Abstract
Kinesin-like calmodulin-binding protein (KCBP), a member of the Kinesin-14 family, is a C-terminal microtubule motor with three unique domains including a myosin tail homology region 4 (MyTH4), a talin-like domain, and a calmodulin-binding domain (CBD). The MyTH4 and talin-like domains (found in some myosins) are not found in other reported kinesins. A calmodulin-binding kinesin called kinesin-C (SpKinC) isolated from sea urchin (Strongylocentrotus purpuratus) is the only reported kinesin with a CBD. Analysis of the completed genomes of Homo sapiens, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and a red alga (Cyanidioschyzon merolae 10D) did not reveal the presence of a KCBP. This prompted us to look at the origin of KCBP and its relationship to SpKinC. To address this, we isolated KCBP from a gymnosperm, Picea abies, and a green alga, Stichococcus bacillaris. In addition, database searches resulted in identification of KCBP in another green alga, Chlamydomonas reinhardtii, and several flowering plants. Gene tree analysis revealed that the motor domain of KCBPs belongs to a clade within the Kinesin-14 (C-terminal motors) family. Only land plants and green algae have a kinesin with the MyTH4 and talin-like domains of KCBP. Further, our analysis indicates that KCBP is highly conserved in green algae and land plants. SpKinC from sea urchin, which has the motor domain similar to KCBP and contains a CBD, lacks the MyTH4 and talin-like regions. Our analysis indicates that the KCBPs, SpKinC, and a subset of the kinesin-like proteins are all more closely related to one another than they are to any other kinesins, but that either KCBP gained the MyTH4 and talin-like domains or SpKinC lost them.
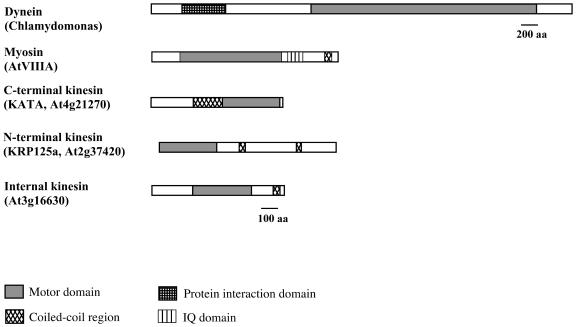
Members of the kinesin superfamily of microtubule (MT) motor proteins have been identified in many taxa ranging from protists to plants and animals (Reddy and Day, 2001; Vale, 2003). The members of the kinesin superfamily have a highly conserved motor domain with ATP- and MT-binding sites. In addition, most kinesins have a less conserved coiled-coil region that is important for dimerization and a nonconserved tail domain that is thought to interact with specific cargo (see Fig. 1). Kinesins bind MTs and a variety of cargoes and perform force-generating tasks such as transport of vesicles and organelles, spindle formation and elongation, chromosome segregation, and MT organization (Leopold et al., 1992; Sawin et al., 1992; Barton and Goldstein, 1996; Moore and Endow, 1996; Wein et al., 1996; Raich et al., 1998; Goldstein and Philip, 1999; Manning and Snyder, 1999; Reddy, 2001). Analysis of completed genome sequences of eukaryotes has resulted in identification of a large number of kinesins (Reddy and Day, 2001; Vale, 2003). Recently, all known kinesins have been grouped into 14 families that were designated as Kinesin-1 to Kinesin-14 (Lawrence et al., 2004). One of these families (Kinesin-14) possesses a C-terminal motor domain with minus-end motility, and the rest of the subfamilies have either an N-terminal or an internal motor domain (Fig. 1) and move toward the plus-end of MTs (Reddy and Day, 2001; Vale, 2003; Lawrence et al., 2004).
Figure 1.
Types of motors found in eukaryotic organisms. Examples are given from Arabidopsis except for dynein that has not been identified in the Arabidopsis genome. Domains are as shown in the key. Dynein, The protein interaction domain is implicated in heavy chain-heavy chain interaction and other protein interactions. Myosin, The myosin IQ domain binds calmodulin. Kinesins, Three major types of kinesins are represented: those with C-terminal, N-terminal, or internal-motor domain.
Plant kinesins were first identified in tobacco (Nicotiana tabacum) pollen tubes (pollen kinesin homolog) and tobacco phragmoplasts (tobacco kinesin related protein 125; Asada et al., 1991; Tiezzi et al., 1992; Cai et al., 1993). Since then, many kinesins have been identified in plants (Mitsui et al., 1993, 1994; Liu et al., 1996; Reddy et al., 1996a; Asada et al., 1997; Tamura et al., 1999; Barroso et al., 2000; Nishihama et al., 2002; Reddy, 2003; Tanaka et al., 2004). In Arabidopsis (Arabidopsis thaliana) alone there are at least 61 kinesins (Reddy and Day, 2001). Using genetic and cell biological approaches, functions of some plant kinesins have been reported (Reddy, 2003; Lee and Liu, 2004). Functional studies with a few kinesins have shown their involvement in mitosis and meiosis (Liu et al., 1996; Bowser and Reddy, 1997; Lee and Liu, 2000; Lee et al., 2001; Chen et al., 2002; Nishihama et al., 2002; Strompen et al., 2002; Yang et al., 2003; Pan et al., 2004; Ambrose et al., 2005), morphogenesis (Oppenheimer et al., 1997; Lu et al., 2005), and oriented deposition of cellulose myofibrils (Zhong et al., 2002). A kinesin that interacts with a geminivirus replication protein has also been reported in Arabidopsis (Kong and Hanley-Bowdoin, 2002). Predicted mitochondrial targeting sequences in two Arabidopsis kinesins mined from the database were shown to direct a green fluorescent protein fusion protein to mitochondria, suggesting that these kinesins are likely to function in these organelles (Itoh et al., 2001). A kinesin containing a calponin homology domain has been shown to interact with actin microfilaments, suggesting the involvement of some kinesins in coordinating actin and MT cytoskeleton (Preuss et al., 2004). The motility properties of only a few of these kinesins have been reported (Asada and Shibaoka, 1994; Song et al., 1997; Marcus et al., 2002).
A calmodulin-binding kinesin (kinesin-like calmodulin-binding protein [KCBP]) has been characterized from several flowering plants (Reddy et al., 1996a, 1996b; Wang et al., 1996; Abdel-Ghany and Reddy, 2000; Preuss et al., 2003). KCBP, like other kinesins, has a motor domain, a stalk, and a tail domain. In addition, it has some unique domains. These include: (1) a calmodulin-binding domain (CBD) at the C terminus that is responsible for Ca2+/calmodulin regulation of its ATPase activity, interaction with MTs, and motor activity (Narasimhulu et al., 1997; Song et al., 1997; Deavours et al., 1998; Narasimhulu and Reddy, 1998), and (2) the myosin tail homology region 4 (MyTH4) and the talin-like region found in some myosins (Reddy and Reddy, 1999). We have shown that the MyTH4 and talin-like region has an MT-binding site (Narasimhulu and Reddy, 1998). Recently, a similar region in animal myosins has also been found to bind MTs (Weber et al., 2004). Biochemical studies together with the crystal structure of the KCBP motor domain with the CBD indicate that calcium/calmodulin negatively regulates KCBP activity by blocking the MT-binding sites on the motor (Reddy and Reddy, 2002; Vinogradova et al., 2004).
Immunolocalization studies and antibody injection using antibodies specific to KCBP indicate that KCBP has a role in cell division (Bowser and Reddy, 1997; Smirnova et al., 1998; Vos et al., 2000). Consistent with its role in cell division, the level of KCBP is cell cycle regulated with a high amount in the mitotic phase (Bowser and Reddy, 1997). KCBP is localized to the plant-specific preprophase band, phragmoplast, and to the spindle apparatus in dividing cells (Bowser and Reddy, 1997; Smirnova et al., 1998; Preuss et al., 2003). KCBP is capable of bundling MTs, suggesting the involvement of KCBP in establishing mitotic MT arrays during different stages of cell division (Kao et al., 2000). Genetic studies have shown that KCBP is essential for trichome morphogenesis, and KCBP mutants (zwichel, zwi) have abnormal trichomes with a shorter stalk and a reduced number of branches (Oppenheimer et al., 1997; Reddy and Day, 2000). In cotton (Gossypium hirsutum) fibers, KCBP was shown to associate with cortical MTs as well as with mitotic MT arrays (Preuss et al., 2003). In addition to calmodulin, KCBP interacts with three other proteins (KCBP-interacting Ca2+ binding protein, ANGUSTIFOLIA, and KCBP-interacting protein kinase), and two of these are also involved in trichome morphogenesis (Day et al., 2000; Folkers et al., 2002; Reddy et al., 2004).
Searches of the National Center for Biotechnology Information (NCBI) and species-specific sequence databases have not identified a homolog of KCBP in the completely sequenced genomes of Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens, Cyanidioschyzon merolae, or Mus musculus (Miki et al., 2001; Schoch et al., 2003; Matsuzaki et al., 2004). However, a calmodulin-binding kinesin called kinesin-C (hereafter called SpKinC), a member of the C-terminal family, has been cloned from the sea urchin (Strongylocentrotus purpuratus; Rogers et al., 1999). The amino acid sequence of CBD of SpKinC shares 35% identity with KCBP. Since SpKinC lacks the MyTH4 and talin-like regions found in plant KCBPs, it is not classified as a KCBP.
The presence of KCBP in flowering plants but not in fungal or animal genomes raises two questions. Is KCBP found in photosynthesizing organisms but not other eukaryote lineages, and what is the level of conservation of KCBP among phylogenetically divergent eukaryotes? To answer these questions, we have cloned and characterized KCBPs from a gymnosperm (Picea abies) and a green alga (Stichococcus bacillaris). In addition, we searched NCBI sequence databases and other databases of phylogenetically divergent photosynthetic and nonphotosynthetic eukaryotes for KCBP (See “Materials and Methods”). Gene tree analysis suggests that the KCBP-like motor domain originated prior to the divergence of plants and animals. However, the addition of the MyTH4 and talin-like domains appears to be limited to the plant lineage. Our data suggest that KCBP is highly conserved in land plants and green algae.
RESULTS AND DISCUSSION
Cloning, Characterization, and Identification of KCBPs
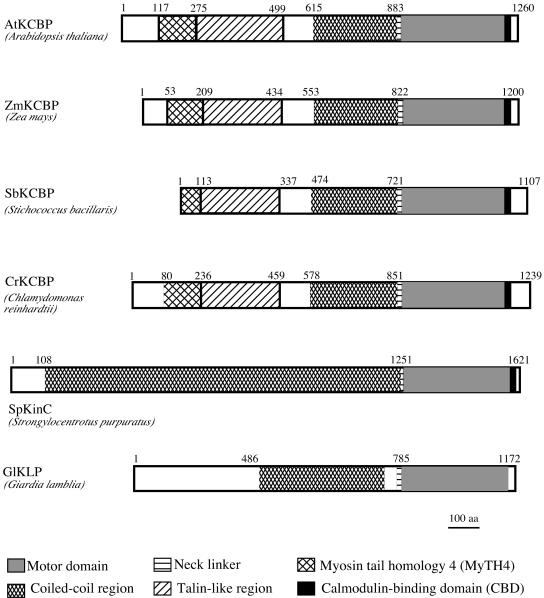
KCBP has been cloned from representative members of monocots and dicots and is highly conserved among angiosperms. In order to examine the presence and the level of conservation of unique domains in KCBP outside the angiosperms, we used PCR and library screening to clone KCBP from a gymnosperm (P. abies) and an alga (S. bacillaris). Primers corresponding to the conserved ATP-binding site of kinesins and the CBD of KCBP were used in PCR with P. abies cDNA library or S. bacillaris genomic DNA. PCR products from each species with sequences highly similar to KCBP from angiosperms were used as probes to screen the P. abies cDNA library and S. bacillaris library. Clones from the screens were isolated and sequenced. The predicted amino acid sequence (PaKCBP) of the longest isolated P. abies cDNA showed from 75% to 80.5% sequence identity to the motor domains of KCBPs from angiosperms. This clone contained the coding region for the motor domain and CBD. An S. bacillaris genomic clone was isolated that coded for a KCBP (SbKCBP, Fig. 2) with the unique KCBP domains (CBD, MyTH4, and talin-like region).
Figure 2.
Schematic diagram of representative KCBPs and proteins with motors most similar to the KCBP motor. Domains are as shown in the key. Amino acid numbers are given at each domain transition. The SbKCBP lacks some amino acids at the N terminus.
Using both the full-length and motor domain sequences of Arabidopsis KCBP (AtKCBP), we searched databases at NCBI, The Institute for Genomic Research (TIGR), and the Joint Genome Institute (JGI) including individual databases of species whose genomes are either completely sequenced or are being completed (see “Materials and Methods” for list of genomes searched), as well as expressed sequence tag (EST) databases, for the presence of KCBP homologs. KCBP sequences were identified only in land plants and green algae genomes (Table I). Using BLAST searches, full-length KCBPs were found in rice (Oryza sativa) and Chlamydomonas reinhardtii at NCBI and JGI, respectively. The rice amino acid sequence (OsKCBP) shows 90% similarity to Zea mays KCBP (ZmKCBP). The C. reinhardtii amino acid sequence (CrKCBP, Fig. 2) has the conserved-motor domain, CBD, coiled coil, MyTH4 region, and talin-like region found in other KCBPs. The sequence was originally generated from an unannotated scaffold (scaffold_633) DNA sequence using a splice site prediction program, two EST sequences, and comparison to the KCBP sequence. JGI has now annotated this sequence, and their annotation is in agreement with our generated sequence except that our sequence contains an additional sequence of about 100 amino acids at the N terminus, which is similar to other KCBPs. A scaffold sequence for a KCBP, which included the MyTH4, talin, coiled-coil, motor, and CBD domains, was also identified for Populus trichocarpa. Its sequence was closest to cotton KCBP. A KCBP-like sequence was found at http://moss.nibb.ac.jp/ for the moss Physcomitrella patens.
Table I.
KCBPs and kinesins with KCBP-like motor domain
| Species | Protein | Accession No./Index No. | Domains Present | Reference |
|---|---|---|---|---|
| Arabidopsis | AtKCBP | AAC49901 | MyTH4, talin, CCa, Neck, MDb, CBD | Reddy et al. (1996a) |
| C. reinhardtii | CrKCBP | BI996794 | MyTH4, talin, CC, Neck, MD, CBD | |
| BI723056 | ||||
| C_1030008c | ||||
| C. intestinalis | CiKLP | Scaffold_28d | Neck, MD (others NDe) | |
| C. paradoxa | CpKLP | AY515315 | MD, possible CBD (others ND) | Abdel-Ghany et al. (2000) |
| G. lamblia | GlKLP | EAA38074 | CC, Neck, MD; No MyTH4, talin, or CBD | |
| Cotton | GhKCBP | AAP41107 | MyTH4, talin, CC, Neck, MD, CBD | Preuss et al. (2003) |
| M. truncatula | MtKCBP | TC79642f | Neck, MD, CBD (others ND) | |
| Tobacco | NtKCBP | AAC49393 | MyTH4, talin, CC, Neck, MD, CBD | Wang et al. (1996) |
| Rice | OsKCBP | CAE03597 | MyTH4, talin, CC, Neck, MD, CBD | |
| P. abies | PaKCBP | AY515262 | Neck, MD, CBD (others ND) | |
| P. ramorum | PrKLP | 141193d | CC, Neck, MD; No MyTH4, talin, or CBD | |
| P. sojae | PsKLP | 84468d | CC, Neck, MD; No MyTH4, talin, or CBD | |
| P. trichocarpa | PtKCBLP | Scaffold LG_XId | MyTH4, talin, CC, MD, CBD | |
| S. tuberosum | StKCBP | AAB37756 | MyTH4, talin, CC, MD, CBD | Reddy et al. (1996b) |
| S. bacillaris | SbKCBP | AY515314 | MyTH4g, talin, CC, Neck, MD, CBD | |
| Sea urchin | SpKinC | AAF04841 | CC, Neck, MD, CBD; No MyTH4, talin | Rogers et al. (1999) |
| T. thermophila | TtKLP | AAQ16681 | Neck, MD (others ND) | |
| T. pseudonana | TpKLP | Scaffold_16d | Neck, MD (others ND) | |
| Z. mays | ZmKCBP | AAG13460 | MyTH4, talin, CC, Neck, MD, CBD | Abdel-Ghany and Reddy (2000) |
Coiled coil.
Motor domain.
Sequence available at http://genome.jgi-psf.org/chlre1/chlre1.home.html.
Sequences available at http://genome.jgi-psf.org/.
Not determined because full-length sequenced is not available.
Sequence available at http://www.tigr.org.
MyTH4 sequence is truncated.
Fungi genomes including yeasts and plasmodium do not have a KCBP. It has been previously reported that KCBP is not present in the human, Drosophila, or C. elegans genomes (Miki et al., 2001; Reddy and Day, 2001; Siddiqui, 2002). Searches of other animal genomes (mouse, rat, dog, etc.) at http://www.ncbi.nih.gov/Genomes/index.html did not identify a KCBP in these species. Searches of the genomes at JGI resulted in identification of scaffold sequences with similarity to the KCBP motor domain (but did not contain the domains unique to KCBP) in the water molds Phytophthora sojae and Phytophthora ramoru and the diatom Thalassiora pseudonana. A scaffold sequence from Ciona intestinalis showed closest similarity to sea urchin SpKINC. The predicted Giardia lamblia amino acid sequence (G. lamblia kinesin-like protein [GlKLP]; Fig. 2) encodes a region of coiled coil followed by a kinesin motor domain that in BLAST searches showed the most similarity to KCBP motor domain from angiosperms. However, it does not contain the MyTH4, talin-like region, or CBD domain. A partial kinesin sequence in the protozoan Tetrahymena thermophila in TIGR also showed similarity to KCBP motor domain but lacked the CBD. Proteins were named KCBP if they were placed in the KCBP clade in our gene tree analysis (see below) and contained MyTH4, talin-like region, and CBD. Proteins that have motor domains most similar to KCBPs, but (1) lack MyTH4 and talin-like regions and (2) are not nested within the KCBP clade are not considered to be KCBPs.
A region that defines kinesin class specificity is the neck region, an approximately 45-amino acid-long segment that emerges from the catalytic core of the motor domain (Vale and Fletterick, 1997). Carboxy-terminal kinesins as a group share a short conserved sequence (ELKGNI) in their neck region. The sequence of the neck region has been shown to be critical for directionality of motor movement (Endow and Waligora, 1998). The neck in KCBPs contains a conserved sequence (EDMKGIRV) that diverges slightly from other Kinesin-14 proteins (Fig. 3A). With the exception of SbKCBP (EDL) and GlKLP (EEM), all of the KCBPs and kinesins with a KCBP-like motor have an initial EDM sequence, whereas it is not conserved in other Kinesin-14 proteins.
Figure 3.
Alignments of conserved-neck region and CBD (see Fig. 2 for the location of these domains). A, A conserved region in the neck (883–894 in AtKCBP) from representative C-terminal kinesins (Kinesin-14 family) was aligned using DNA Star Megalign. The last four sequences are from non-KCBP Kinesin-14 family members. B, The region corresponding to the CBD of AtKCBP (1,217–1,239) was aligned with the representative C-terminal kinesins using DNA Star Megalign. Identical amino acids are shown by reverse lettering. At, Arabidopsis; Gh, cotton; Nt, tobacco; Mt, Medicago truncatcula; St, S. tuberosum; Pt, P. trichocarpa; Os, rice; Zm, Z. mays; Pa, P. abies; Cr, C. reinhardtii; Sb, S. bacillaris; Sp, sea urchin; Ci, C. intestinalis; Pr, P. ramorum; Tt, T. thermophila; Gl, G. lamblia; Ce, C. elegans; Dm, D. melanogaster.
A comparison of the sequence following the motor domain of known KCBPs and the KCBP-like proteins reported here is shown in Figure 3B. All reported KCBPs have a conserved CBD (70%–87% similar to AtKCBP) in this region. PaKCBP and CrKCBP show a high level of similarity to angiosperm CBDs (65% and 61%, respectively), while the SbKCBP sequence is more diverged (48%). A bacterially produced protein encompassing the CBD and motor domain of PaKCBP was shown to bind 35S calmodulin in the presence of Ca2+ but not in the presence of EGTA (data not shown). The sequence for the moss (P. patens) KCBP-like protein (PpKLP) shows 58% similarity to the CBD of AtKCBP. The sea urchin kinesin (SpKinC) showing similarity to the KCBP motor domain also contains a CBD, but the sequence is less conserved than the CBDs of angiosperm KCBPs (30% identical to AtKCBP). However, the bacterially expressed SpKinC has been shown to bind calmodulin (Rogers et al., 1999). A kinesin from C. intestinalis (CiKLP) also has a CBD that is 30% identical to AtKCBP and is similar to SpKinC (Fig. 3B). Although the binding of CiKLP to calmodulin has not been shown, it is likely that it also binds to calmodulin. The sequences following the motor domain in the T. thermophila and G. lamblia kinesin sequences have little similarity to the CBD of angiosperm KCBPs (9% and 13% identity to AtKCBP).
Gene Tree Analysis of KCBP-Like Proteins
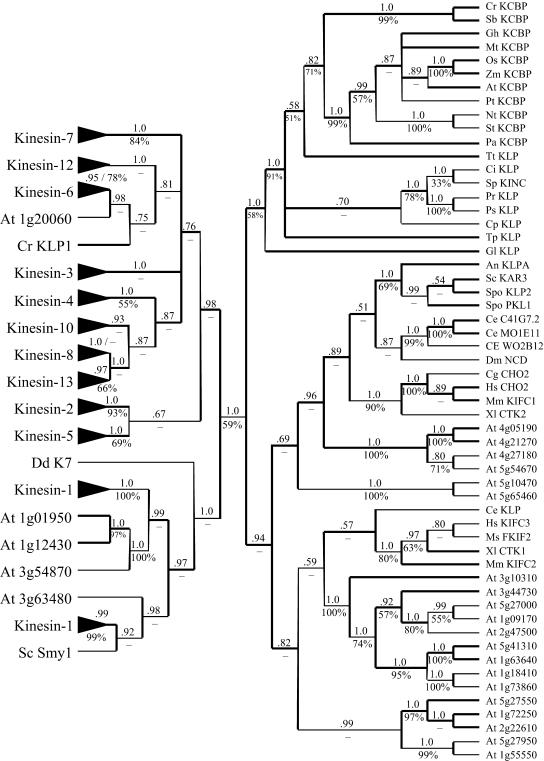
Supplemental data, consisting of the DIALIGN alignment, the data matrices, and the complete Bayesian and parsimony trees, are available online (Supplemental Figs. 1–5). The grouping of Arabidopsis kinesins in the complete trees is consistent with the earlier tree presented by Reddy and Day (2001). Both the Bayesian and parsimony analyses support all KCBPs as more closely related to one another than they are to any other kinesin family. The unrooted Bayesian tree for the kinesin superfamily, based on their motor domains, is presented in Figure 4. All families other than Kinesin-14 (C terminal) are represented as single branches in the abbreviated tree. The members of each group are provided in the supplemental figures. Consistent with the inferred phylogeny of plants (Melkonian and Surek, 1995; Lewis and McCourt, 2004) the KCBPs of the two chlorophyte algae are resolved as a clade, sister to KCBPs from the seed plants, and all angiosperm KCBPs are resolved as a clade sister to the P. abies KCBP. TtKLP is resolved as the sister group to the plant KCBPs, but it is poorly supported as such in both the parsimony and Bayesian analyses; no conclusion can be made about its inclusion or exclusion from the KCBP lineage. Sequences from the water molds (PrKLP and PsKLP), the sea squirt (CiKLP), sea urchin (SpKinC), and cyanophora (CpKLP) and the diatom (TpKLP) are resolved as closely related to the KCBP group. Based on the inferred phylogeny of the eukaryotes (Baldauf, 2003), the resolution of GlKLP as the sister group of the remaining KCBP-like proteins suggests that the KCBP-like motor was present in the ancestral eukaryotic lineage prior to the diversification of the major extant groups of eukaryotes.
Figure 4.
Unrooted Bayesian tree for the kinesin family based on their motor domains. Bayesian posterior probabilities for clades are listed above the branches; parsimony jackknife values are listed below the branches. Clades resolved on the Bayesian tree that were not resolved on the parsimony jackknife tree are indicated by the “–” symbol below the respective branches. Clades of non-Kinesin-14 sequences that have been previously named are indicated by the “▸” symbols. See the supplemental figures for members of these clades included in this analysis. Kinesin-14 family, which contains C-terminal kinesins, is shown on the right side of the figure. Kinesin-13 family members contain an internal-motor domain. The members of the other families have an N-terminal motor domain. At, Arabidopsis; An, Aspergillus nidulans; Ce, C. elegans; Cg, Cricetulus griseus; Ci; C. intestinalis, Cp, Cyanophora paradoxa; Cr, C. reinhardtii; Dm, D. melanogaster; Gh, cotton; Gl, G. lamblia; Hs, H. sapiens; Mm, M. musculus; Mt, M. truncatula; Nt, tobacco; Os, rice; Pa, P. abies; Pr, P. ramorum; Ps, P. sojae; Pt, P. trichocarpa; St, S. tuberosum; Sc, S. cerevisiae; Sb, S. bacillaris; Sp, sea urchin; Spo, Schizosaccharomyces pombe; Tp, T. pseudonana; Tt, T. thermophila; Xl, Xenopus laevis.
Evolution of KCBP
Kull et al. (1998) used several criteria to evaluate the evolutionary relationships between motor proteins and concluded that the motor domain of kinesins and myosins may have evolved from an ancestral protomotor by divergent evolution. A hypothetical ancestor was proposed that was possibly shared with G proteins. The three proteins are proposed to have diverged from a common core nucleotide-binding motif (Kull et al., 1998). Distinct families of kinesins are thought to have evolved from the protokinesin. Using G. lamblia as a representative of the early derived extant eukaryotes, Iwabe and Miyata (2002) addressed the question of whether the subfamilies might have arisen early in eukaryotic evolution. They cloned 13 G. lamblia kinesins and four kinesins from four different metazoan species and analyzed them along with the motor domain of 142 kinesins in the databases. They concluded that most gene duplications that gave rise to different kinesin subfamilies had been completed before the earliest divergence of extant eukaryotes (Iwabe and Miyata, 2002). Based on their study of the kinesins in filamentous ascomycetes, Schoch et al. (2003) concluded that the founding members of most kinesin subfamilies were present before fungi, metazoans, and plants diverged. Gene tree analysis of the uncoordinated-104 family suggested that the divergence of the members of the kinesin family by gene duplications seems to have occurred intermittently. Iwabe and Miyata (2002) suggest three active periods: before the separation of G. lamblia, after the separation from fungi and plants but before the parazoan-eumetazoan split, and early in the evolution of vertebrates before the cyclostome-gnathostome split. A study of the kinesins in the Arabidopsis genome suggests duplication events in the kinesin families in plants after their separation from animals (Reddy and Day, 2001).
KCBP orthologs with the MyTH4, talin-like region, and CBD have been isolated from phylogenetically divergent land plants and green algae including Arabidopsis, tobacco, Solanum tuberosum, Z. mays, cotton (Reddy et al., 1996a, 1996b; Wang et al., 1996; Abdel-Ghany and Reddy, 2000; Preuss et al., 2003) and S. bacillaris (a cholorophyte alga). Sequences retrieved from databases for the alga C. reinhardtii and rice also contain these domains. The genome for the red alga C. merolae has been sequenced and five kinesins were reported (Matsuzaki et al., 2004; http://merolae.biol.s.u-tokyo.ac.jp/). However, none of them contain domains unique to KCBP. ESTs for several partial sequences from flowering plants and from the moss P. patens contain the motor domain and CBD that is similar to KCBP. However, kinesins from Tetrahymena and G. lamblia that showed the closest similarity to KCBP motor domain did not have a conserved CBD.
The only reported KCBP-like motor domain outside photosynthetic organisms is SpKinC isolated from sea urchin (Rogers et al., 1999). Searches of the genomes of fungi, invertebrates, and vertebrates have not identified a KCBP ortholog (Miki et al., 2001; Iwabe and Miyata, 2002; Schoch et al., 2003). However, our database searches revealed the presence of a kinesin with a KCBP-like motor domain in two protozoans, T. thermophila and G. lamblia. The G. lamblia kinesin was not included in the study by Iwabe and Miyata (2002). Gene tree analysis using the motor domain sequence resolved the sea urchin, Ciona intesnialis, P. ramorum, P. sojae, and G. lamblia proteins as members of the KCBP subfamily of the C-terminal kinesins (Fig. 4). While the motor domains of SpKinC, GlKLP, PrKLP, and PsKLP show a close relationship, the proteins do not contain the conserved N-terminal MyTH4 and talin-like regions found in KCBPs. The sequences for CiKLP, TpKLP, and TtKLP are not full length so the presence of these domains cannot be determined.
Our results show that complete KCBP with the N-terminal MyTH4 and talin-like regions, C-terminal motor, and CBD is found only in land plants and green algae. These results are consistent with the phylogeny of the eukaryotes, with the land plants and the green algae supported as a monophyletic group. (Baldauf, 2003; Philippe et al., 2004). In a review of kinesins functioning in intracellular transport, Vale (2003) suggested that kinesin motors expanded in higher eukaryotes through gene duplication, alternative splicing, and the addition of associated subunits. KCBP appears to be an example of a kinesin motor to which the MyTH4 and talin-like domains were added after the divergence of the green algae. These observations suggest that KCBP, with its characteristic domains, may have evolved early in the plant lineage from a single common ancestor to perform plant-specific functions.
KCBP Function
Functional studies with KCBP in flowering plants have shown its involvement in trichome morphogenesis and cell division (Bowser and Reddy, 1997; Oppenheimer et al., 1997; Smirnova et al., 1998). Since unicellular algae do not have trichomes, the presence of KCBP in these taxa suggests that it is likely to play a role in cell division. In dividing cells of flowering plants, the KCBP is localized to preprophase band, mitotic spindle, and phragmoplast (Bowser and Reddy, 1997; Smirnova et al., 1998; Preuss et al., 2003). However, the mode of cell division in some members of green algae (e.g. Chlamydomonas and Stichococcus) differs considerably from flowering plants in that they form a phycoplast in place of the phragmoplast during cytokinesis (Pickett-Heaps, 1976; Stewart and Mattox, 1984). The presence of KCBP in these taxa indicates that it may have a role in the formation of phycoplast also. Localization of KCBP in members of green algae that form either a phycoplast or phragmoplast should provide insights into the function(s) of this protein.
Relationship between KCBP and SpKinC
All reported KCBPs from land plants and green algae and SpKinC from sea urchin have a CBD at the C terminus that is responsible for regulating the interaction of the motor domain with MTs in a Ca2+-dependent manner (Song et al., 1997; Deavours et al., 1998; Narasimhulu and Reddy, 1998; Rogers et al., 1999; Kao et al., 2000). Over 250 KLPs have been characterized from various eukaryotes. However, kinesin-heavy chains other than KCBP and SpKinC are not known to bind calmodulin (Matthies et al., 1993; Reddy et al., 1996b; Rogers et al., 1999). In the kinesin gene tree (Fig. 4), SpKinC is resolved as nested within the clade of KCBP-like proteins. Outside the motor domain, there is no sequence similarity between KCBPs and SpKinC, whereas, as stated above, the KCBP tail is highly conserved among land plants and green algal KCBPs. Two evolutionary propositions may explain the relationship between KCBPs and SpKinC. First, KCBP and SpKinC are not closely related and the CBD was added to both of them independently (convergent evolution) during the course of evolution (e.g. as a result of gene transfer [Smith et al., 1992] or domain swaps [Doolittle and Bork, 1993]) to confer Ca2+/calmodulin regulation. The arguments in support of this proposition include the fact that the conserved MyTH4 and talin-like regions present in the N-terminal region of all characterized KCBPs are not present in SpKinC (Fig. 1). A second argument in support of this proposition is that KCBP orthologs are not found in the completely sequenced genomes of C. elegans, D. melanogaster, and S. cerevisiae, or in the draft human genome. Also, fusion of the CBD from Arabidopsis to the motor domain of C-terminal or N-terminal kinesin of Drosophila confers calmodulin-binding property to these motifs (Reddy and Reddy, 2002).
The second proposition is that the common ancestor of KCBPs and SpKinC with a CBD existed before the divergence of plants and animals, which is believed to have occurred about 1.5 billion years ago (Wang et al., 1999). The differences in domain organization between KCBP and SpKinC could have occurred through domain insertion or deletion at the amino-terminal region of the motor in order to carry specific cargoes or to confer plant-specific functions. One argument in support of this notion is that the motor domain of SpKinC is resolved in a well-supported clade that includes the KCBPs in both the Bayesian and parsimony analyses (Fig. 4). In addition, the class-specific coiled-coil linker, a short motif that links the catalytic core of the motor domain to the coiled-coil region, is mostly identical in SpKinC and KCBPs and is diverged from other C-terminal KLPs (Fig. 2A). The class-specific cluster of residues in the kinesin motor domain is found in the neck linker and not within the catalytic core (Case et al., 2000). However, based on the inferred phylogeny of the metazoa (Halanych and Passamaneck, 2001) this proposition would require at least three independent losses (one in the S. cerevisiae lineage, a second in the Ecdysozoa prior to the divergence of the arthropods and nematodes, and a third in the chordates [and perhaps fourth in the oomycetes]). Although, we cannot exclude either of these propositions based on our results, we favor the second proposition based on the resolution in our gene tree and the presence of a CBD in both KCBPs and SpKinC.
MATERIALS AND METHODS
Materials
Stichococcus bacillaris was grown in sterilized lake water enriched with Algo-Gro concentrate solution (Carolina Biological, Burlington, NC) at 18°C in a 16:8-h light/dark cycle. After 10 d, cells were collected by centrifugation and kept at −80°C for DNA isolation. Picea abies seeds, kindly provided by Dr. Peter Hedley (Scottish Crop Research Institute, Invergowire, UK) were germinated and grown on moist vermiculite in a growth chamber at 25°C for 3 weeks in white light (16 h light, 8 h dark). Seedlings were collected and stored at −80°C until use.
KCBP Motor Domain Amplification and Cloning
The KCBP motor domains of S. bacillaris and P. abies were amplified by PCR using either a cDNA library or genomic DNA as templates. Two degenerate primers, sense (5′-ATT/C/ATTT/CGCITAT/CGGICAA/GAC-3′) and antisense (5′-CIGCT/CTGT/CTCT/CTTCCAG/ATA-3′), based on conserved motifs in the ATP and CBD in KCBP, were used to amplify approximately 2 kb and approximately 450 bp from S. bacillaris genomic DNA and P. abies cDNA, respectively. Sense and antisense primers contained restriction sites for EcoRI and BamHI, respectively. PCR reactions were performed in a final volume of 50 μL. The reaction mixtures were preheated at 94°C for 4 min and cooled to 50°C, and Taq polymerase was added to initiate the amplification reactions. Thirty-five cycles of amplification were performed in an Eppendorf Mastercycler Gradient (Eppendorf Scientific, Westbury, NY) followed by a final extension for 10 min. Each amplification cycle consisted of 1 min of denaturation at 94°C, 1 min of annealing at 50°C, and 2 min of extension at 72°C. After separation on an agarose gel, the PCR products were purified with Qiagen PCR purification kit (Qiagen USA, Valencia, CA), cloned into EcoRI/BamHI sites of Bluescript-KS+ vector (Stratagene, La Jolla, CA), verified by sequencing, and used as probes for library screening.
Construction of S. bacillaris Genomic Library
S. bacillaris genomic DNA was prepared from frozen cells using the urea-phenol-containing buffer method (Golovkin and Reddy, 1996). High-Mr DNA was partially digested with Sau3A1 and size fractionated on a 0.8% agarose gel. Fragments of 9 to 20 kb were excised from the gel and purified using low-melting-point-agarose method and ligated into Lambda DASH II vector predigested with BamHI (Stratagene). The ligated DNA was packaged using Gigapack III packaging extract and transferred into Escherichia coli XLI-blue MRA(P2).
Library Screening
A P. abies cDNA library (kindly provided by Dr. Peter Engstrom) had been constructed from female strobilus RNA using a cDNA synthesis kit (Pharmacia Biotech, Piscataway, NJ) in λgt10 cloning vector. For screening, approximately 5 × 105 recombinant plaques from the P. abies cDNA library and the S. bacillaris genomic libraries were transferred to Hybond N+ (Amersham, Piscataway, NJ) membranes according to the manufacturer's procedures. Membranes were hybridized to a 32P-labeled, PCR-amplified product at 60°C in a solution containing 0.5 m sodium phosphate, pH 7.2, and 0.7% w/v SDS. These membranes were washed twice in 2× SSC, 0.1% SDS for 10 min at room temperature, then twice with 1× SSC, 0.1% SDS for 15 min at 60°C. After three rounds of screening, phage DNA from each clone was prepared using Qiagen Lambda purification kit and sequenced.
DNA Sequencing and Sequence Analysis
PCR-amplified products as well as cDNA and genomic clones were sequenced using double-stranded DNAs as templates. Primer walking was used to obtain the complete sequences. Sequence analysis was performed using Sequencer (Genescale, Ann Arbor, MI) and Mac Vector sequence analysis software (International Biotechnologies, New Haven, CT). Searches for sequence similarity using nucleotide- and predicted-amino acid sequences were performed using the BLAST network service provided by the National Library of Medicine (http://www.ncbi.nlm.nih.gov). Splice sites were predicted using the NetplantGene (http://genome.cbs.dtu.dk/services/NetPGene/). Alignments were performed using MegAlign (DNASTAR, Madison, WI). Protein secondary structure and domain analyses were performed using Simple Modular Architecture Research Tool (SMART v.3.1, http://smart.embl-heidelberg.de/) and InterProScan (http://www.ebi.ac.uk/InterProScan/).
Database Searches
Searches were done using AtKCBP full-length sequence at NCBI (www.ncbi.nlm.nih.gov/BLAST/) and the databases available at www.ncbi.nlm.nih.gov/Genomes/index.html (Aspergillus, bee, cat, chicken, chimp, cow, dictyostelium, dog, frog, fruit fly, human, malaria, microbes, mosquito, mouse, nematode, pig, plant genomes, rat, sea urchin, sheep, and zebrafish). The dictyostelium database search site included the following species (*completed genomes): Cryptosporidium hominis, Cryptosporidium parvum, Plasmodium berghei strain ANKA, Plasmodium chabaudi, *Plasmodium falciparum 3D7, Plasmodium yoelii yoelii, Theileria annulata, Toxoplasma gondii, Giardia lamblia ATCC 50803, Entamoeba histolytica, Entamoeba histolytica HM-1:IMSS, Aspergillus fumigatus, Aspergillus nidulans FGSC A4, Aspergillus terreus ATCC 20542, Coccidioides immitis RS, Coccidioides posadasii C735, Gibberella zeae PH-1, Magnaporthe grisea 70-15, Neurospora crassa, Candida albicans SC5314, *Candida glabrata CBS138, *Debaryomyces hansenii CBS767, *Eremothecium gossypii, *Kluyveromyces lactis NRRL Y-1140, Eremothecium gossypii, Kluyveromyces waltii NCYC 2644, Naumovia castellii NRRL Y-12630, Saccharomyces bayanus 623-6C, Saccharomyces bayanus MCYC 623, *Saccharomyces cerevisiae, Saccharomyces kluyveri NRRL Y-12651, Saccharomyces kudriavzevii IFO 1802, Saccharomyces mikatae IFO 1815, Saccharomyces paradoxus NRRL Y-17217, *Yarrowia lipolytica CLIB99, *Schizosaccharomyces pombe, Coprinopsis cinerea okayama7#130, Cryptococcus neoformans var grubii H99, Cryptococcus neoformans var neoformans B-3501A, Cryptococcus neoformans var neoformans JEC21, Cryptococcus neoformans var neoformans JEC21, Phanerochaete chrysosporium RP-78, Ustilago maydis 521, *Encephalitozoon cuniculi, Anopheles gambiae str. PEST, Apis mellifera, Bombyx mori, *Drosophila melanogaster, Drosophila pseudoobscura, Drosophila yakuba, *Sea squirt (Ciona intestinalis), Ciona savignyi, Gallus gallus, Takifugu rubripes, Tetraodon nigroviridis, Brugia malayi, Caenorhabditis briggsae, *Caenorhabditis elegans, *Leishmania major, Trypanosoma brucei, and Trypanosoma cruzi. Databases representing animal, protista, fungi, and plant species at TIGR Sequence Search (http://tigrblast.tigr.org/tgi/) were searched using the Arabidopsis (Arabidopsis thaliana) KCBP motor domain sequence. The sequences at TIGR are tentative consensus and singleton EST sequences. Searches at the JGI (http://genome.jgi-psf.org/) included the following organisms: C. reinhardtii, C. intestinalis, Fugu rubripes, Homo sapiens, P. chrysosporium, Phytophthora ramorum, Phytophthora sojae, Populus trichocarpa Thalassiosira pseudonana, and Xenopus tropicalis.
Gene Tree Construction
The sequences included in this analysis are those from Reddy and Day (2001) matrix, with the addition of GhKCBP, MtKCBP, ZmKCBP, OsKCBP, PaKCBP, SbKCBP, CrKCBP, PtKCBP, CpKLP1, CiKLP, PrKLP, PsKLP, TpKLP, TtKLP, and GlKLP. A total of 189 sequences were included in this study. Alignment of amino acid kinesin motor domain sequences was performed online (http://bibiserv.techfak.uni-bielefeld.de/dialign/submission.html) using DIALIGN 2.2.1 (Morgenstern, 1999) with the default settings (threshold = 0; regions of maximum similarity = 5). In contrast to programs such as ClustalX (Thompson et al., 1997), which perform global alignments, DIALIGN finds regions of local similarity without necessarily aligning the entire sequences with one another (Morgenstern et al., 1998). DIALIGN has been shown to perform well relative to other alignment programs in aligning conserved domains within rapidly evolving (with respect to both indels and substitutions) regions (Thompson et al., 1997; Lassmann and Sonnhammer, 2002).
Amino acids from individual sequences that DIALIGN did not align were replaced with gaps. All gap-only positions were then removed from the alignment using ClustalX. The resulting alignment of 780 positions was then exported into MacClade 4.03 (Maddison and Maddison, 2001), which was used to exclude seven regions (511 positions) from all genes. Zurawski and Clegg's (1987) alignment criterion, in which gaps are considered as characters and similarity is maximized, was applied to exclude regions from individual genes (65 cells) that appeared to be ambiguously aligned, and realign regions that appeared to be suboptimally aligned. Of the 269 included amino acid characters, 234 (87%) were parsimony informative. Among the parsimony-informative characters, 1,291 cells (2.5%) were either missing data or gaps. Six parsimony-informative gap characters in conserved domains that were unambiguously aligned according to DIALIGN (i.e. flanked by aligned amino acids at both the amino and carboxy termini) were coded as additional characters using modified complex indel coding (Simmons and Ochoterena, 2000; K. Müller, unpublished data) for the parsimony analysis.
Gene tree analyses were performed using amino acid characters. Although amino acid characters have problems with a type of convergence (Simmons, 2000; Simmons et al., 2002a) and composite coding (Simmons and Freudenstein, 2002) that nucleotide characters are not subject to, they are expected to perform relatively better when high genetic distances occur among closely related terminals included in the analysis (Simmons et al., 2002b), as is the case here. This expectation is based on silent substitutions undergoing saturation (i.e. multiple hits along individual branches).
Gene tree inference was performed using both parsimony and Bayesian (Rannala and Yang, 1996; Yang and Rannala, 1997) approaches. Parsimony tree searches were performed using PAUP* (Phylogenetic Analysis Using Parsimony, version 4.0, Sinaur Associates, Sunderland, MA; Swofford, 1998) with all characters assigned equal weights. Jackknife analyses (Farris et al., 1996) were performed using 1,000 replicates with each replicate consisting of 10 tree-bisection-reconnection heuristic searches and a maximum of 10 trees held per search. Following Farris et al. (1996), the deletion probability for each character was set at 36.7879% and Jac resampling was emulated, resulting in support values nearly equivalent to those provided by the bootstrap (Felsenstein, 1985).
Bayesian tree searches were performed using MrBayes 3.0b4 (Huelsenbeck and Ronquist, 2001) with a mixed-amino acid model. Two independent Bayesian analyses were performed, with four chains per analysis, and each analysis consisting of seven million generations with trees sampled every 100 generations. Both analyses reached the same stationarity within the first 500,000 generations. The combined 130,000 trees from the two analyses that were sampled at stationarity were used to infer the posterior probabilities for individual clades. Majority-rule consensus trees were calculated using PAUP*. Note that although Bayesian analyses appear to be more efficient than parsimony analyses (Simmons and Miya, 2004), they also can produce inflated support values (Suzuki et al., 2002; Cummings et al., 2003; Simmons et al., 2004). Maximum likelihood analyses (Felsenstein, 1973) were not performed because they are not currently computationally tractable for a matrix of this size (Sanderson and Kim, 2000).
The rooting of the kinesin family used by Goodson et al. (1994), Kim and Endow (2000), and Reddy and Day (2001) is arbitrary. Outgroups sequences are to be selected such that all members of the ingroup are more closely related to one another than any one of them is to the outgroups (i.e. the ingroup should be monophyletic relative to the outgroup; Nixon and Carpenter, 1993). Although ScSMY1 is “a highly divergent kinesin protein” (Kim and Endow, 2000), this does not satisfy the criterion of selecting an outgroup. See Lawrence et al. (2002) for an alternative rooting, wherein ScSMY1 is nested within the Kinesin-I family. Following Hirokawa (1998), Miki et al. (2001), Iwabe and Miyata (2002), and Schoch et al. (2003), our gene trees are presented as unrooted.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
Supplemental Material
Supplemental data, consisting of the DIALIGN alignments, the data matrices and the complete Bayesian and parsimony jackknife trees are available online.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY515262 (PaKCBP) and AY515314 (StKCBP).
Acknowledgments
We thank Dr. Peter Engstrom for providing the spruce cDNA library. The C. reinhardtii sequence data were produced by the U.S. Department of Energy Joint Genome Institute, http://www.jgi.doe.gov/ and are provided for use in this publication only.
This work was supported by a grant from the National Science Foundation (to A.S.N.R.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.060913.
References
- Abdel-Ghany SE, Reddy ASN (2000) A novel calcium/calmodulin-regulated kinesin-like protein is highly conserved between monocots and dicots. DNA Cell Biol 19: 567–578 [DOI] [PubMed] [Google Scholar]
- Abdel-Ghany SE, Kurgrens P, Reddy ASN (2000) CpKLP1: a calmodulin-binding kinesin-like protein from Cyanophora paradoxa (Glaucophyta). J Phycol 36: 686–692 [DOI] [PubMed] [Google Scholar]
- Ambrose JC, Li W, Marcus A, Ma H, Cyr R (2005) A minus-end directed kinesin with +TIP activity is involved in spindle morphogenesis. Mol Biol Cell 16: 1584–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada T, Kuriyama R, Shibaoka H (1997) TKRP125, a kinesin-related protein involved in the centrosome-independent organization of the cytokinetic apparatus in tobacco BY-2 cells. J Cell Sci 110: 179–189 [DOI] [PubMed] [Google Scholar]
- Asada T, Shibaoka H (1994) Isolation of polypeptides with microtubule-translocating activity from phragmoplasts of tobacco BY-2 cells. J Cell Sci 107: 2249–2257 [DOI] [PubMed] [Google Scholar]
- Asada T, Sonobe S, Shibaoka H (1991) Microtubule translocation in the cytokinetic apparatus of cultured tobacco cells. Nature 350: 238–241 [Google Scholar]
- Baldauf SL (2003) The deep roots of eukaryotes. Science 300: 1703–1706 [DOI] [PubMed] [Google Scholar]
- Barroso C, Chan J, Allan V, Doonan J, Hussey P, Lloyd C (2000) Two kinesin-related proteins associated with the cold-stable cytoskeleton of carrot cells: characterization of a novel kinesin, DcKRP120-2. Plant J 24: 859–868 [DOI] [PubMed] [Google Scholar]
- Barton NR, Goldstein LSB (1996) Going mobile: microtubule motors and chromosome segregation. Proc Natl Acad Sci USA 93: 1735–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowser J, Reddy ASN (1997) Localization of a kinesin-like calmodulin-binding protein in dividing cells of Arabidopsis and tobacco. Plant J 12: 1429–1438 [DOI] [PubMed] [Google Scholar]
- Cai G, Bartalesi A, Del Casino C, Moscatelli A, Tiezzi A, Cresti M (1993) The kinesin-immunoreactive homologue from Nicotiana tabacum pollen tubes: biochemical properties and subcellular localization. Planta 191: 496–506 [Google Scholar]
- Case RB, Rice S, Hart CL, Ly B, Vale RD (2000) Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr Biol 10: 157–160 [DOI] [PubMed] [Google Scholar]
- Chen C, Marcus A, Li W, Hu Y, Calzada JP, Grossniklaus U, Cyr RJ, Ma H (2002) The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 129: 2401–2409 [DOI] [PubMed] [Google Scholar]
- Cummings MP, Handley SA, Myers DS, Reed DL, Rokas A, Winka K (2003) Comparing bootstrap and posterior probability values in the four-taxon case. Syst Biol 52: 477–487 [DOI] [PubMed] [Google Scholar]
- Day IS, Miller C, Golovkin M, Reddy ASN (2000) Interaction of a kinesin-like calmodulin-binding protein with a protein kinase. J Biol Chem 275: 13737–13745 [DOI] [PubMed] [Google Scholar]
- Deavours BE, Reddy ASN, Walker RA (1998) Ca2+/calmodulin regulation of the Arabidopsis kinesin-like calmodulin-binding protein. Cell Motil Cytoskeleton 40: 408–416 [DOI] [PubMed] [Google Scholar]
- Doolittle RF, Bork P (1993) Evolutionary mobile modules in proteins. Sci Am 269: 50–56 [DOI] [PubMed] [Google Scholar]
- Endow SA, Waligora KW (1998) Determinants of kinesin motor polarity. Science 281: 1200–1202 [DOI] [PubMed] [Google Scholar]
- Farris JS, Albert VA, Kallersjo M, Lipscomb D, Kluge AG (1996) Parsimony jackknifing outperforms neighbor-joining. Cladistics 12: 99–124 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1973) Maximum likelihood and minimum-steps methods for estimating evolutionary trees from data on discrete characters. Syst Zool 22: 240–249 [Google Scholar]
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Folkers U, Kirik V, Schobinger U, Falk S, Krishnakumar S, Pollock MA, Oppenheimer DG, Day I, Reddy ASN, Jurgens G, et al (2002) The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J 21: 1280–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LSB, Philip AV (1999) The road less traveled: emerging principles of kinesin motor utilization. Annu Rev Cell Dev Biol 15: 141–183 [DOI] [PubMed] [Google Scholar]
- Golovkin M, Reddy ASN (1996) Structure and expression of a plant U1 snRNP 70K gene: alternative splicing of U1 snRNP 70K pre-mRNAs produces two different transcripts. Plant Cell 8: 1421–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson HV, Kang SJ, Endow SA (1994) Molecular phylogeny of the kinesin family of microtubule motor proteins. J Cell Sci 107: 1875–1884 [DOI] [PubMed] [Google Scholar]
- Halanych KM, Passamaneck Y (2001) A brief review of the metazoan phylogeny and future prospects in Hox-research. Am Zool 42: 629–639 [Google Scholar]
- Hirokawa N (1998) Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279: 519–526 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Itoh R, Fujiwara M, Yoshida S (2001) Kinesin-related proteins with a mitochondrial targeting signal. Plant Physiol 127: 724–726 [PMC free article] [PubMed] [Google Scholar]
- Iwabe N, Miyata T (2002) Kinesin-related genes from diplomonad, sponge, amphioxus, and cyclostomes: divergence pattern of kinesin family and evolution of giardial membrane-bounded organella. Mol Biol Evol 19: 1524–1533 [DOI] [PubMed] [Google Scholar]
- Kao Y-L, Deavours BE, Phelps KK, Walker R, Reddy ASN (2000) Bundling of microtubules by motor and tail domains of a kinesin-like calmodulin-binding protein from Arabidopsis: regulation by Ca2+/calmodulin. Biochem Biophys Res Commun 267: 201–207 [DOI] [PubMed] [Google Scholar]
- Kim AJ, Endow SA (2000) A kinesin family tree. J Cell Sci 113: 3681–3682 [DOI] [PubMed] [Google Scholar]
- Kong LJ, Hanley-Bowdoin L (2002) A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Vale RD, Fletterick RJ (1998) The case for a common ancestor: kinesin and myosin motor proteins and G proteins. J Muscle Res Cell Motil 19: 877–886 [DOI] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL (2002) Quality assessment of multiple alignment programs. FEBS Lett 529: 126–130 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, et al (2004) A standardized kinesin nomenclature. J Cell Biol 167: 19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Malmberg RL, Muszynski MG, Dawe RK (2002) Maximum likelihood methods reveal conservation of function among closely related kinesin families. J Mol Evol 54: 42–53 [DOI] [PubMed] [Google Scholar]
- Lee YR, Giang HM, Liu B (2001) A novel plant kinesin-related protein specifically associates with the phragmoplast organelles. Plant Cell 13: 2427–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Liu B (2004) Cytoskeletal motors in Arabidopsis: sixty-one kinesins and seventeen myosins. Plant Physiol 136: 3877–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-RJ, Liu B (2000) Identification of a phragmoplast-associated kinesin-related protein in higher plants. Curr Biol 10: 797–800 [DOI] [PubMed] [Google Scholar]
- Leopold PL, McDowall AW, Pfister KK, Bloom GS, Brady ST (1992) Association of kinesin with characterized membrane-bounded organelles. Cell Motil Cytoskeleton 23: 19–33 [DOI] [PubMed] [Google Scholar]
- Lewis LA, McCourt RM (2004) Green algae and the origin of land plants. Am J Bot 91: 1535–1556 [DOI] [PubMed] [Google Scholar]
- Liu B, Cyr RJ, Palevitz BA (1996) A kinesin-like protein, KatAp, in the cells of Arabidopsis and other plants. Plant Cell 8: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Lee YR, Pan R, Maloof JN, Liu B (2005) An internal motor Kinesin is associated with the Golgi apparatus and plays a role in trichome morphogenesis in Arabidopsis. Mol Biol Cell 16: 811–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP (2001) MacClade: Analysis of Phylogeny and Character Evolution Version 4.03. Sinauer, Sunderland, MA
- Manning BD, Snyder M (1999) Drivers and passengers wanted! The role of kinesin-associated proteins. Trends Cell Biol 10: 281–289 [DOI] [PubMed] [Google Scholar]
- Marcus AI, Ambrose JC, Blickley L, Hancock WO, Cyr RJ (2002) Arabidopsis thaliana protein, ATK1, is a minus-end directed kinesin that exhibits non-processive movement. Cell Motil Cytoskeleton 52: 144–150 [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin IT, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Yoshida Y, et al (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428: 653–657 [DOI] [PubMed] [Google Scholar]
- Matthies HJG, Miller RJ, Palfrey HC (1993) Calmodulin binding to and cAMP-dependent phosphorylation of kinesin light chains modulate kinesin ATPase activity. J Biol Chem 268: 11176–11187 [PubMed] [Google Scholar]
- Melkonian M, Surek B (1995) Phylogeny of the chlorophyta: congruence between ultrastructural and molecular evidence. Bull Soc Zool Fr 120: 191–208 [Google Scholar]
- Miki H, Setou M, Kaneshiro K, Hirokawa N (2001) All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci USA 98: 7004–7011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui H, Nakatani K, Yamaguchi-Shinozaki K, Shinozaki K, Nishikawa K, Takahashi H (1994) Sequencing and characterization of the kinesin-related genes katB and katC of Arabidopsis thaliana. Plant Mol Biol 25: 865–876 [DOI] [PubMed] [Google Scholar]
- Mitsui H, Yamaguchi-Shinozaki K, Shinozaki K, Nishikawa K, Takahashi H (1993) Identification of a gene family (kat) encoding kinesin-like proteins in Arabidopsis thaliana and the characterization of secondary structure of KatA. Mol Gen Genet 238: 362–368 [DOI] [PubMed] [Google Scholar]
- Moore JD, Endow SA (1996) Kinesin proteins: a phylum of motors for microtubule-based motility. Bioessays 18: 207–219 [DOI] [PubMed] [Google Scholar]
- Morgenstern B (1999) DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15: 211–218 [DOI] [PubMed] [Google Scholar]
- Morgenstern B, Frech K, Dress A, Werner T (1998) DIALIGN: finding local similarities by multiple sequence alignment. Bioinformatics 14: 290–294 [DOI] [PubMed] [Google Scholar]
- Narasimhulu SB, Kao Y-L, Reddy ASN (1997) Interaction of Arabidopsis kinesin-like calmodulin-binding protein with tubulin subunits: modulation by Ca2+-calmodulin. Plant J 12: 1139–1149 [DOI] [PubMed] [Google Scholar]
- Narasimhulu SB, Reddy ASN (1998) Characterization of microtubule binding domains in the Arabidopsis kinesin-like calmodulin-binding protein. Plant Cell 10: 957–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R, Soyano T, Ishikawa M, Araki S, Tanaka H, Asada T, Irie K, Ito M, Terada M, Banno H, et al (2002) Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109: 87–99 [DOI] [PubMed] [Google Scholar]
- Nixon KC, Carpenter JM (1993) On outgroups. Cladistics 9: 413–426 [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Pollock MA, Vacik J, Szymanski DB, Ericson B, Feldmann K, Marks D (1997) Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc Natl Acad Sci USA 94: 6261–6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R, Lee YR, Liu B (2004) Localization of two homologous Arabidopsis kinesin-related proteins in the phragmoplast. Planta 220: 156–164 [DOI] [PubMed] [Google Scholar]
- Philippe H, Snell EA, Bapteste E, Lopez P, Holland PWH, Casane D (2004) Phylogenomics of eukaryotes: impact of missing data on large alignments. Mol Biol Evol 21: 1740–1752 [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps J (1976) Cell division in eukaryotic algae. Bioscience 6: 445–450 [Google Scholar]
- Preuss ML, Delmer DP, Liu B (2003) The cotton kinesin-like calmodulin-binding protein associates with cortical microtubules in cotton fibers. Plant Physiol 132: 154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Kovar DR, Lee YR, Staiger CJ, Delmer DP, Liu B (2004) A plant-specific kinesin binds to actin microfilaments and interacts with cortical microtubules in cotton fibers. Plant Physiol 136: 3945–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich WB, Moran AN, Rothman JH, Hardin J (1998) Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol Biol Cell 9: 2037–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Yang Z (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol 43: 304–311 [DOI] [PubMed] [Google Scholar]
- Reddy ASN (2001) Molecular motors and their functions in plants. Intl Rev Cytol 204: 97–178 [DOI] [PubMed] [Google Scholar]
- Reddy ASN (2003) Molecular motors in plant cells. In M Schliwa, ed, Molecular Motors. Wiley-VCH, GmBH & Co., Weinheim, Germany, pp 433–469
- Reddy ASN, Day IS (2000) The role of the cytoskeleton and a molecular motor in trichome morphogenesis. Trends Plant Sci 5: 503–505 [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Day IS (2001) Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics 2: 2.1–2.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN, Narasimhulu SB, Safadi F, Golovkin M (1996. b) A plant kinesin heavy chain-like protein is a calmodulin-binding protein. Plant J 10: 9–21 [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Safadi F, Narasimhulu SB, Golovkin M, Hu X (1996. a) A novel plant calmodulin-binding protein with a kinesin heavy chain motor domain. J Biol Chem 271: 7052–7060 [DOI] [PubMed] [Google Scholar]
- Reddy VS, Day IS, Thomas T, Reddy AS (2004) KIC, a novel Ca2+ binding protein with one EF-hand motif, interacts with a microtubule motor protein and regulates trichome morphogenesis. Plant Cell 16: 185–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VS, Reddy ASN (1999) A plant calmodulin-binding motor is part kinesin and part myosin. Bioinformatics 15: 1055–1057 [DOI] [PubMed] [Google Scholar]
- Reddy VS, Reddy ASN (2002) The calmodulin-binding domain from a plant kinesin functions as a modular domain in conferring Ca2+-CaM regulation to animal plus- and minus-end kinesins. J Biol Chem 277: 48058–48065 [DOI] [PubMed] [Google Scholar]
- Rogers GC, Hart CL, Wedman KP, Scholey JM (1999) Identification of kinesin-C, a calmodulin-binding carboxy-terminal kinesin in animal (Strongylocentrotus purpuratus) cells. J Mol Biol 294: 1–8 [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, Kim J (2000) Parametric phylogenetics? Syst Biol 49: 817–829 [DOI] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ (1992) Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359: 540–543 [DOI] [PubMed] [Google Scholar]
- Schoch CL, Aist JR, Yoder OC, Gillian Turgeon B (2003) A complete inventory of fungal kinesins in representative filamentous ascomycetes. Fungal Genet Biol 39: 1–15 [DOI] [PubMed] [Google Scholar]
- Siddiqui SS (2002) Metazoan motor models: kinesin superfamily in C. elegans. Traffic 3: 20–28 [DOI] [PubMed] [Google Scholar]
- Simmons MP (2000) A fundamental problem with amino-acid-sequence characters for phylogenetic analyses. Cladistics 16: 274–282 [DOI] [PubMed] [Google Scholar]
- Simmons MP, Freudenstein JV (2002) Artifacts of coding amino acids and other composite characters for phylogenetic analysis. Cladistics 18: 354–365 [DOI] [PubMed] [Google Scholar]
- Simmons MP, Miya M (2004) Efficiently resolving the basal clades of a phylogenetic tree using Bayesian and parsimony approaches: a case study using mitogenomic data from 100 higher teleost fishes. Mol Phylogenet Evol 31: 351–362 [DOI] [PubMed] [Google Scholar]
- Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49: 369–381 [PubMed] [Google Scholar]
- Simmons MP, Ochoterena H, Freudenstein JV (2002. a) Conflict between amino acid and nucleotide characters. Cladistics 18: 200–206 [DOI] [PubMed] [Google Scholar]
- Simmons MP, Ochoterena H, Freudenstein JV (2002. b) Amino acid vs. nucleotide characters: challenging preconceived notions. Mol Phylogenet Evol 24: 78–90 [DOI] [PubMed] [Google Scholar]
- Simmons MP, Pickett KM, Miya M (2004) How meaningful are Bayesian posterior probabilities? Mol Biol Evol 21: 188–199 [DOI] [PubMed] [Google Scholar]
- Smirnova E, Reddy ASN, Bowser J, Bajer AS (1998) A minus end-directed kinesin-like motor protein, KCBP, localizes to anaphase spindle poles in Haemanthus endosperm. Cell Motil Cytoskeleton 41: 271–280 [DOI] [PubMed] [Google Scholar]
- Smith MW, Feng DF, Doolittle RF (1992) Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem Sci 17: 489–493 [DOI] [PubMed] [Google Scholar]
- Song H, Golovkin M, Reddy ASN, Endow SA (1997) In vitro motility of AtKCBP, a calmodulin-binding kinesin-like protein of Arabidopsis. Proc Natl Acad Sci USA 94: 322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart KD, Mattox KR (1984) The case for a polyphyletic origin of mitochondria: morphological and molecular comparisons. J Mol Evol 21: 54–57 [DOI] [PubMed] [Google Scholar]
- Strompen G, El Kasmi F, Richter S, Lukowitz W, Assaad FF, Jurgens G, Mayer U (2002) The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr Biol 12: 153–158 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Glazko GV, Nei M (2002) Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proc Natl Acad Sci USA 99: 16138–16143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL (1998) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, MA
- Tamura K, Nakatani K, Mitsui H, Ohashi Y, Takahashi H (1999) Characterization of katD, a kinesin-like protein gene specifically expressed in floral tissues of Arabidopsis thaliana. Gene 230: 23–32 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ishikawa M, Kitamura S, Takahashi Y, Soyano T, Machida C, Machida Y (2004) The AtNACK1/HINKEL and STUD/TETRASPORE/AtNACK2 genes, which encode functionally redundant kinesins, are essential for cytokinesis in Arabidopsis. Genes Cells 9: 1199–1211 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiezzi A, Moscatelli A, Cai G, Bartalesi A, Cresti M (1992) An immunoreactive homolog of mammalian kinesin in Nicotiana tabacum pollen tubes. Cell Motil Cytoskeleton 21: 132–137 [DOI] [PubMed] [Google Scholar]
- Vale RD (2003) The molecular motor toolbox for intracellular transport. Cell 112: 467–480 [DOI] [PubMed] [Google Scholar]
- Vale RD, Fletterick RJ (1997) The design plan of kinesin motors. Annu Rev Cell Dev Biol 13: 745–777 [DOI] [PubMed] [Google Scholar]
- Vinogradova M, Reddy VS, Reddy ASN, Fletterick RJ (2004) Crystal structure of KCBP: regulation by calcium/calmodulin. J Biol Chem 279: 23504–23509 [DOI] [PubMed] [Google Scholar]
- Vos JW, Safadi F, Reddy AS, Hepler PK (2000) The kinesin-like calmodulin binding protein is differentially involved in cell division. Plant Cell 12: 979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DY, Kumar S, Hedges SB (1999) Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc R Soc Lond B Biol Sci 266: 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Takezawa D, Narasimhulu SB, Reddy ASN, Poovaiah BW (1996) A novel kinesin-like protein with a calmodulin-binding domain. Plant Mol Biol 31: 87–100 [DOI] [PubMed] [Google Scholar]
- Weber KL, Sokac AM, Berg JS, Cheney RE, Bement WM (2004) A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature 431: 325–329 [DOI] [PubMed] [Google Scholar]
- Wein H, Foss M, Brady B, Cande WZ (1996) DSK1, a novel kinesin-related protein from the diatom Cylindrotheca fusiformis that is involved in anaphase spindle elongation. J Cell Biol 133: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Spielman M, Coles JP, Li Y, Ghelani S, Bourdon V, Brown RC, Lemmon BE, Scott RJ, Dickinson HG (2003) TETRASPORE encodes a kinesin required for male meiotic cytokinesis in Arabidopsis. Plant J 34: 229–240 [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B (1997) Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Mol Biol Evol 14: 717–724 [DOI] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Morrison WH, Ye ZH (2002) A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14: 3101–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G, Clegg MT (1987) Evolution of higher-plant chloroplast DNA-encoded genes: implications for structure-function and phylogenetic studies. Annu Rev Plant Physiol 38: 391–418 [Google Scholar]