the two methods.
We also compared our SAGE data with proteins identified in a proteomic study at BGI by using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS; Supplemental Fig. 1), in which a significant amount of rice proteins were identified in panicles (338), leaves (249), and roots (416) of 93-11. Although the confirmation rate (percentage of proteins identified in FL-cDNA collections) between the protein and the FL-cDNA collections was rather limited due to the fact that all the proteome data were acquired from the early flowering stage and a single cultivar (93-11) and other data were acquired from the same generally defined tissues but at different growing stages, genes expressed in particular tissues were comparable and the confirmation rates ranged from 19% to 27% in all tissues (panicles, 88/338; leaves, 48/249; and roots, 111/416). Among the FL-cDNA-confirmed proteins, 51 (58%), 21 (44%), and 63 (57%) were detected in our SAGE tags (also confirmed with the same FL-cDNA dataset) in panicles, leaves, and roots, respectively. The remaining proteins identified with MALDI-TOF MS method were not found in the corresponding SAGE libraries, largely due to biases in the acquisition method applied and sampling depth limitations in each type of experiments.
Differential Gene Expression in Panicles: LYP9 versus 93-11 and PA64s
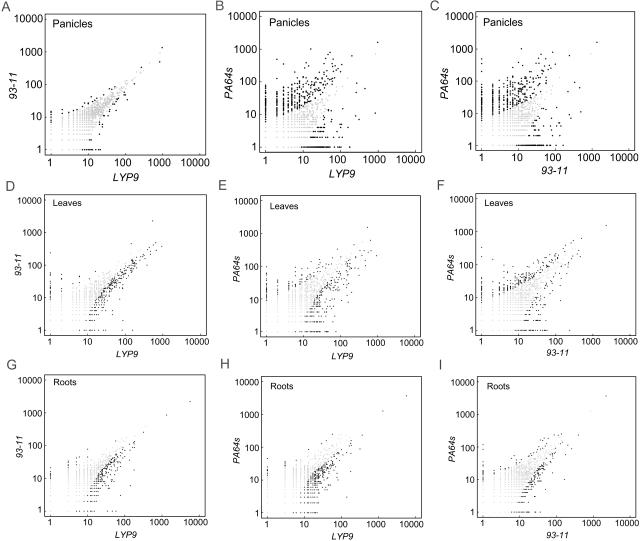
We obtained 68,549 tags from panicles of LYP9, which represented 23,178 unique transcripts (33.8%). Comparable numbers of unique tags were also acquired from the two parental lines: 22,890 out of a sum of 69,550 from PA64s and 11,878 out of a sum of 47,086 from 93-11. Scatterplot analysis indicated that the expression profile in panicles of LYP9 closely resembled that of its paternal line rather than the maternal line with a correlation coefficient of 0.95 (Fig. 4, A–C). The result is consistent with field observations on several phenotypic characteristics of 93-11. Table V lists some of the differentially expressed genes in panicles of the triad. There were only 84 genes categorized as significantly differentially expressed (P < 0.01) between the hybrid and its paternal strain due to their rather similar expression profiles, in comparison to 907 and 914 genes found between the hybrid and its maternal line and between the two parental lines, respectively.
Figure 4.
Scatterplots of tag frequencies compared in a pair-wise fashion among three sampled tissues. Black dots represent tags that are significantly differential expression (P < 0.01) between two libraries.
Table V.
Differentially expressed tags in panicles of LYP9 (P < 0.01)
| Tag
|
Copy No.
|
Ratioa
|
P-Value
|
Accession No.
|
Gene Description
|
|||
|---|---|---|---|---|---|---|---|---|
| P1 | N1 | L1 | L1 versus N1 | L1 versus P1 | ||||
| Up-Regulated Tags | ||||||||
| AGCTAAATAT | 1 | 0 | 21 | 21.0 | 0 | 1.67E-04 | NA | NA |
| TGTTTGTGCC | 0 | 0 | 19 | 19.0 | 0 | 7.67E-05 | NA | NA |
| GGCTCGGATC | 0 | 0 | 16 | 16.0 | 1.00E-05 | 2.53E-04 | AK066070 | Z. mays chlorophyll a/b-binding apoprotein CP24 (Lhcb6-1) |
| GAGACGCGCT | 0 | 0 | 16 | 16.0 | 1.00E-05 | 2.53E-04 | NA | NA |
| GTAGCGCCAG | 1 | 0 | 13 | 13.0 | 1.10E-04 | 6.65E-03 | AK069926 | Common tobacco RNA binding protein homolog |
| CCAAGGATAG | 0 | 0 | 13 | 13.0 | 1.10E-04 | 1.14E-03 | NA | NA |
| TATGTTGTTT | 0 | 0 | 12 | 12.0 | 2.00E-04 | 1.89E-03 | AK059894 | Chr. 1 |
| TTTGCTTGTT | 0 | 0 | 11 | 11.0 | 4.13E-04 | 3.23E-03 | AK107933 | Chr. 4 |
| CCGTTTTTGG | 0 | 0 | 11 | 11.0 | 4.13E-04 | 3.23E-03 | NA | NA |
| TTCGAGCGAA | 0 | 3 | 20 | 10.0 | 2.17E-04 | 4.34E-05 | NA | NA |
| AGGCGTTTAA | 0 | 0 | 9 | 9.0 | 1.87E-03 | 9.33E-03 | NA | NA |
| TAAGAGATGC | 0 | 2 | 12 | 8.0 | 5.22E-03 | 1.89E-03 | AK068865 | At. oligopeptide transporter family protein similar to Z. mays iron-phytosiderophore transporter protein yellow stripe 1 |
| CATTATTATC | 2 | 5 | 24 | 6.9 | 2.87E-04 | 2.13E-04 | AK069317 | MADS box-like protein similar to At. AP3 |
| GAGAACTGAG | 0 | 6 | 24 | 6.9 | 6.70E-04 | 3.34E-06 | NA | NA |
| CCTTGTGGTA | 0 | 7 | 20 | 5.0 | 8.90E-03 | 4.34E-05 | NA | NA |
| AAGTGCGTAC | 1 | 11 | 26 | 4.3 | 8.90E-03 | 6.68E-06 | AK064842 | Ribosomal protein L35 (NH77 gene) |
| GGGGAATATG | 0 | 48 | 92 | 3.8 | 5.33E-05 | 0 | AK060318 | At. 2OG-Fe(II) oxygenase family protein similar to FHT |
| AATAAAGTAG | 11 | 494 | 835 | 3.3 | 0 | 0 | AK071196 | Chitinase (Chia1d) |
| GTTTTGAATA | 1 | 105 | 173 | 3.3 | 1.00E-05 | 0 | AK069922 | Z. mays group 3 pollen allergen |
| GTTGGGACGT | 2 | 38 | 65 | 3.3 | 4.13E-03 | 0 | NA | NA |
| ATCGATTAGT | 1 | 58 | 93 | 3.2 | 2.06E-03 | 0 | AK069940 | Chr.8 |
| TTACCTGTAA | 6 | 30 | 56 | 3.1 | 2.79E-03 | 0 | NA | NA |
| Down-Regulated Tags | ||||||||
| ACAAGTTTTT | 794 | 85 | 52 | 16.9 | 4.21E-03 | 0 | AK060847 | Rubisco activase small isoform precursor (OsrcaA2) |
| TGAAATTCCT | 19 | 14 | 3 | 11.0 | 6.95E-03 | 1.00E-05 | AK058509 | Peroxiredoxin |
| ATTGAGTTGC | 32 | 27 | 8 | 7.4 | 1.11E-03 | 0.00E+00 | AK067703 | OsRad6 similar to At. UBC2 |
| CGTTCGCTAG | 83 | 38 | 17 | 7.1 | 3.87E-03 | 0 | NA | NA |
| ACATCTATTT | 13 | 17 | 5 | 6.0 | 8.37E-03 | 6.69E-03 | AK058290 | At. peptidyl-prolyl cis-trans isomerase PPIC-type family protein |
| ATGGTGCTGT | 18 | 21 | 7 | 5.6 | 6.61E-03 | 1.61E-03 | NA | NA |
| AAGCGGCCGC | 1,617 | 1,352 | 970 | 3.1 | 0 | 0 | NA | NA |
Ratios are calculated as ratio = L1/[(P1 + N1)/2] for up-regulated tags and [(P1 + N1)/2]/L1 for down-regulated tags. For calculation of ratios, a tag value of 1 is used to avoid division by zero. NA indicates that gene annotations are not available. At. stands for Arabidopsis.
We annotated 29 tags that were significantly differentially expressed and shared by the hybrid and its parental lines with high confidence. Among them, 22 tags were found up-regulated and seven tags were defined as down-regulated in LYP9 (Table V). Among the up-regulated genes, there was a MADS box-like protein (AK069317), similar to the Arabidopsis floral homeotic protein APETALA3 (AP3). Another up-regulated gene (AK060318) is a protein similar to an Arabidopsis oxidoreductase, which belongs to 2-oxoglutarate- and ferrous iron-dependent oxygenase family and similar to flavanone 3-hydroxylase (FHT). FHT and several other members of 2-oxoglutarate- and ferrous-iron-dependent oxygenase family are involved in the biosynthesis of flavonoids including flavonols, anthocyanins, and catechins (Turnbull et al., 2004). In addition, a gene of this up-regulated group (AK068865) encodes a protein homologous to an Arabidopsis oligopeptide transporter family protein and also similar to Zea mays iron-phytosiderophore transporter protein yellow stripe1 (Ys1). Ys1 encodes a Fe(III)-phytosiderophore transporter protein (Roberts et al., 2004). A final example is OsChia1d (AK071196), a chitinase that belongs to pathogenesis-related proteins. The OsChia1d gene is highly expressed in floral organs but not or at an extremely low level in vegetative organs (Takakura et al., 2000). It appeared that most of these up-regulated genes were related to the growth and development of panicles.
Most of the down-regulated genes in the hybrid were found related to protein processing (maturation and degradation). Examples are OsRCAA2 (AK060847) that encodes Rubisco activase small isoform precursor, OsRad6 (AK067703) similar to the Arabidopsis ubiquitin-conjugating enzyme (UBC2), and peptidyl-prolyl cis-trans isomerase (PPIase; AK058290). PPIase catalyzes rotations of X-Pro peptide bonds from a cis to trans conformation, a rate-limiting step in protein folding, and is very important since over 90% of proteins contain trans prolyl imide bonds. Plant PPIase-type family proteins, such as cyclophilins, are likely to be important proteins involved in a wide variety of cellular processes (Romano et al., 2004). Another example of down-regulated genes is thioredoxin (Trx) peroxidase (AK058509), a protein belonging to the alkyl hydroperoxide reductase and thiol-specific antioxidant family (Jung et al., 2002). The relationship among the up- and down-regulated genes in the hybrid panicles was not obvious from our preliminary analysis.
Differential Gene Expression in Leaves: LYP9 versus 93-11 and PA64s
From the leaf libraries, we identified a sum of 135,561 tags composed of 36,226 from LYP9, 52,515 from 93-11, and 46,820 from PA64s; among three data groups, 9,873, 15,398, and 13,927 were characterized as unique tags, respectively. Pair-wise comparisons yielded similar numbers of differentially expressed genes at a significant P-value (P < 0.01): 458 from LYP9 versus 93-11, 596 from LYP9 versus PA64s, and 510 from 93-11 versus PA64s (Fig. 4, D–F). Not only were 243 tags found between the hybrid and both parental lines, but also an overwhelming majority of them were classified as up-regulated genes in the hybrid, 228 tags (92%) out of the total (Supplemental Table I). Only 15 tags were found down-regulated in LYP9 (Table VI).
Table VI.
Differentially expressed tags in leaves of LYP9 (P < 0.01)
| Tag
|
Tag No.
|
Ratioa
|
P-Value
|
Accession No.
|
Gene Description
|
|||
|---|---|---|---|---|---|---|---|---|
| P2 | N2 | L2 | L2 versus P2 | L2 versus N2 | ||||
| Up-Regulated Tags (≥8.5-fold) | ||||||||
| GAGAAATCTG | 1 | 1 | 26 | 26.0 | 0 | 0 | AK100585 | Small GTP-binding protein RACBP (RACB) |
| TCTGGTTCTT | 1 | 0 | 26 | 26.0 | 0 | 0 | NM | NM |
| CGGGTGCGCG | 0 | 4 | 46 | 18.4 | 0 | 0 | AK109382 | At. probable NADP-dependent oxidoreductase P1 |
| AGTTTGATTT | 1 | 0 | 15 | 15.0 | 4.99E-05 | 0 | NM | NM |
| CTCAGTTCAC | 1 | 1 | 13 | 13.0 | 1.87E-04 | 8.67E-05 | AK062671 | At. calmodulin-related protein, putative |
| GTTTCCTATG | 3 | 1 | 26 | 13.0 | 0 | 0 | AK062869 | None |
| ATGGACAATG | 1 | 1 | 13 | 13.0 | 1.87E-04 | 8.67E-05 | AK072658 | At. clone 123128 mRNA |
| CCGGCCGTCT | 1 | 1 | 13 | 13.0 | 1.87E-04 | 8.67E-05 | NM | NM |
| GGAAATGTAA | 1 | 1 | 13 | 13.0 | 1.87E-04 | 8.67E-05 | NM | NM |
| CCAATGGCTT | 1 | 1 | 12 | 12.0 | 3.93E-04 | 1.50E-04 | NM | NM |
| GTTGAGATGG | 0 | 1 | 11 | 11.0 | 1.30E-04 | 3.50E-04 | AK102265 | Wheat porphobilinogen deaminase |
| CGCGTGTTGT | 0 | 1 | 11 | 11.0 | 1.30E-04 | 3.50E-04 | AK063647 | Putative psk4 |
| CGCGCCGCCG | 1 | 1 | 11 | 11.0 | 7.77E-04 | 3.50E-04 | AK105817 | At. myb family transcription factor |
| CGTCACTGGT | 0 | 1 | 11 | 11.0 | 1.30E-04 | 3.50E-04 | AK066613 | At. unknown protein |
| CATCTTGTCT | 2 | 4 | 30 | 10.0 | 0 | 0 | AK070467 | Trypsin inhibitor |
| ACATTGCCTG | 0 | 0 | 9 | 9.0 | 5.13E-04 | 3.03E-04 | AK062116 | At. PYRase family protein |
| ATCAGCTTGT | 1 | 1 | 9 | 9.0 | 3.65E-03 | 1.89E-03 | AK108154 | Phytoene synthase radicle isoform |
| AGCCACCTGA | 0 | 1 | 9 | 9.0 | 5.13E-04 | 1.89E-03 | AK068727 | ATP-dependent Clp protease ATP-binding subunit precursor (CLPD1) |
| ATCCGATAAT | 0 | 1 | 9 | 9.0 | 5.13E-04 | 1.89E-03 | NM | NM |
| GGAATCCGCC | 0 | 0 | 9 | 9.0 | 5.13E-04 | 3.03E-04 | NM | NM |
| ACCACCTCCG | 1 | 0 | 9 | 9.0 | 3.65E-03 | 3.03E-04 | NM | NM |
| GTCGTCGCGC | 1 | 2 | 13 | 8.7 | 1.87E-04 | 3.80E-04 | AK063719 | None |
| ATTTAAGTTC | 1 | 3 | 17 | 8.5 | 1.00E-05 | 9.67E-05 | AK061602 | Ipomoea nil PnFL-2 (unknown protein) |
| CCCATTGTGTb | 1 | 3 | 17 | 8.5 | 1.00E-05 | 9.67E-05 | AK059278 | At. calcium-binding EF hand family protein |
| AK103409 | At. calcium-binding EF hand family protein | |||||||
| Down-Regulated Tags | ||||||||
| GATGGAACGG | 41 | 13 | 1 | 27.0 | 0 | 6.59E-03 | AK098940 | At. peptidase M48 family protein contains Pfam domain |
| GATGAGTGGG | 25 | 10 | 0 | 17.5 | 0 | 5.21E-03 | AK064774 | At. ATG1 |
| AAAAAAAAAA | 93 | 46 | 6 | 11.6 | 0 | 3.34E-06 | AK107232 | None |
| TTATTATCTT | 21 | 27 | 4 | 6.0 | 3.53E-03 | 7.87E-04 | AK099513 | Deschampsia antarctica polyubiquitin 2 |
| AGCAGGCAAG | 1,546 | 2,284 | 535 | 3.6 | 0 | 0 | AK059621 | Pea Chloroplast 4.5S, 5S, 16S, and 23S |
| TAAAAAAAAAb | 17 | 13 | 1 | 15.0 | 4.93E-04 | 6.59E-03 | AK059235 | Potato cytochrome P450 (CYP71D4) |
| AK110892 | Tobacco UDP-Glc:salicylic acid glucosyltransferase (SA-GTase) | |||||||
| AGTTGGAGGCb | 25 | 16 | 2 | 10.3 | 6.00E-05 | 6.56E-03 | AK060121 | At. rubredoxin family protein contains Pfam profile |
| AK107112 | Ceratopteris richardii transcription factor (CerMADS1) | |||||||
| GAAAAAAAAA | 21 | 20 | 1 | 20.5 | 3.67E-05 | 2.27E-04 | NA | NA |
| CCCAAGGACA | 28 | 10 | 0 | 19.0 | 0 | 5.21E-03 | NA | NA |
| GGATTACATC | 19 | 19 | 1 | 19.0 | 1.87E-04 | 4.23E-04 | NA | NA |
| CCATACTCCC | 38 | 37 | 2 | 18.8 | 0 | 0 | NA | NA |
| TACTTCAAAA | 21 | 16 | 1 | 18.5 | 3.67E-05 | 1.73E-03 | NA | NA |
| TCAAGGACAC | 16 | 13 | 1 | 14.5 | 8.17E-04 | 6.59E-03 | NA | NA |
| GTGATGCGGC | 11 | 15 | 0 | 13.0 | 1.85E-03 | 3.30E-04 | NA | NA |
| TTGTCCAAAA | 68 | 44 | 6 | 9.3 | 0 | 6.68E-06 | NA | NA |
Ratios are calculated as ratio = L2/[(P2 + N2)/2] for up-regulated tags and [(P2 + N2)/2]/L2 for down-regulated tags. For calculation of ratios, a tag value of 1 is used to avoid division by zero. NA indicates that gene annotations are not available.
Tag matches more than one full-length cDNA. At. stands for Arabidopsis.
The most extraordinarily up-regulated genes in LYP9, in comparison to their expression in the parental lines (8.5-fold or greater), are listed in Table VI. Some of the up-regulated genes appeared related to chlorophyll or carotene synthesis pathways, albeit the majority of them were highly expressed in all the lines examined and only quantitative differences between the hybrid and its parental lines were noticed. One example is a porphobilinogen deaminase (AK102265) involved in the early steps of chlorophyll synthesis. Another gene (AK108154) encodes a phytoene synthase, the first of four specific enzymes essential for beta-carotene biosynthesis in plants. OsClpD1 (AK068727) encodes an ATP-binding subunit precursor of ATP-dependent Clp protease, one of the newly identified proteolytic systems in plant organelles that incorporate the activity of molecular chaperones to target specific polypeptide substrates and avoid inadvertent degradation of others, and expresses in chloroplasts (Zheng et al., 2002). OsClpD1 was also found expressed at a much-elevated level in the hybrid as opposed to in both parental lines. The most dramatically up-regulated gene (AK100585) encodes the small GTP-binding protein (RACBP), a member of Rac or Rop (Rhos in plants) GTPases family. In plants, Rac/Rop GTPases refer to a large number of Rac-like GTPases representing the Rho subfamily of the Ras-related small GTPase superfamily (Winge et al., 1997; Yang, 2002). They are molecular switches in signal transduction of many cellular processes, such as regulating hormone levels and subcellular Ca2+ gradients, organizing cytoskeletons, and producing reactive oxygen intermediates. The second most up-regulated gene (AK109382) is the NADP-dependent oxidoreductase, a component of the demonstrated antioxidative systems in plant peroxisomes. The expression level of OsPsk4 (AK063647), which encodes a precursor of phytosulfokine (PSK), increased more than 11-fold in the hybrid leaves. Another interesting up-regulated gene (AK062116) is the pyrrolidone-carboxylate peptidase (PYRase), an exopeptidase that selectively removes pyrrolidone carboxylic acid from some pyrrolidone carboxylic acid peptides and proteins and has been found in a variety of bacteria and in plant, animal, and human tissues. However, the exact role of PYRases in plants remains unclear. Other obviously up-regulated genes appeared in the categories of transcription factors, signal cascade proteins, and stress-induced proteins.
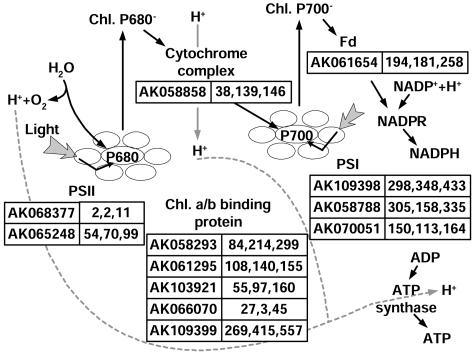
One group of up-regulated genes was found involved in photosynthesis (Fig. 5). Five of them (AK061295, AK066070, AK103921, AK058293, and AK109399) encode chlorophyll a/b binding proteins; two (AK068377 and AK065248) and three (AK070051, AK058788, and AK109398) of them are PSII and PSI component genes, respectively. We also identified various genes involved in the antioxidant system, including ascorbate peroxidase (APX; AK070842), catalase (AK066378), Trx (AK059196), and glutaredoxin (AK105335). There are two major systems to maintain thiols essentially in the reduced state in the cytosol, the Trx and the glutathione/glutaredoxin systems (Gelhaye et al., 2003). A complete list of genes that up-regulated in hybrid rice is summarized in Supplemental Table I.
Figure 5.
Expression patterns of up-regulated genes in hybrid leaves, which are involved in the light reaction of photosynthesis pathways. The numbers of tags in PA64s, 93-1, and the hybrid (LYP9) are listed.
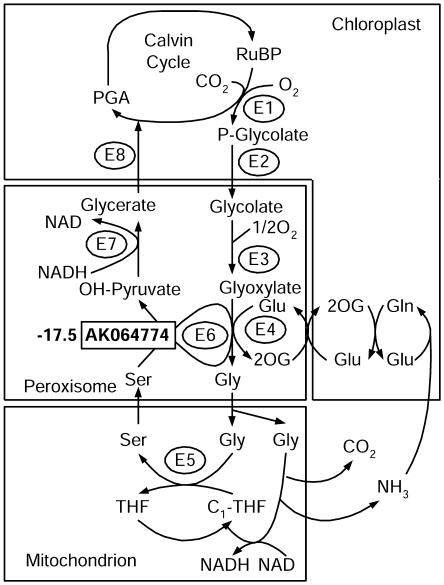
We identified a limited number of significantly down-regulated genes in LYP9 (Table VI). The most obvious gene (AK098940) encodes a protein similar to Arabidopsis peptidase M48 family proteins, which functions in protein degradation. The second most down-regulated gene (AK064774) is a rice homolog of the Arabidopsis Ala:glyoxylate aminotransferase 1 (AGT1; Fig. 6).
Figure 6.
Expression pattern of a down-regulated gene in hybrid rice, which is involved in the photorespiratory pathway. The average ratio of tag expression between the hybrid (LYP9) and its parental cultivars (means) is listed. Photorespiration enzymes: E1, Rubisco; E2, phosphoglycolate phosphatase; E3, glycolate oxidase; E4, Glu:glyoxylate aminotransferase; E5, Gly decarboxylase/Ser transhydroxymethytransferase; E6, Ser:glyoxylate aminotransferase; E7, hydroxypyruvate reductase; E8, glycerate kinase. PGA, 3-phosphoglycerate; P-glycolate, 2-phosphoglycolate; RuBP, ribulose 1,5-bisphosphate.
Differential Gene Expression in Roots: LYP9 versus 93-11 and PA64s
From the root-specific SAGE libraries, we assembled 10,871 (from a total of 28,902 tags), 18,084 (from 48,238), and 19,602 (from 67,793) unique tags from LYP9, 93-11, and PA64s, respectively. Similar expression patterns were seen in roots between the hybrid and its parental lines based on the scatterplots (Fig. 4, G–I). A total of 348 differentially expressed tags were obtained by comparing the result from the hybrid to that of its parental lines, and the overwhelming majority of them (345 tags) were found up-regulated (Supplemental Table II). Only three genes appeared down-regulated in LYP9 (Table VII) and one of them (AK069098) was identified as Ramy1, a zinc-induced protein.
Table VII.
Differentially expressed tags in roots of LYP9 (P < 0.01)
| Tag
|
Copy No.
|
Ratioa
|
P-Value
|
Accession No.
|
Gene Description
|
|||
|---|---|---|---|---|---|---|---|---|
| P3 | N3 | L3 | L3 versus P3 | L3 versus N3 | ||||
| Up-Regulated Tags (≥6.5-fold) | ||||||||
| AAGCGGCCGA | 3 | 6 | 140 | 31.1 | 0 | 0 | NA | NA |
| TACTTGCGTA | 0 | 0 | 13 | 13.0 | 0 | 6.68E-06 | NA | NA |
| GAGAAAATTA | 1 | 0 | 12 | 12.0 | 3.34E-06 | 6.68E-06 | AK058284 | Z. mays photosystem II subunit PsbS precursor |
| CTTAGATACA | 0 | 0 | 12 | 12.0 | 3.34E-06 | 6.68E-06 | NA | NA |
| AAGCGGCCGG | 7 | 1 | 46 | 11.5 | 0 | 0 | NA | NA |
| AAGCGGCCGT | 12 | 3 | 76 | 10.1 | 0 | 0 | NA | NA |
| GATATATGGA | 0 | 1 | 10 | 10.0 | 3.34E-06 | 4.27E-04 | AK059750 | Hordeum vulgare photosystem I protein (PSI-L) |
| CCATTCTCCA | 0 | 1 | 10 | 10.0 | 3.34E-06 | 4.27E-04 | NA | NA |
| AGCAGGCAAG | 18 | 18 | 178 | 9.9 | 0 | 0 | AK059621 | Pea Chloroplast 4.5S, 5S, 16S and 23S |
| GAAAACTTGT | 1 | 3 | 19 | 9.5 | 0 | 0 | AK060103 | None |
| ACTTGCTGTG | 0 | 1 | 8 | 8.0 | 6.00E-05 | 2.24E-03 | AK072299 | At. expressed protein |
| TTATTTTGTT | 0 | 1 | 8 | 8.0 | 6.00E-05 | 2.24E-03 | AK070868 | None |
| AGACAAATGT | 1 | 1 | 8 | 8.0 | 3.87E-04 | 2.24E-03 | NA | NA |
| ATTATTGTTT | 0 | 1 | 8 | 8.0 | 6.00E-05 | 2.24E-03 | NA | NA |
| GAATTCGTCA | 0 | 1 | 8 | 8.0 | 6.00E-05 | 2.24E-03 | NA | NA |
| GACTTTGTAC | 0 | 1 | 8 | 8.0 | 6.00E-05 | 2.24E-03 | NA | NA |
| GCGGCCGCTC | 0 | 1 | 8 | 8.0 | 6.00E-05 | 2.24E-03 | NA | NA |
| ATAATTATTG | 1 | 0 | 8 | 8.0 | 3.87E-04 | 4.10E-04 | NA | NA |
| AGCAGGCAAC | 0 | 0 | 8 | 8.0 | 6.00E-05 | 4.10E-04 | NA | NA |
| CATTGGCTGA | 0 | 0 | 8 | 8.0 | 6.00E-05 | 4.10E-04 | NA | NA |
| CTCTTAGTTG | 0 | 0 | 8 | 8.0 | 6.00E-05 | 4.10E-04 | NA | NA |
| TGTTACCGTG | 0 | 0 | 8 | 8.0 | 6.00E-05 | 4.10E-04 | NA | NA |
| TTCCTAGTGT | 2 | 3 | 19 | 7.6 | 0 | 0 | NA | NA |
| TTTAGACGTG | 0 | 3 | 15 | 7.5 | 0 | 8.33E-05 | NA | NA |
| AACCGGCCGC | 2 | 2 | 15 | 7.5 | 0 | 6.68E-06 | NA | NA |
| AGTATATTTC | 5 | 5 | 36 | 7.2 | 0 | 0 | NA | NA |
| TTCGGGTGCA | 6 | 5 | 38 | 6.9 | 0 | 0 | AK061611 | Small subunit of ribulose-1,5-bisphosphate carboxylase |
| TAACTACGCT | 2 | 3 | 17 | 6.8 | 3.34E-06 | 1.00E-05 | AK102185 | At. glycosyl hydrolase family 17 protein similar to elicitor inducible chitinase |
| AATCTTTTCT | 7 | 8 | 51 | 6.8 | 0 | 0 | NA | NA |
| ACCATCCTGC | 0 | 2 | 10 | 6.7 | 3.34E-06 | 1.61E-03 | AK103503 | Photosystem II D1 protein |
| GGTTAATTAG | 2 | 1 | 10 | 6.7 | 2.33E-04 | 4.27E-04 | AK069642 | V. vinifera putative ripening-related protein (grip28) |
| GTAAATTATC | 1 | 2 | 10 | 6.7 | 3.67E-05 | 1.61E-03 | NA | NA |
| TTCTAGTTGG | 1 | 2 | 10 | 6.7 | 3.67E-05 | 1.61E-03 | NA | NA |
| GCTGTACTTG | 2 | 1 | 10 | 6.7 | 2.33E-04 | 4.27E-04 | NA | NA |
| TATTATACTA | 2 | 1 | 10 | 6.7 | 2.33E-04 | 4.27E-04 | NA | NA |
| TGTGTACGTGb | 1 | 5 | 20 | 6.7 | 0 | 6.68E-06 | AK060602 | S. oleracea PsbY precursor |
| AK064211 | At. putative flavanone 3-beta-hydroxylase | |||||||
| AACTGTGTTG | 2 | 2 | 13 | 6.5 | 1.00E-05 | 1.27E-04 | AK058989 | At. myb-like DNA-binding domain and Zinc finger |
| GGATGATTTG | 3 | 0 | 13 | 6.5 | 3.67E-05 | 6.68E-06 | AK073356 | Putative universal stress protein USP1 |
| Down-Regulated Tags | ||||||||
| TTCGGGGCTC | 21 | 11 | 0 | 16.0 | 6.03E-04 | 5.67E-03 | NA | NA |
| CTGGGAGATG | 16 | 12 | 0 | 14.0 | 3.23E-03 | 3.84E-03 | NA | NA |
| TAACAGCGAG | 249 | 139 | 57 | 3.4 | 6.67E-06 | 8.36E-03 | AK069098 | Ramy1, zinc-induced protein |
Ratios are calculated as ratio = L3/[(P3 + N3)/2] for up-regulated tags and [(P3 + N3)/2]/L3 for down-regulated tags. For calculation of ratios, a tag value of 1 is used to avoid division by zero. NA indicates that gene annotations are not available.
Tag matches more than one full-length cDNA. At. stands for Arabidopsis.
Four of the most up-regulated genes in the hybrid roots are related to photosynthesis (Table VII), which encode PSII and PSI subunits (AK058284, AK059750, AK061611, and AK103503). An elicitor inducible chitinase (AK102185), similar to Arabidopsis glycosyl hydrolase family 17 proteins, was found among the up-regulated genes (also seen earlier in the hybrid panicles), together with another chitinase, OsChia1d. Other interesting genes among the group were a zinc finger protein (AK058989) and those involving in amino acid metabolism, glycolysis/TCA cycle, antioxidant system, signal cascade, and stress-induced reaction. A complete list of genes that up-regulated in hybrid rice is shown in Supplemental Table II.
DISCUSSIONS
SAGE Is a Powerful Tool in Exploiting Gene Expression Profiles in Rice
In this study, we performed a broad survey on gene expression profiles of a hybrid rice strain and its parental lines with nearly one-half million SAGE tags, coupled with comparative analysis on other available data from FL-cDNA, EST, protein, and other SAGE studies. Despite the fact we analyzed only the tags confirmed by FL-cDNA data, the rest, almost a comparable amount to the predicted number of genes for the rice genome, is freely released to the rice research community and public databases for future analysis as well as gene discovery and annotation. Another notion is that there are some incomplete confirmations of SAGE results with other types of data, such as those of EST sampling, reverse transcription-PCR, and microarrays. The reasons are rather complex. In our comparative analysis with the EST result, the incomplete overlapping of the data is largely due to the source materials, which are collected at different developmental stages of the panicle, one from the flowering stage and the other from the pollen-maturing stage before flowering. Most often are systematic biases created by different experimental protocols, such as mRNA preparations, cloning procedures, primer designs, and freshness of experimental materials. Nevertheless, SAGE provides an inexpensive choice for not only expression profiling but also the discovery of rare transcripts largely attributable to large samples sizes, especially when ample and multifaceted samples are to be handled at the same time.
Many Up-Regulated Genes Discovered in the Hybrid Leaves and Roots Have Functions in Promoting C- and N-Assimilation
Plant productivity or yields are usually dependent on the source-and-sink relationship, i.e. the capacity of source, largely the leaf, to fix carbon dioxide (CO2) and the capacity of developing sink tissues or organs to assimilate and convert the fixed carbon into the dry matter. The synergy between CO2 and nitrate (NO3−) assimilations as well as their dynamics is of key importance for crop productivity. In leaves, an accelerated CO2 assimilation involves intensive gene regulations in the chloroplasts, particularly the light harvesting chlorophyll-protein complexes, electron transports, and NADPH-reducing components of thylakoids, the CO2 assimilating enzyme Rubisco, and other enzymes required for CO2 assimilation in the stroma (Lawlor, 2002). In our study, a large number of genes involved in photosynthesis were identified as up-regulated in the hybrid, such as genes encoding pigment synthesis enzymes, chlorophyll binding proteins, Rubisco, and other members and regulators of the photosynthetic system. This result strongly suggests that the increased expression of photosynthesis-promoting genes in leaves may be an important factor in heterosis. In addition, as the output of the photosynthetic process, Suc and its storage form, the starch, are synthesized, partitioned between storage carbohydrates, and exported to sink tissues. A number of genes encoded β-amylase and Suc transporters were classified as up-regulated genes in the hybrid, perhaps to facilitate the source-to-sink trafficking between relevant compartments involved in energy generation, conservation, and consumption. The notion is also supported by the fact that the expression level of a starch phosphorylase involved in converting starch into Suc was found up-regulated in a wheat (Triticum aestivum) hybrid (Wu et al., 2003). Among other up-regulated, functionally known genes identified in our study was PSK, a peptide growth factor important in regulating cell proliferation and differentiation in higher plants. PSK was originally isolated from conditioned medium from mesophyll culture of asparagus (Matsubayashi and Sakagami, 1996). One of its two isoforms (PSK-α and PSK-β), PSK-α has been found enhancing growth and chlorophyll content of Arabidopsis seedlings under high nighttime temperature conditions (Yamakawa et al., 1999). The elevated expression of psk4 found in our study suggests that PSK may play an important role in regulating chlorophyll synthesis.
We also noticed some of the up-regulated genes were related to nitrogen assimilation, especially in the hybrid roots (see Supplemental Table II). Nitrogen uptake in the root depends on volume of soil exploited and rooting density, which affect the efficiency of absorbing nitrogen (Lawlor, 2002). Aside from genes related to photosynthesis, a large number of up-regulated, root-associated genes in the hybrid appeared related to respiration (i.e. glycolysis, TCA cycle, and respiratory chain), suggesting a higher cell growth rate in its roots. Examples are several up-regulated genes identified as associated to root hair initiation, such as expansion genes, originally demonstrated in Arabidopsis (Cho and Cosgrove, 2002), and cytoskeleton components, crucial for root hair tip growth (Mathur et al., 2003). Other categorized up-regulated genes have been indicated playing roles in nitrogen uptake, including ammonium transporters (Lin et al., 2000; Quaggiotti et al., 2003; Suenaga et al., 2003), carbonic anhydrases (Galvez et al., 2000; Buvana and Kannaiyan, 2002), and Rubisco activase (Okubara et al., 1999).
Increased photosynthesis and respiration in the hybrid may generate an enhanced production of reactive oxygen species (ROS). Therefore, an improved ROS-scavenging mechanism is required in the hybrid to ensure plant survival, growth, and productivity. Various antioxidative enzymes with crucial functions in protecting cellular components under ROS-generating stress conditions, such as APX, glutathione reductase, glutathione S-transferase, superoxide dismutase, and catalase (Noctor and Foyer, 1998; Noctor et al., 1998) were also up-regulated in the hybrid roots. A hexaploid wheat thylakoid-bound APX mutant was found exhibiting impaired electron transport, photosynthetic activity, and biomass accumulation (Danna et al., 2003). We also discovered that several classes of stress-induced genes were up-regulated in the hybrid roots, suggesting that a stress-stimulated cellular response may also be relevant to hybrid vigor. The examples are chitinases and metallothioneins. Plant chitinases appear responsible for the formation of elicitors (N-acetylchitooligosaccharides) from fungal cell walls to activate their own synthesis and other types of defense-related responses (Inui et al., 1996; Nishizawa et al., 1999). They have been indicated in our study as three chitinase genes identified as highly expressed in rice seedling based on a SAGE study (Matsumura et al., 1999). Members of chitinases have been shown to compensate developmental arrest during embryogenesis of a mutant carrot (De Jong et al., 1992). Metallothioneins showed elevated expression in our study and another strawberry study (Nam et al., 1999). Furthermore, they have been found in floral development, regulated by two MADS box-containing genes, AP3 and PI (PISTILLATA; Zik and Irish, 2003).
The genes mentioned above are merely representatives of the total genes found in this study as differentially expressed and their increased (or decreased) expressions are believed to have a potential effect on growth. The precise molecular mechanisms are not readily revealed by just expression level documentation, especially when regulatory genes are involved and the effect may also simply be a down-stream consequence of differential expression of a set of key regulatory proteins that are regulated by protein modifications, such as phosphorylation. It is, however, noteworthy that the complementation at transcriptome level is rather massive, suggesting that the underlying mechanisms may not be as simple as expected from studies of limited number of genes (Birchler et al., 2003). In addition, our proteomics data from comparative analysis on the rice embryos from the same triad have identified at least several percents of the proteins appeared uniquely (or highly expressed to the extent that the counterparts are deemed absent) expressed in the parental lines, and nearly one-half of them are discovered in the hybrid embryo (data not shown).
Diverse Classes of Up-Regulated Genes Identified in the Hybrid May Also Play Roles in Promoting Panicle Development
Pollen maturing stage is one of the most important developmental stages of rice panicles. The main characteristics of this stage are rapid growth of stamens and pistil and maturation of pollens. Several classes of genes were recognized as up-regulated genes in the hybrid panicles, and they are related to defense against phytopathogens (Paiva, 2000), mineral utilization, and flavonoid biosynthesis for flower coloring (Kazuma et al., 2003), pollen germination, and tube growth (van Eldik et al., 1997). Graminaceous plants secrete mugineic acid family phytosiderophores from their roots to solubilize Fe in the soil. Nicotianamine (NA) aminotransferase converts NA to mugineic acid family phytosiderophores (Shojima et al., 1990). Transgenic tobacco (Nicotiana tabacum) plants that constitutively express the barley (Hordeum vulgare) nicotianamine aminotransferase gene had flowers abnormally shaped and sterile due to disrupted distribution of Fe, Zn, and Mn (Takahashi et al., 2003). One of the rice YS1-like genes (OsYSLs), OsYSL2, encoding a rice metal-NA transporter, has been found responsible for the translocation of Fe and Mn from leaf into the developing grain (Koike et al., 2004). MADS-box genes encode a family of transcription factors that control diverse developmental processes in flowering plants, including the root, flower, and fruit. Many members of the MADS-box gene family work as floral organ identity genes (floral homeotic selector genes; Becker and Theissen, 2003). Several classes of them have been identified in Arabidopsis, including AP1, AG (AGAMOUS), PI, and AP3 (Yanofsky et al., 1990; Mandel et al., 1992; Mizukami and Ma, 1992; Goto and Meyerowitz, 1994; Jack et al., 1994). The continuous expression of AP3 is required to specify floral organ identity (Bowman et al., 1989) and required for conferring cell type identity as well as organ shape and size in the stamens (Jenik and Irish, 2001). Our results indicated that YS1-like genes, AP3 genes, and genes involved in flavonoid biosynthesis might all play an important role for the enhanced development of hybrid panicles.
The Down-Regulated Genes in the Hybrid Rice
Only a limited number of genes were identified as down-regulated in the hybrid and only a few were annotated. These down-regulated genes are mostly either related to photorespiratory or protein processing pathways, most noticeably in panicles and leaves of the hybrid. For instance, the most down-regulated genes in the hybrid leaves were identified as a peptidase and the AGT1; both were previously found and characterized in Arabidopsis. AGT1, localized in peroxisomes of Arabidopsis, is a key photorespiratory enzyme that has the highest specific activity with the Ser:glyoxylate aminotransferase reaction (Liepman and Olsen, 2001). Previous studies have shown that plant mutants lacking Ser:glyoxylate aminotransferase activity were not viable when grown in the atmospheric condition but survived when photorespiration was suppressed by increasing CO2 concentration in a growing environment (Somerville and Ogren, 1980; Murray et al., 1987). The enhancement of photorespiratory pathway severely diminishes the efficiency of CO2 assimilation and the yield of C3-crops (such as rice, wheat, soybean [Glycine max], and potato [Solanum tuberosum]) and improvement of crop yield of C3 plants can be achieved in an atmosphere containing elevated CO2 level, resulting in increased CO2 assimilation and suppressed photorespiration (Reynolds et al., 2000). Our results suggested that a suppressed photorespiration process and an accelerated photosynthesis in the hybrid leaves were fundamental factors in improving crop yield of the hybrid rice.
Another class of down-regulated genes seemed related to protein processing. The examples are several genes involving in both protein maturation and degradation, including peptidyl-prolyl cis-trans isomerase, glucosyltransferase, peptidase, and UBC2. A few transcription factors are also noticeable in the lists of down-regulated genes. Although we have been unable to plot plausible functional scenarios on precise roles of these genes at the present time, the finding undoubtedly provides useful clues for future detailed investigations, especially when the number of these apparently down-regulated genes is rather limited.
As a final note to this report, we believe that much broader expression-profiling surveys on hybrid rice strains and their parental lines should be encouraged in order to paint better pictures of the biological process of heterosis. Two fronts are to be especially explored: the methodology and the sampling. Among an increasing number of techniques used for large-scale gene expression studies, SAGE and microarray techniques are most inexpensive and efficient. However, microarrays are often constrained by gene discovery and prediction procedures, even when genome sequences become available, unless probes representing sequences of an entire genome are mounted on a set of microarrays; a few such attempts have been reported recently by using genomic sequence tags (Bertone et al., 2004; Hilson et al., 2004). The drawbacks of the SAGE method are its rather lengthy cloning processes that may sometimes introduce biased sampling and its tag length that may result in sequence ambiguity when matched to similar sequences, such as those of gene families. As to the aspect of sampling, it is indisputable that thorough investigations on gene expression profiling over an entire growth period and on most of the anatomic parts of a well-established hybrid and its parental lines are of essence in unveiling molecular details of hybrid vigor.
MATERIALS AND METHODS
Plant Materials and SAGE Library Construction
Rice (Oryza sativa) seeds were sown in green houses until reaching the seedling stage. The seedlings were transferred subsequently to an outdoor rice paddy. LYP9 and 93-11 were cultivated under typical conditions, and PA64s was maintained under male-sterility condition. First leaves were harvested at the milky stage of the grain ripening phase and panicles were from the top one-third portion at the pollen-maturing stage. Roots were collected at the first tillering stage. Total RNAs were prepared according to a LiCl-precipitation protocol (Lobreaux et al., 1992).
Poly(A+) RNA isolation, cDNA synthesis, and SAGE library construction were performed according to a published protocol (Lee et al., 2001). Briefly, mRNAs were purified with the Oligotex mRNA Mini kit (Qiagen USA, Valencia, CA) and double-stranded cDNAs were synthesized from the fractioned poly(A+)-containing mRNAs with 5′-biotinylated 3′-anchored oligo(dT) primers (5′-biotin-ATCTAGAGCGGCCGCdT16[A/G/CA/CG/CC]-3′). cDNAs were digested with NlaIII and collected with Streptavidin-coated magnet beads (Roche Diagnostic, Mannheim, Germany). After ligation with the two SAGE linkers, cDNAs were PCR-amplified with the sense SAGE primer 1 (5′-GGATTTGCTGGTGCAGTACA-3′) or SAGE primer 2 (5′-CTGCTCGAATTCAAGCTTCT-3′) and paired with the antisense primer (5′-ACTATCTAGAGCGGCCGCTT-3′). The antisense primer was located at the 3′ end of all cDNAs generated from the anchored oligo(dT) primers. Amplified cDNAs were released from the beads by BsmFI-digestion and subsequently purified on a polyacrylamide gel. Tag-containing cDNA fragments were blunt-ended, ligated, amplified, and redigested with NlaIII. Digested tags were concatemerized at their NlaIII overhangs. The ligation mixture was heated at 65°C for 15 min and separated on an 8% (w/v) polyacrylamide gel. DNA fragments between 500 and 1,000 bp in size were extracted and cloned into the SphI site of the pZero-1 vector (Invitrogen, Carlsbad, CA). Sequencing was carried out with the DYEnamic ET Terminator Cycle sequencing kit and Megabase 1000 sequencer (Amersham Biosciences, Uppsala). Sequence data were analyzed with SAGE 2000 (a software package kindly provided by Dr. Kenneth Kinzler's laboratory, Johns Hopkins University, Baltimore) that automatically detected and counted tags from the sequence data.
SAGE Data Analysis
The reference FL-cDNA dataset is a collection of 28,469 FL-cDNAs from KOME (Kikuchi, et al., 2003; ftp://cdna01.dna.affrc.go.jp/pub/data/20031024/). After removal of redundancy, 20,259 nonredundant sequences (nr-KOME-cDNAs) were used in our analysis (Yu et al., 2005). To generate virtual SAGE tags from the reference dataset, a 10-base tag was extracted from the immediate downstream sequence of a 3′-most NlaIII site (CATG), yielding 20,010 virtual tags, and only 1% of them matched more than one rice genomic sequence in the whole genome assembly of 93-11 (Yu et al., 2005).
Since the numbers of tags extracted from nine SAGE libraries were not exactly equal, a normalization (to 50,000 tags) procedure was performed (Porter et al., 2001). Pearson correlation coefficients were calculated for each pair-wise comparison with normalized tag values for each library (Hough et al., 2000). A P value of less than 0.01 for a difference in tag numbers between two libraries was said as significant, based on Monte Carlo simulation analysis and by using SAGE 2000 software (Zhang et al., 1997).
Other Rice Data
EST sequences (BGI ESTs) used for our analysis were published previously (Zhou et al., 2003). Although there were data from three cDNA libraries (Ls, Ps, and Ns) generated from rice leaves, Ls and Ps were constructed from leaf materials at the trefoil stage from LYP9 and PA64s, respectively, and Ns was made from cDNAs isolated from leaves of 93-11 at the first tillering stage. BGI proteomics data were acquired from fresh rice tissue samples at an early flowering stage by 2-D electrophoresis followed by protein identification with MALDI-TOF MS.
Acknowledgments
The authors acknowledge technical assistance provided by Jun Zhou in sequencing and Yong Tao in data analysis.
This work was supported by the Natural Science Foundation of China (grant no. 30370330 to J.Y., J.W., and H.Y.).
The online version of this article contains Web-only data.
References
- Becker A, Theissen G (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al (2004) Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Birchler JA, Auger DL, Riddle NC (2003) In search of the molecular basis of heterosis. Plant Cell 15: 2236–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvana R, Kannaiyan S (2002) Influence of cell wall degrading enzymes on colonization of N2 fixing bacterium, Azorhizobium caulinodans in rice. Indian J Exp Biol 40: 369–372 [PubMed] [Google Scholar]
- Chen JJ, Rowley JD, Wang SM (2000) Generation of longer cDNA fragments from serial analysis of gene expression tags for gene identification. Proc Natl Acad Sci USA 97: 349–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14: 3237–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrast R, Scott HS, Papasavvas MP, Rossier C, Antonarakis ES, Barras C, Davisson MT, Schmidt C, Estivill X, Dierssen M, et al (2000) The mouse brain transcriptome by SAGE: differences in gene expression between P30 brains of the partial trisomy 16 mouse model of Down syndrome (Ts65Dn) and normals. Genome Res 10: 2006–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna CH, Bartoli CG, Sacco F, Ingala LR, Santa-Maria GE, Guiamet JJ, Ugalde RA (2003) Thylakoid-bound ascorbate peroxidase mutant exhibits impaired electron transport and photosynthetic activity. Plant Physiol 132: 2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong AJ, Cordewener J, Lo Schiavo F, Terzi M, Vandekerckhove J, Van Kammen A, De Vries SC (1992) A carrot somatic embryo mutant is rescued by chitinase. Plant Cell 4: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman DR, Lorenz WW, Przybyla AE, Wolfe NL, Dean JF (2003) SAGE analysis of transcriptome responses in Arabidopsis roots exposed to 2,4,6-trinitrotoluene. Plant Physiol 133: 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fizames C, Munos S, Cazettes C, Nacry P, Boucherez J, Gaymard F, Piquemal D, Delorme V, Commes T, Doumas P, et al (2004) The Arabidopsis root transcriptome by serial analysis of gene expression. Gene identification using the genome sequence. Plant Physiol 134: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez S, Hirsch AM, Wycoff KL, Hunt S, Layzell DB, Kondorosi A, Crespi M (2000) Oxygen regulation of a nodule-located carbonic anhydrase in alfalfa. Plant Physiol 124: 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP (2003) Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction. FEBS Lett 555: 443–448 [DOI] [PubMed] [Google Scholar]
- Gibbings JG, Cook BP, Dufault MR, Madden SL, Khuri S, Turnbull CJ, Dunwell JM (2003) Global transcript analysis of rice leaf and seed using SAGE technology. Plant Biotechnol J 1: 271–285 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev 8: 1548–1560 [DOI] [PubMed] [Google Scholar]
- Gowda M, Jantasuriyarat C, Dean RA, Wang GL (2004) Robust-LongSAGE (RL-SAGE): a substantially improved LongSAGE method for gene discovery and transcriptome analysis. Plant Physiol 134: 890–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H (2003) Serial analysis of gene expression and cancer. Curr Opin Oncol 15: 44–49 [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14: 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P (2000) Prospects for crop improvement through the genetic manipulation of photosynthesis: morphological and biochemical aspects of light capture. J Exp Bot 51: 475–485 [DOI] [PubMed] [Google Scholar]
- Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, Cho KR, Riggins GJ, Morin PJ (2000) Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res 60: 6281–6287 [PubMed] [Google Scholar]
- Inui H, Yamaguchi Y, Ishigami Y, Kawaguchi S, Yamada T, Ihara H, Hirano S (1996) Three extracellular chitinases in suspension-cultured rice cells elicited by N-acetylchitooligosaccharides. Biosci Biotechnol Biochem 60: 1956–1961 [DOI] [PubMed] [Google Scholar]
- Jack T, Fox GL, Meyerowitz EM (1994) Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76: 703–716 [DOI] [PubMed] [Google Scholar]
- Jenik PD, Irish VF (2001) The Arabidopsis floral homeotic gene APETALA3 differentially regulates intercellular signaling required for petal and stamen development. Development 128: 13–23 [DOI] [PubMed] [Google Scholar]
- Jung BG, Lee KO, Lee SS, Chi YH, Jang HH, Kang SS, Lee K, Lim D, Yoon SC, Yun DJ, et al (2002) A Chinese cabbage cDNA with high sequence identity to phospholipid hydroperoxide glutathione peroxidases encodes a novel isoform of thioredoxin-dependent peroxidase. J Biol Chem 277: 12572–12578 [DOI] [PubMed] [Google Scholar]
- Kazuma K, Noda N, Suzuki M (2003) Flavonoid composition related to petal color in different lines of Clitoria ternatea. Phytochemistry 64: 1133–1139 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2004) OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J 39: 415–424 [DOI] [PubMed] [Google Scholar]
- Lawlor DW (2002) Carbon and nitrogen assimilation in relation to yield: mechanisms are the key to understanding production systems. J Exp Bot 53: 773–787 [PubMed] [Google Scholar]
- Lee JY, Lee DH (2003) Use of serial analysis of gene expression technology to reveal changes in gene expression in Arabidopsis pollen undergoing cold stress. Plant Physiol 132: 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Zhou G, Clark T, Chen J, Rowley JD, Wang SM (2001) The pattern of gene expression in human CD15+ myeloid progenitor cells. Proc Natl Acad Sci USA 98: 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Olsen LJ (2001) Peroxisomal alanine:glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J 25: 487–498 [DOI] [PubMed] [Google Scholar]
- Lin CM, Koh S, Stacey G, Yu SM, Lin TY, Tsay YF (2000) Cloning and functional characterization of a constitutively expressed nitrate transporter gene, OsNRT1, from rice. Plant Physiol 122: 379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobreaux S, Massenet O, Briat JF (1992) Iron induces ferritin synthesis in maize plantlets. Plant Mol Biol 19: 563–575 [DOI] [PubMed] [Google Scholar]
- Lu C, Zhou J (2000) Breeding and utilization of two-line interspecific hybrid rice Liangyou Peijiu. Hybrid Rice 15: 4–5 [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hulskamp M (2003) Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15: 1632–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y (1996) Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci USA 93: 7623–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H, Nirasawa S, Terauchi R (1999) Technical advance: transcript profiling in rice (Oryza sativa L.) seedlings using serial analysis of gene expression (SAGE). Plant J 20: 719–726 [DOI] [PubMed] [Google Scholar]
- Matsumura H, Reich S, Ito A, Saitoh H, Kamoun S, Winter P, Kahl G, Reuter M, Kruger DH, Terauchi R (2003) Gene expression analysis of plant host-pathogen interactions by SuperSAGE. Proc Natl Acad Sci USA 100: 15718–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Ma H (1992) Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71: 119–131 [DOI] [PubMed] [Google Scholar]
- Murray AJS, Blackwell RD, Joy KW, Lea PJ (1987) Photorespiratory N-donors, aminotransferase specificity and photosynthesis in a mutant of Barley deficient in serine:glyoxylate aminotransferase activity. Planta 172: 106–113 [DOI] [PubMed] [Google Scholar]
- Nam YW, Tichit L, Leperlier M, Cuerq B, Marty I, Lelievre JM (1999) Isolation and characterization of mRNAs differentially expressed during ripening of wild strawberry (Fragaria vesca L.) fruits. Plant Mol Biol 39: 629–636 [DOI] [PubMed] [Google Scholar]
- Nishizawa Y, Kawakami A, Hibi T, He DY, Shibuya N, Minami E (1999) Regulation of the chitinase gene expression in suspension-cultured rice cells by N-acetylchitooligosaccharides: differences in the signal transduction pathways leading to the activation of elicitor-responsive genes. Plant Mol Biol 39: 907–914 [DOI] [PubMed] [Google Scholar]
- Noctor G, Arisi AC, Jouanin L, Foyer CH (1998) Manipulation of glutathione and amino acid biosynthesis in the chloroplast. Plant Physiol 118: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Foyer CH (1998) Ascorbate And Glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 249–279 [DOI] [PubMed] [Google Scholar]
- Okubara PA, Pawlowski K, Murphy TM, Berry AM (1999) Symbiotic root nodules of the actinorhizal plant Datisca glomerata express Rubisco activase mRNA. Plant Physiol 120: 411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva NL (2000) An introduction to the biosynthesis of chemicals used in plant-microbe communication. J Plant Growth Regul 19: 131–143 [DOI] [PubMed] [Google Scholar]
- Porter DA, Krop IE, Nasser S, Sgroi D, Kaelin CM, Marks JR, Riggins G, Polyak K (2001) A SAGE (serial analysis of gene expression) view of breast tumor progression. Cancer Res 61: 5697–5702 [PubMed] [Google Scholar]
- Quaggiotti S, Ruperti B, Borsa P, Destro T, Malagoli M (2003) Expression of a putative high-affinity NO3- transporter and of an H+-ATPase in relation to whole plant nitrate transport physiology in two maize genotypes differently responsive to low nitrogen availability. J Exp Bot 54: 1023–1031 [DOI] [PubMed] [Google Scholar]
- Reynolds MP, van Ginkel M, Ribaut JM (2000) Avenues for genetic modification of radiation use efficiency in wheat. J Exp Bot 51 Spec No: 459–473 [DOI] [PubMed] [Google Scholar]
- Roberts LA, Pierson AJ, Panaviene Z, Walker EL (2004) Yellow stripe1. Expanded roles for the maize iron-phytosiderophore transporter. Plant Physiol 135: 112–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano PG, Horton P, Gray JE (2004) The Arabidopsis cyclophilin gene family. Plant Physiol 134: 1268–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojima S, Nishizawa NK, Fushiya S, Nozoe S, Irifune T, Mori S (1990) Biosynthesis of phytosiderophores. Plant Physiol 93: 1497–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull GH (1952) Beginnings of the heterosis concept. In JW Gowen, ed, Heterosis. Iowa State College Press, Ames, pp 14–48
- Somerville CR, Ogren WL (1980) Photorespiration mutants of Arabidopsis thaliana deficient in serine-glyoxylate aminotransferase activity. Proc Natl Acad Sci USA 77: 2684–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga A, Moriya K, Sonoda Y, Ikeda A, Von Wiren N, Hayakawa T, Yamaguchi J, Yamaya T (2003) Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol 44: 206–211 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Terada Y, Nakai I, Nakanishi H, Yoshimura E, Mori S, Nishizawa NK (2003) Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell 15: 1263–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakura Y, Ito T, Saito H, Inoue T, Komari T, Kuwata S (2000) Flower-predominant expression of a gene encoding a novel class I chitinase in rice (Oryza sativa L.). Plant Mol Biol 42: 883–897 [DOI] [PubMed] [Google Scholar]
- Turnbull JJ, Nakajima J, Welford RW, Yamazaki M, Saito K, Schofield CJ (2004) Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis: anthocyanidin synthase, flavonol synthase, and flavanone 3beta-hydroxylase. J Biol Chem 279: 1206–1216 [DOI] [PubMed] [Google Scholar]
- van Eldik GJ, Reijnen WH, Ruiter RK, van Herpen MM, Schrauwen JA, Wullems GJ (1997) Regulation of flavonol biosynthesis during anther and pistil development, and during pollen tube growth in Solanum tuberosum. Plant J 11: 105–113 [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW (1995) Serial analysis of gene expression. Science 270: 484–487 [DOI] [PubMed] [Google Scholar]
- Winge P, Brembu T, Bones AM (1997) Cloning and characterization of rac-like cDNAs from Arabidopsis thaliana. Plant Mol Biol 35: 483–495 [DOI] [PubMed] [Google Scholar]
- Wu J, Maehara T, Shimokawa T, Yamamoto S, Harada C, Takazaki Y, Ono N, Mukai Y, Koike K, Yazaki J, et al (2002) A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell 14: 525–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LM, Ni ZF, Meng FR, Lin Z, Sun QX (2003) Cloning and characterization of leaf cDNAs that are differentially expressed between wheat hybrids and their parents. Mol Genet Genomics 270: 281–286 [DOI] [PubMed] [Google Scholar]
- Xiao J, Li J, Yuan L, Tanksley SD (1995) Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140: 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa S, Matsubayashi Y, Sakagami Y, Kamada H, Satoh S (1999) Promotive effects of the peptidyl plant growth factor, phytosulfokine-alpha, on the growth and chlorophyll content of Arabidopsis seedlings under high night-time temperature conditions. Biosci Biotechnol Biochem 63: 2240–2243 [DOI] [PubMed] [Google Scholar]
- Yang Z (2002) Small GTPases: versatile signaling switches in plants. Plant Cell (Suppl) 14: S375–S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW (1997) Gene expression profiles in normal and cancer cells. Science 276: 1268–1272 [DOI] [PubMed] [Google Scholar]
- Zheng B, Halperin T, Hruskova-Heidingsfeldova O, Adam Z, Clarke AK (2002) Characterization of chloroplast Clp proteins in Arabidopsis: localization, tissue specificity and stress responses. Physiol Plant 114: 92–101 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Tang J, Walker MG, Zhang X, Wang J, Hu S, Xu H, Deng Y, Dong J, Ye L, et al (2003) Gene identification and expression analysis of 86,136 Expressed Sequence Tags (EST) from the rice genome. Genomics Proteomics Bioinformatics 1: 26–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zik M, Irish VF (2003) Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15: 207–222 [DOI] [PMC free article] [PubMed] [Google Scholar]