Abstract
PDZ (Postsynaptic density protein, Disc large, Zona occludens) domains are protein–protein interaction modules that predominate in submembranous scaffolding proteins. Recently, we showed that the PDZ domains of syntenin-1 also interact with phosphatidylinositol 4,5-bisphosphate (PIP2) and that this interaction controls the recruitment of the protein to the plasma membrane. Here we evaluate the general importance of PIP2–PDZ domain interactions. We report that most PDZ proteins bind weakly to PIP2, but that syntenin-2, the closest homolog of syntenin-1, binds with high affinity to PIP2 via its PDZ domains. Surprisingly, these domains target syntenin-2 to nuclear PIP2 pools, in nuclear speckles and nucleoli. Targeting to these sites is abolished by treatments known to affect these PIP2 pools. Mutational and domain-swapping experiments indicate that high-affinity binding to PIP2 requires both PDZ domains of syntenin-2, but that its first PDZ domain contains the nuclear PIP2 targeting determinants. Depletion of syntenin-2 disrupts the nuclear speckles–PIP2 pattern and affects cell survival and cell division. These findings show that PIP2–PDZ domain interactions can directly contribute to subnuclear assembly processes.
Keywords: nuclear speckles, nucleoli, PDZ domain, phosphoinositides, syntenin
Introduction
PDZ (Postsynaptic density protein, Disc large, Zona occludens) domains are protein–protein interaction modules of 90 amino acids (aa), containing six β strands and two α helices. It is well established that PDZ domains generally interact with the C-terminal end of the cytoplasmic tail of transmembrane proteins. The C-terminus of the interacting peptide docks in a cavity formed by the βB strand, the αB helix and the carboxylate binding loop of the PDZ domain. The first residue of the αB helix (αB1 residue) confers specificity for the peptide ligand and forms the basis for PDZ domain classification (Sheng and Sala, 2001; Hung and Sheng, 2002). The PDZ protein syntenin-1 was originally identified as a ligand for the syndecans (Grootjans et al, 1997). Recently, we reported that the PDZ domains of syntenin-1 bind to phosphatidylinositol 4,5-bisphosphate (PIP2) and that PIP2, as well as syndecans, regulates the targeting of syntenin-1 to the plasma membrane (Zimmermann et al, 2002). As most proteins with PDZ domains act as submembranous scaffolding proteins, controlling the correct subcellular targeting of supramolecular signaling complexes (Nourry et al, 2003), this raises the question of whether PDZ domains in general function as phosphoinositide-binding modules.
Phosphoinositides are phosphorylated forms of phosphatidylinositol (PI), present at relatively low levels within cells compared to other phospholipids. They have emerged as essential players in a variety of cellular processes, ranging from cell surface receptor signaling, actin cytoskeleton remodeling and compartmentalization of vesicular trafficking to cell growth and proliferation. Phosphoinositides can function as precursors of second messengers or directly as docking sites for target proteins (Toker, 2002; Czech, 2003; De Matteis and Godi, 2004). Protein domains known to bind phosphoinositides are FYVE, PX, PH, ENTH, ANTH, Tubby and FERM domains (Lemmon, 2003), and were identified in proteins that are principally cytoplasmic or membrane bound (Gozani et al, 2003).
It is now established that phosphoinositides are also present in the nucleus, but their function there remains poorly understood (Irvine, 2003). PIP2, in particular, concentrates in nuclear speckles and nucleoli (Osborne et al, 2001). Nuclear speckles are dynamic punctate subnuclear structures that are enriched in pre-messenger RNA (pre-mRNA) splicing factors and are located in the interchromatin regions of the nucleoplasm (Lamond and Spector, 2003). Nuclear PIP2 regulates chromatin remodeling and gene transcription, and in vitro studies indicate that nuclear PIP2 might modulate pre-mRNA splicing (Yu et al, 1998; Zhao et al, 1998; Osborne et al, 2001; Jones and Divecha, 2004). Gozani et al (2003) showed that the chromatin-associated protein ING2 binds nuclear phosphatidylinositol 5-phosphate (PI(5)P), via its PHD finger, and suggested that PHD fingers, domains present in a large number of chromatin regulatory factors, function as nuclear phosphoinositide receptors. No other protein domains recognizing nuclear phosphoinositides have been identified so far.
Here, further investigating the importance of phosphoinositide–PDZ interactions, we found that the PDZ protein syntenin-2, a protein closely related to syntenin-1, also directly interacts with PIP2 in a PDZ-dependent mode. This interaction specifically targets the protein to nuclear PIP2 pools. Depletion of syntenin-2, by siRNA, disrupts the nuclear speckles–PIP2 pattern and affects cell survival and cell division.
Results
Most PDZ proteins bind weakly to PIP2-containing lipid layers
To evaluate whether binding to PIP2 might be a general property of PDZ domains, we investigated whether the PDZ domains of nine different proteins could directly and specifically interact with lipid layers containing PIP2. We selected PDZ proteins containing multiple PDZ domains and PDZ domains organized in tandem as in syntenin-1, and fused these to GST. GST was included as negative control, while GST-tagged full-length (FL) syntenin-1 (human form) and GST fused to the PH domain of phospholipase C (PLC)δ1 (GST-PH-PLCδ) were used as positive controls (Zimmermann et al, 2002). In surface plasmon resonance (SPR) assays (Figure 1A), a segment of PSD95 containing the three PDZ domains of the protein (GST-PDZ1-3-PSD95, rat form aa 65–393) or a segment of nNOS containing the single PDZ domain of the protein (GST-PDZ1-nNOS, rat form aa 1–299) did not interact with PIP2. The tandem PDZ domains of EBP50 (GST-PDZ1-2-EBP50, human form aa 1–245), the tandem PDZ domains of Mint3 (GST-PDZ1-2-Mint3, mouse form aa 391–556), a segment of Bazooka containing the three PDZ domains of the protein (GST-PDZ1-3-Bazooka, aa 291–737), and the FL PDZ proteins Magi-3 (GST-FL-Magi-3, human form), Dlt (GST-FL-Dlt) and PAR-6 (GST-FL-PAR-6, human form) bound with low affinity to PIP2-containing lipid layers, as the sensogram showed a clear and specific association and dissociation phase, and the binding was concentration dependent; however, these interactions were at the limits of the sensitivity of the assay (data not shown). Based on the characteristics of our biosensor, we conclude that the KD of these interactions is around or above 10−4 M.
Figure 1.
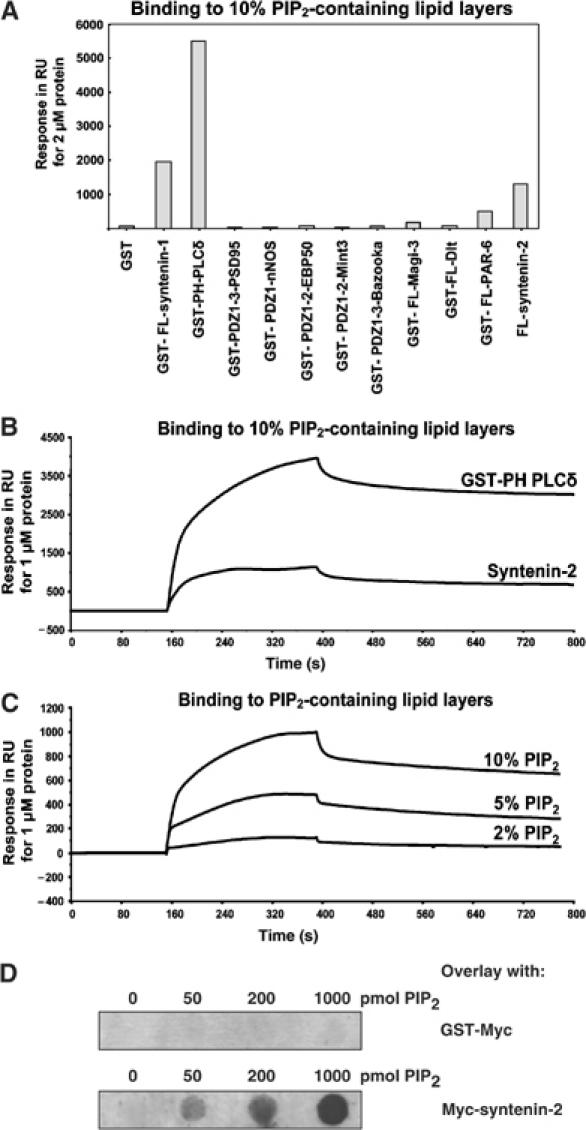
PIP2 binding of selected PDZ proteins. (A) Binding of PDZ proteins or PDZ domains to PIP2 in SPR analysis. Purified recombinant proteins were perfused at 2 μM over a sensorchip coated with vesicles containing 10% PIP2. Values correspond to response units (RU) measured 4 min after perfusion of the proteins. (B) SPR sensograms showing the interaction of syntenin-2 with lipid layers containing 10% PIP2. The syntenin-2 sensogram is compared to the sensogram for GST-PH-PLCδ (positive control for PIP2 binding). The purified recombinant proteins were perfused at 1 μM. (C) SPR sensogram showing that the binding of syntenin-2 to PIP2 increases as the PIP2 concentration in the lipid layers increases. (D) PIP2 binding of syntenin-2 in overlay assay. Increasing amounts of PIP2 (as indicated) were spotted on a nitrocellulose membrane and incubated with GST-Myc as a negative control (upper panel) or Myc-tagged syntenin-2 (lower panel).
Syntenin-2 binds with high affinity to PIP2
In contrast, the PDZ protein syntenin-2 bound strongly to lipid layers containing 10% PIP2 (Figure 1A and B). To evaluate the PIP2 binding of syntenin-2, we used a nontagged form of the protein, produced by self-splicing from an intein fusion protein. Decreasing the concentration of PIP2 in the lipid layers decreased the binding of syntenin-2, illustrating that the binding was dependent on PIP2 (Figure 1C). Apparent KD values for PIP2 binding were calculated for FL syntenin-2 and for GST-PH-PLCδ, and determined to be 1.5 × 10−6 and 0.2 × 10−6 M, respectively. Dot-blot assays, whereby increasing amounts of PIP2 were immobilized before overlay with syntenin-2, indicated that PIP2 does not need to be incorporated in lipid layers for binding to syntenin-2 (Figure 1D). We also investigated whether syntenin-2 significantly bound to phosphoinositides other than PIP2 (Supplementary data 1). Binding of syntenin-2 to other phosphoinositides was at the limit of biosensor sensitivity, except for PI(3,4,5)P3 for which the apparent KD was determined to be 2.0 × 10−6 M.
Syntenin-2 is targeted to the plasma membrane, nucleoli and nuclear speckles via its PDZ domains
Specific anti-syntenin-2 antibodies were raised to study the expression and subcellular distribution of endogenous syntenin-2. Western blots of total protein extracts showed that MCF-7 and Caco-2 cells express syntenin-2 (Figure 2A). In MCF-7 cells, the main signal distributed to subnuclear structures and within the cytoplasm (Figure 2Ba and b). In Caco-2 cells, the syntenin-2 signal was mainly observed within the cytoplasm and at the plasma membrane (Figure 2Bc), but a weak signal was also present in subnuclear structures (Figure 2Bd). The endogenous syntenin-2 signal in the nucleus codistributed with nucleoli (Figure 2Ca, b and a-b), a subnuclear compartment that contains PIP2 according to electron microscopy studies (Osborne et al, 2001), but not with PIP2 in nuclear speckles (Figure 2Ca, c and a-c), as traced by the antibody 2C11 (Osborne et al, 2001). To specify the respective roles of the different syntenin-2 domains in this subcellular targeting, we transfected MCF-7 cells with expression vectors encoding eGFP fusions to the different domains of syntenin-2 and analyzed the subcellular locations of these proteins by confocal microscopy (Figure 2D). eGFP itself, included as a control, was diffusely distributed over the cell. When the first or the second PDZ domain of syntenin-2 was fused to eGFP in isolation, the fluorescence was distributed over the cytosol and the nucleosol, with some enrichment in the nucleosol for PDZ1. When the tandem of PDZ1 and PDZ2 was fused to eGFP, the fluorescence was concentrated in the nucleus, in discrete subnuclear structures, and at the plasma membrane. Similar distributions were observed for constructs where either the N-terminal or the C-terminal domain was added to the PDZ tandem. Nuclear enrichment predominated over plasma membrane and cytosolic distribution, with 78% (±3%) of the cells displaying fluorescence exclusively in the nucleus. Surprisingly, when FL syntenin-2 was fused to eGFP, the fluorescence was primarily localized in the cytoplasm and excluded from the nucleus. A similar exclusion was observed for a Myc-tagged FL syntenin-2 (data not shown), suggesting that nuclear exclusion was not due to the size of the eGFP fusion. However, overexpressed nontagged syntenin-2 (detected with anti-syntenin-2 antibodies, and scoring expression above endogenous levels) was detected in the nucleus of MCF-7 cells (Supplementary data 2A), indicating that tagging the FL protein interferes with its nuclear concentration. The reason for this interference is unknown, but it could be alleviated by treating the cells with the nuclear export inhibitor, leptomycin B (Supplementary data 2B). We concluded that, although other domains might influence the subcellular distribution of syntenin-2, the PDZ domains together are necessary and sufficient for targeting syntenin-2 to the plasma membrane and subnuclear structures. In contrast to endogenous syntenin-2, all tagged constructs of syntenin-2 containing both PDZ domains were concentrated in nuclear speckles, in addition to nucleoli, as illustrated for the eYFP-PDZ1-2-Cter construct (Figure 2E). Investigating possible reasons for this discrepancy revealed that our anti-syntenin-2 antibodies are not suited for detecting the protein in nuclear speckles (Supplementary data 3). We concluded that syntenin-2 concentrates at three PIP2-rich regions—the plasma membrane, in nucleoli and nuclear speckles—by virtue of its PDZ domains.
Figure 2.
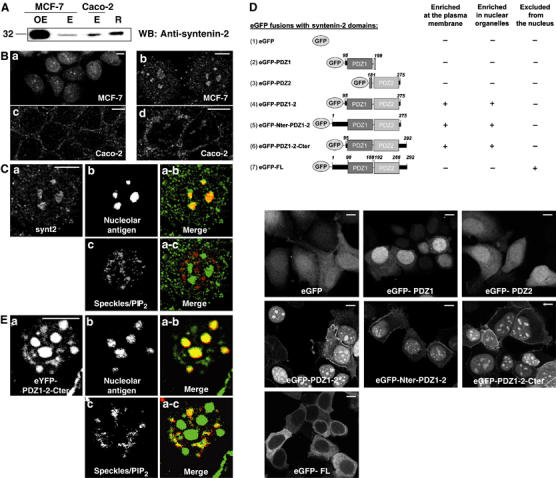
Contribution of the syntenin-2 PDZ domains in subcellular targeting. (A) Overexpressed nontagged (OE) and endogenous (E) syntenin-2 levels were analyzed by Western blotting of total cell extracts originating from MCF-7 or Caco-2 cells as indicated. Purified recombinant nontagged syntenin-2 produced in bacteria (R) was included as a positive control. (B) Immunofluorescence micrographs of MCF-7 (a, b) and Caco-2 cells (c, d) showing the distribution of endogenous syntenin-2. (C) Confocal immunofluorescence micrographs of the nucleus of an MCF-7 cell showing the distribution of endogenous syntenin-2 (a), nucleolar antigen (b) and nuclear speckles/PIP2 (c) as detected with the 2C11 antibody that does not detect PIP2 in nucleoli (Osborne et al, 2001). In the merge panels, syntenin-2 is in green and nucleolar antigen (a-b) or nuclear speckles/PIP2 (a-c) are in red. (D) Schematic representation of the different domains of syntenin-2 that were transiently overexpressed as eGFP fusion proteins in MCF-7 cells. The limit of each domain is indicated by the amino-acid number. The subcellular distribution of the different eGFP fusion proteins was assessed by confocal microscopy, as illustrated. (E) Confocal immunofluorescence micrographs of the nucleus of an MCF-7 cell showing the distribution of eYFP fused to the segment of syntenin-2 that contains the two PDZ domains plus the C-terminal domain (a), nucleolar antigen (b) and nuclear speckles/PIP2 (c). In the merge panels, syntenin-2 is in green and nucleolar antigen (a-b) or nuclear speckles/PIP2 (a-c) are in red. Size bars are 10 μm.
Plasma membrane, nuclear speckles and nucleoli targeting of syntenin-2 PDZ domains relies on PIP2
To evaluate whether PIP2–PDZ domain interactions govern these subcellular distributions of syntenin-2, we performed colocalization studies with probes for PIP2 and tested whether treatments known to affect PIP2 levels also influence the distribution of syntenin-2. For these studies, we used the segment of syntenin-2 containing both PDZ domains plus the C-terminal domain, for simplicity further referred to as ‘tandem PDZ'.
In the small percentage of transfected cells where the eYFP-tandem PDZ was detected at the plasma membrane, it significantly colocalized with eCFP fused to the PH domain of PLCδ, a probe for plasma membrane PIP2 pools (Figure 3A, compare plasma membrane staining in a and d). As revealed by time-lapse video microscopy, plasma membrane PIP2 hydrolysis induced by ionomycin treatment (Varnai and Balla, 1998) caused simultaneous delocalization of eYFP-tandem PDZ and eCFP-PH-PLCδ (Figure 3A). Moreover, increasing the plasma membrane PIP2 pools, by overexpressing a phosphatidylinositol 4-phosphate 5 kinase, or by overexpressing Arf6, a small GTPase that recruits and activates phosphatidylinositol 4-phosphate 5 kinase at the plasma membrane (Honda et al, 1999), caused enrichment of the tandem PDZ at the plasma membrane (data not shown). These results support a role for PIP2 in recruiting the tandem PDZ domains of syntenin-2 to the plasma membrane. Since syntenin-2 interacts also significantly with PI(3,4,5)P3 (see above), we wondered whether PI(3,4,5)P3 plays a role in the plasma membrane targeting of the tandem PDZ domains. Neither serum stimulation, a treatment that produces a local enrichment of PI(3,4,5)P3 at the plasma membrane (as indicated by membrane recruitment of the PH domain of Akt, a PI(3,4,5)P3 probe), nor addition of wortmannin, which prevents synthesis of PI(3,4,5)P3, influenced the localization of the tandem PDZ (data not shown). Therefore, we conclude that PI(3,4,5)P3 does not significantly contribute to the plasma membrane localization of syntenin-2. This is consistent with the fact that PIP2 is 25-fold more abundant in cells compared to other phosphoinositides and with the view that a protein will not target rare phosphoinositides unless it displays stringent affinity for this particular phosphoinositide (Lemmon, 2003).
Figure 3.
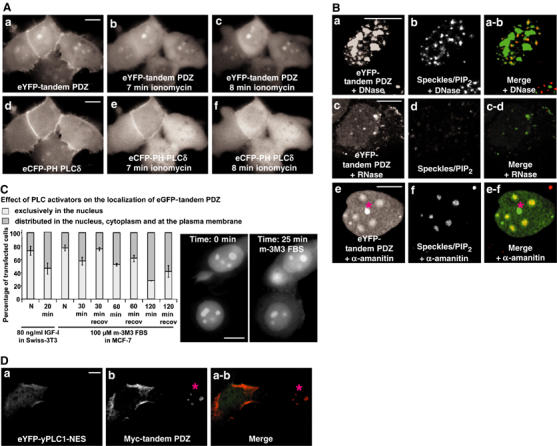
Modulation of cellular PIP2 levels affects the distribution of syntenin-2 PDZ domains. (A) Time-lapse microscopy of MCF-7 cells expressing the tandem PDZ domains of syntenin-2 (tandem PDZ) fused to eYFP (a–c), and the PH domain of PLCδ fused to eCFP (d–f) before (a, d), 7 min after (b, e) or 8 min after (c, f) ionomycin-induced hydrolysis of plasma membrane PIP2. (B) MCF-7 cells expressing the tandem PDZ fused to eYFP were treated with DNase I (a, b), RNase A (c, d) or α-amanitin (e, f). The subnuclear distributions of eYFP-tandem PDZ (a, c, e) and nuclear speckles/PIP2 (b, d, f) were analyzed by confocal microscopy. In the merge panels, eYFP-tandem PDZ is in green and nuclear speckles/PIP2 are in red. The structure below the asterisk in panels e and e-f most likely corresponds to a nucleolar fragment. (C) Distribution of the tandem PDZ after activation of nuclear PLCs. MCF-7 cells expressing the eGFP-tandem PDZ were scored for nuclear enrichment of the fluorescence. Diagram, left part: Swiss 3T3 cells were treated with IGF-I for 20 min and compared to nontreated cells (N). Diagram, right part: MCF-7 cells were treated with the PLC activator m-3M3 FBS for different periods, as indicated, and compared to nontreated cells (N), or cells that were allowed to recover in serum-supplemented medium for 1 h after removal of the activator (recov). The distribution of the fluorescence was scored on three separate experiments, with 100 cells counted for each condition. Bars indicate standard deviations. Pictures: Time-lapse micrographs of MCF-7 cells before and after 25 min incubation with m-3M3 FBS. Note that the treatment induces cytosolic distribution of the fluorescence. (D) Confocal micrograph of MCF-7 cells expressing the eYFP-yeast PLC1 with a mutated NES ‘eYFP-yPLC1-NES' (a) and a Myc fusion to the tandem PDZ (b). eYFP-yPLC1-NES was detected by eYFP fluorescence and the Myc-tandem PDZ using anti-Myc antibodies (9E10). In the merge (a-b), eYFP-yPLC1-NES is in green and the Myc-tandem PDZ is in red. Note the nuclear concentration of the Myc-tandem PDZ in the cell on the right side (below the asterisk) and the cytosolic/plasma membrane distribution in the cell expressing yPLC1-NES, on the left side. Size bars are 10 μm.
RNA, but not DNA, is known to be essential for the association of PIP2 with nuclear speckles (Osborne et al, 2001). RNase A treatment (Figure 3Bc-d) reduced the nuclear localization of the eYFP-tandem PDZ, while DNase I treatment (Figure 3Ba-b) had no effect. Speckles are dynamic structures, and their size, shape and number varies in response to environmental signals that influence the available pools of active splicing and transcription factors. When transcription is inhibited, splicing factors and PIP2 accumulate in enlarged and rounded speckles (Boronenkov et al, 1998). Treating cells with the transcriptional inhibitor α-amanitin, at doses that allow specific inhibition of RNA polymerase II, caused reorganization of the speckles/PIP2 structures (Figure 3Bf, compare the number and the size of the speckles in f and b) and of the distribution of eYFP-tandem PDZ (Figure 3Be). This indicates that the tandem PDZ and PIP2 are physically and dynamically associated in nuclear speckles. A pool of the eYFP-tandem PDZ did not colocalize with the speckles/PIP2 in these experiments (asterisk in Figure 3Be and e-f), and most probably represents a nucleolar fragment as inhibition of transcription can cause breakdown or loss of the nucleolar structure (Haaf and Ward, 1996). To determine whether the nuclear localization of the tandem PDZ can be modulated by nuclear PIP2 breakdown, we induced cells to activate nuclear phospholipases. In Swiss-3T3 cells, IGF-I treatment activates nuclear PIP2 breakdown via activation of PLCβ1 (Cocco et al, 1988; Divecha et al, 1991). Treating Swiss-3T3 cells that expressed the eGFP-tandem PDZ with 80 ng/ml of IGF-I for 20 min caused an enrichment of the fluorescence in the cytoplasm and at the plasma membrane (Figure 3C, left part of the diagram). The eGFP-tandem PDZ was also translocated from the nucleus to the cytoplasm and to the plasma membrane in MCF-7 cells when the cells were exposed to 100 μM m-3M3 FBS, a general PLC activator (Bae et al, 2003) (Figure 3C, right part of the diagram). Time-lapse microscopy revealed a clear translocation after 25 min (Figure 3C, micrographs). A complete release from the nuclear organelles was observed after 2 h treatment, but by that time nucleoli were not distinguishable anymore by differential interference contrast (DIC) (data not shown). This effect was reversible, as removal of the PLC activator and incubation of the cells in serum-rich medium for 1 h led to recovery of nuclear enrichment (Figure 3C, recov). Finally, expression of the yeast phospholipase PLC1 (mutated in its nuclear export signal (NES) to allow nuclear localization) shifted the distribution of the tandem PDZ from nucleoli/speckles toward the cytoplasm and the plasma membrane (Figure 3D, compare the distribution of the tandem PDZ in the left cell, positive for PLC1, with the distribution in the right cell, negative for PLC1). In these experiments, nucleoli could still be traced by antinucleolar antigen staining (Supplementary data 4). Nevertheless, this staining was weaker than in non-PLC1-transfected cells, suggesting that PLC overexpression affects nucleolar structure. Altogether, these experiments indicate that nuclear PIP2 is required for the subnuclear enrichment of syntenin-2 tandem PDZ.
Both PDZ domains of syntenin-2 contribute to PIP2 binding, but PDZ1 plays a crucial role in PIP2 subnuclear targeting
We then analyzed the relative contributions of the individual PDZ domains to the PIP2 binding of syntenin-2. In isolation, both PDZ domains bound specifically and similarly to lipid layers containing 10% PIP2 (Figure 4A). Yet, the binding of the wild-type form of the isolated PDZ domains was weak in comparison to the binding of the tandem PDZ (compare Figure 4B with Figure 4A and note differences in protein concentrations and RU values). We then constructed PDZ domains that are unable to interact with PIP2, using an approach similar to that used for syntenin-1 (Zimmermann et al, 2002). Two lysine to alanine mutations, affecting a lysine in the carboxylate binding loop and a second lysine close to the αB1 residue, were introduced in the PDZ1 domain (K113A and K167A, i.e. mut PDZ1) and in the PDZ2 domain (K197A and K244A, i.e. mut PDZ2). As intended, introduction of these point mutations into the isolated PDZ domains of syntenin-2 abolished their PIP2 binding (Figure 4A). Mutating the PDZ domains separately in the PDZ tandem of syntenin-2 reduced the binding to PIP2, but could not abolish binding. Only the PDZ tandem of syntenin-2 in which both PDZ domains were mutated failed to bind PIP2-containing lipid layers (Figure 4B). Interestingly, such mutant syntenin-2 still interacts with the cytoplasmic domain of tetraspanin L6A, the sole peptide ligand identified so far for the syntenin-2 PDZ domains (Borrell-Pages et al, 2000), and is still able to self-associate, as described for the wild-type protein (Koroll et al, 2001) (data not shown). Next, we explored the impact of these mutations on the subcellular localizations of the tandem PDZ in MCF-7 cells. When expressed as eYFP fusion protein, the tandem PDZ of syntenin-2 with two mutant PDZ domains (eYFP-mut PDZ1+mut PDZ2) (Figure 4Cd) was not enriched at the plasma membrane, nor in speckles or in nucleoli. This was in contrast to the wild-type tandem PDZ (eYFP-wt PDZ1+wt PDZ2) (Figure 4Ca). Mutation of only one of the PDZ domains of the tandem (eYFP-wt PDZ1+mut PDZ2 or eYFP-mut PDZ1+wt PDZ2) did not abolish PIP2 subcellular targeting (Figure 4Cb and c).
Figure 4.
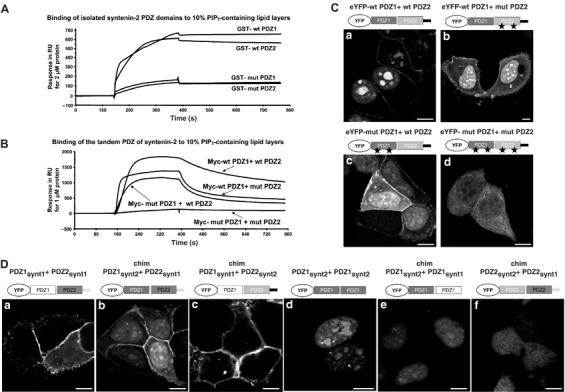
Respective contribution of syntenin-2 PDZ domains in PIP2 binding and targeting to specific subcellular PIP2 pools. (A) SPR sensograms showing the binding of the individual PDZ domains of syntenin-2 to 10% PIP2-containing lipid layers. Wild-type (wt) and mutant (mut) PDZ domains were purified as GST fusion proteins and perfused at 2 μM. Mutant PDZ domains (mut PDZ1 and mut PDZ2) were made by changing two lysine residues into alanine residues. (B) SPR sensograms showing the interaction of the tandem PDZ of syntenin-2 with 10% PIP2-containing lipid layers. Wild-type and mutant PDZ domains were purified as Myc fusion proteins and perfused at 1 μM over the sensorchip. (C) Subcellular localization of wild-type and mutant tandem PDZ of syntenin-2 in MCF-7 cells was analyzed by confocal microscopy. Proteins were expressed as eYFP fusions. (D) Confocal micrographs of MCF-7 cells expressing different combinations of the PDZ domains of syntenin-1 and syntenin-2 as indicated. Size bars are 10 μm.
Since the tandem PDZ of syntenin-1 also binds with high affinity to PIP2 (Zimmermann et al, 2002) but never concentrates in subnuclear PIP2-rich regions (Figure 4Da), the relative contributions of the PDZ domains to plasma membrane and nuclear PIP2 targeting were also tested by domain-swapping experiments. We constructed chimeric proteins where the first PDZ domain of syntenin-2 was fused to the second PDZ domain of syntenin-1 (eYFP-chim-PDZ1synt2+PDZ2synt1; chim: chimera) or the inverse (eYFP-chim-PDZ1synt1+PDZ2synt2), and tested the subcellular localization of these chimeras by confocal microscopy. Although the first chimera localized to all PIP2-rich regions (Figure 4Db), the second chimera was restricted to the plasma membrane (Figure 4Dc), suggesting that the PDZ1 of syntenin-2 determines subnuclear PIP2 targeting. Consistently, a construct containing two syntenin-2 PDZ1 domains in tandem (eYFP-chim-PDZ1synt2+PDZ1synt2) and also a chimera containing the PDZ1 domain of syntenin-2 fused to the PDZ1 of syntenin-1 (eYFP-chim-PDZ1synt2+PDZ1synt1) were also targeted to PIP2-rich regions in the nucleus (Figure 4Dd and e). Nevertheless, the structural combination of one PDZ1 domain with one PDZ2 domain seems important for syntenins to target PIP2 pools. Indeed, the tandem PDZ of syntenin-2 (Figure 4Ca) represented the best combination for nuclear PIP2 targeting, while a chimera between the first PDZ domain of syntenin-1 and the second PDZ domain of syntenin-2 (Figure 4Dc) represented the best combination for plasma membrane PIP2 targeting. Moreover, the combination of two PDZ2 domains, one originating from syntenin-2 and the other from syntenin-1 (eYFP-chim-PDZ2synt2+PDZ2synt1), was not targeted to any PIP2 pool but diffusely distributed (Figure 4Df) and diffuse distribution was also observed with a construct of two PDZ1 domains of syntenin-1 (data not shown).
Syntenin-2 loss-of-function disrupts nuclear speckles–PIP2 pattern and affects cell division and survival
To clarify the function of syntenin-2, we investigated the effect of a syntenin-2 depletion in cultured cells. Transfection of MCF-7 or U-2 OS cells with siRNA for syntenin-2 (RNAi) strongly reduced the levels of syntenin-2 protein, compared to cell transfection with nontargeting RNA (NT) (Figure 5A). While we did not observe substantial differences in the morphology of the nucleoli between cells that were treated with NT or RNAi, loss of syntenin-2 expression had a drastic consequence on nuclear PIP2 as observed by 2C11 staining. While in NT-treated cells, the 2C11 pattern was similar to that in nontransfected cells, 70% of the RNAi-treated cells displayed dispersed smaller speckles (Figure 5B, compare b with a and d with c). To further investigate the potential consequences of a syntenin-2 depletion for nuclear processes, U-2 OS cells, stably expressing histone 2B fused to red fluorescent protein, were transfected with RNAi or NT and followed by time-lapse video microscopy over 20 h. Cell death, accompanied by cell surface blebbing and chromatin compaction, was observed two-fold more frequently in RNAi-treated cells than in NT-treated cells (Figure 5C). Cell division and BrdU staining were reduced by a factor of 3 and 2 respectively in RNAi-treated cells (Figure 5C). These results indicate that syntenin-2 may function as an organizer of nuclear PIP2 and that syntenin-2 expression levels can affect cell viability and the rate of cell division.
Figure 5.
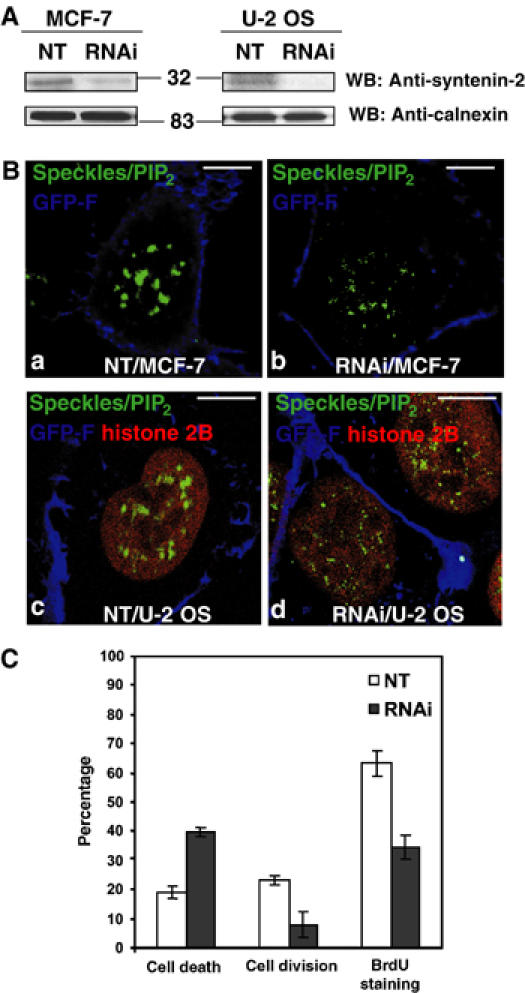
Suppression of syntenin-2 affects the nuclear PIP2 staining pattern, cell viability and cell division. (A) Western blot analysis of total cell extracts from MCF-7 cells (left panels) or U-2 OS cells (right panels), treated with an NT or an siRNA targeting syntenin-2 (RNAi). Note that endogenous levels of syntenin-2 decrease in RNAi-treated cells (upper panels) while calnexin signals are not affected (lower panels). Numbers indicate molecular weight markers in kDa. (B) Confocal micrographs of MCF-7 cells (a, b) and U-2 OS cells stably expressing RFP-histone 2B (c, d), transfected with an NT (a, c) or an siRNA targeting syntenin-2 (b, d) together with an expression vector for farnesylated eGFP (GFP-F) as a marker of transfected cells (blue signal at the plasma membrane). The nuclear speckles/PIP2 pattern was visualized using 2C11 antibody (green). RFP-histone 2B in U-2 OS is in red. Size bars are 10 μm. (C) Cell death and cell division in U-2 OS cells stably expressing RFP-histone 2B, treated with an NT or an siRNA for syntenin-2 (RNAi), were quantified in three independent experiments, by multipositioning time-lapse video microscopy, looking at the fate of a minimum of 50 transfected cells per condition in each experiment. Incorporation of BrdU by U-2 OS cells was quantified in three independent experiments, by wide-field fluorescence microscopy, on 200 cells per condition in each experiment. Bars indicate standard deviations.
Discussion
PDZ domains: phosphoinositide-binding domains?
Most PDZ proteins tested in this study displayed very weak affinity for PIP2. Also, phosphoinositides other than PIP2 interact only weakly with some of these PDZ proteins in vitro (data not shown). Moreover, when these weakly interacting PDZ proteins, or their PDZ domains, were overexpressed as fluorescent fusion proteins, these were neither targeted to discrete subcellular PIP2 pools nor to compartments rich in other phosphoinositides (data not shown). This is reminiscent of what is known for most PH domains (Lemmon, 2003; Yu et al, 2004), which also tend to bind PIP2 rather weakly, are quite nonspecific in their phosphoinositide binding and are by themselves not able to target their host proteins to phosphoinositide-rich domains. The functional importance of such weak interactions has not yet been established. In organisms, PDZ proteins generally oligomerize directly or indirectly, forming larger complexes containing multiple membrane anchors with affinity for transmembrane receptors (Bilder et al, 2003; Roh and Margolis, 2003). In such complexes, avidity effects could become important. As cultured cells and overexpression experiments might not reproduce the correct environmental contexts (cell polarity, associated subunits and presence and concentration of peptide ligands) of these PDZ proteins, one cannot exclude that in physiological situations, PIP2–PDZ domain interactions might augment the membrane-binding capacity of these PDZ proteins and modulate their plasma membrane targeting, but further work is needed to clarify this issue.
High-affinity binding to PIP2 by the PDZ domains of syntenins
This screen identified syntenin-2 as a second PDZ protein that binds with high affinity to PIP2 (apparent KD=1.5 μM) via its PDZ domains. This suggests that high-affinity binding to PIP2 might be a unique characteristic of syntenin PDZ domains. Although syntenin-2 is highly related to syntenin-1 in its overall domain organization and sequence, the known peptide ligands for the PDZ domains of syntenin-1 do not interact with syntenin-2 (Koroll et al, 2001), and vice versa, arguing that functionally these proteins are not redundant. It is therefore remarkable that the interaction of syntenin-2 with PIP2 is conserved. For both syntenin-1 and syntenin-2, the PDZ domains taken in isolation are not sufficient for targeting to PIP2-rich regions in cells. The tandem is required. Consistently, the in vitro interaction with PIP2 of the tandem PDZ domains of both syntenins is also stronger than that of the PDZ domains taken in isolation. Nevertheless, in syntenin-1, the first PDZ domain clearly displays higher affinity for PIP2 than does the second PDZ domain (Zimmermann et al, 2002), while in syntenin-2, the relative contribution to PIP2 binding is evenly distributed over both PDZ domains. Mutating the PDZ1 domain (introducing K119A in the carboxylate binding loop and replacing 171SDK173 by HEQ in the αB helix) in the tandem repeat already totally abolishes syntenin-1 binding to PIP2 (Zimmermann et al, 2002) but only moderately affects syntenin-1–syndecan-2 binding (Grootjans et al, 2000). Mutation of both PDZ domains of the tandem (introducing K to A mutations in the carboxylate binding loop and close to the αB1 residue in PDZ1 and PDZ2) is required to abolish syntenin-2–PIP2 interaction, but preserves binding to the peptide ligand tetraspanin L6A and the capacity to self-associate. This means that although some of the residues implicated in lipid and peptide binding overlap, the two activities can be dissociated. At this stage, it is difficult to delineate the structural determinants in PDZ domains that mediate high-affinity binding to PIP2. By amino-acid alignments, comparing PDZ domains displaying high affinity for PIP2 (the first PDZ domain of syntenin-1 and the two PDZ domains of syntenin-2) to PDZ domains displaying low or no affinity for PIP2, we failed to identify a unifying ‘PIP2-binding motif'. Kachel et al (2003) studied the interaction of the PDZ2b domain of PTP-Bas with PIP2 and PI(3,4,5)P3 by NMR, and identified 10 residues making contacts with the lipids (Kachel et al, 2003). Sequence alignments of the PDZ2b domain of PTP-Bas and the PDZ domains of the syntenins also failed to identify a consensus sequence for PIP2 binding. It should be noted, however, that the PDZ domains of the human ortholog of PTP-Bas display only weak PIP2 interaction in the experimental settings used in the present study (unpublished observation). Clearly, our studies indicated that positively charged residues are implicated, as in many other phosphoinositide-binding motifs. Nevertheless, competition experiments with inositol (1,4,5)-triphosphate (IP3) and water-soluble PIP2 also showed that the inositol phosphate headgroup is not sufficient for the syntenin-2 interaction with PIP2 (data not shown), suggesting that hydrophobic interactions could also play a role. Moreover, our data also imply that one has to consider the tandem of the PDZ1-2 domains of the syntenins as one functional unit rather than as two independent modules interacting with PIP2. Structural studies of syntenin–PIP2 complexes are ongoing and will hopefully clarify which residues are implicated in the interaction with PIP2.
Targeting to plasma membrane versus nuclear PIP2 pools
Our data suggest that subcellular targeting to PIP2 pools only occurs for constructs with an affinity for PIP2 that is above a certain threshold, and that this threshold can only be reached when the PDZ domains of syntenins are in tandem. Nevertheless, the tandem PDZ domains of syntenin-1 are exclusively targeted to PIP2 pools at the plasma membrane (Figure 4Da) while the tandem PDZ domains of syntenin-2 are predominantly targeted to nuclear PIP2 pools (Figure 4Ca). Endogenous syntenin-1 and syntenin-1 tandem PDZ can also be found in the nucleus, but they never concentrate in speckles or nucleoli (Zimmermann et al, 2001). Domain-swapping experiments indicate that the first but not the second PDZ domain of syntenin-2 contains the nuclear PIP2 targeting determinants (Figure 4Db–e). Several hypotheses can be put forward to explain this property. Most obvious is that targeting to nucleoli and nuclear speckles might necessitate an additional peptide anchorage for syntenin-2 PDZ domains in these structures, but so far we failed to identify such ligand. Alternatively, as detergent-insoluble pools of PIP2 are found in the nucleus (Divecha et al, 1993; Maraldi et al, 1999), the first PDZ domain of syntenin-2 might be better suited to recognize non-membrane-associated nuclear PIP2. Finally, interaction with unconventional ligands like RNA remains an unexplored possibility.
Understanding the biology of phosphoinositides has tremendously benefited from the identification of protein domains displaying specific and high-affinity binding for the different phosphoinositides and that can be used as probes in living cells (Varnai et al, 2002). The PH domain of PLCδ has been shown to be an excellent probe for recognizing PIP2 at the plasma membrane (Varnai and Balla, 1998), but cannot target PIP2 pools in the nucleus (Okada et al, 2002). So far, a probe detecting nuclear pools of PIP2 was lacking. The tandem PDZ domains of syntenin-2 represent such a probe.
Syntenin-2 as an organizer of nuclear PIP2
Understanding the role of PIP2 in nuclear compartmentalization and as a signaling molecule in these compartments is an emerging challenge. PIP2, by interacting with histone H1, might counteract transcriptional repression (Yu et al, 1998). By controlling the nuclear actin cytoskeleton, it might influence chromatin remodeling and pre-mRNA processing (Zhao et al, 1998; Skare et al, 2003). Moreover, PIP2 also undergoes PLC cleavage in the nucleus (Divecha et al, 1991). Subsequent generation of DAG and activation of PKC leads to phosphorylation events, resulting for example in lamin breakdown during mitosis (Haas and Jost, 1993). On the other hand, IP3 can serve as precursor of inositol polyphosphate, necessary for efficient mRNA export (York et al, 1999).
Multiple PIPK isoforms, diacylglycerol kinase θ and particular phospholipases are concentrated at nuclear speckles, indicating that PI signaling pathways are localized at this site (Martelli et al, 2004). Conceivably, syntenin-2 could function as a scaffold that brings PIP2 in proximity of components of the inositol cycle machinery or other speckles components (RNA, proteins, etc.). One possibility could have been that syntenin-2 links PIP2 with the nuclear enzyme PLCβ1, which contains a PDZ binding domain interacting with NHERF (Tang et al, 2000). Unfortunately, direct syntenin-2–PLCβ1 binding assays failed to support this idea (unpublished observation). Macromolecular interactions form the basis for speckle morphogenesis (Lamond and Spector, 2003), and when one structural component is missing, the architecture of the whole complex might be affected. Downregulation of syntenin-2 by RNAi causes a dispersion of the nuclear speckles/PIP2 (Figure 5B). Syntenin-2 could support and stabilize speckle structure by its propensity to self-associate and to bind PIP2. Syntenin-2 might protect PIP2 from degradation, and in such case the dispersed PIP2 pattern in the RNAi-treated cells could be due to reduced levels of PIP2. Speckles are also dynamic structures and their components travel to other nuclear locations. In this context, syntenin-2 could be part of a proteolipid complex that functions as a PIP2 transporter. Our data show that syntenin-2 is able to shuttle between several subcellular compartments. The disruption of the PIP2–syntenin-2 complex in our RNAi experiments may disturb one or several crucial processes, and lead to cell death and reduced cell divisions under these conditions. The characterization of other components interacting with syntenin-2 in the nucleus and of the cues that influence its localization should provide further elements to understand its function.
Materials and methods
Molecular biology
Untagged and Myc-tagged (EQKLISEEDL) forms of syntenin-2 cDNAs were subcloned in the pTYB1 vector (New England Biolabs, USA). The various phosphoinositide probes, the PDZ domains and the PDZ protein cDNAs were cloned in pGEX vectors. All proteins were expressed as intein fusion or GST fusion proteins in ER2566 cells and purified as described before (Grootjans et al, 2000). The purified proteins were dialyzed in HBS buffer (20 mM Hepes pH 7.4, 150 mM NaCl and 1 mM EDTA). Eukaryotic expression vectors encoding eGFP, eCFP and eYFP fluorescent proteins were obtained from Clontech (BD Biosciences, USA). The eukaryotic expression vector pCDNA3.1/Zeo(+) for expression of untagged and Myc-tagged proteins was obtained from Invitrogen (CA, USA). The yeast PLC1 open reading frame was amplified from yeast genomic DNA and TA cloned into pGEM-T-EASY (Promega, USA). The nuclear export sequence 188-KLKAH-192 was changed into KRKRH. Mutated yeast PLC1 was subcloned in the mammalian expression vector peYFP-C. All constructs were certified by sequencing. More detailed information is available upon request.
Lipid-binding assays
Lipid manufacturers, PIP2 overlays, preparation of vesicles and detailed protocols for SPR experiments can be found in Supplementary data 5. Briefly, reconstituted composite vesicles containing PC, PE, PS and PI-PIPs prepared in appropriate molar ratios (30:40:20:10) were coated on L1-chips. Coated chips were perfused with purified proteins in HBS for association. Sensograms were corrected for background association to 10% PI vesicles. For apparent KD measurements, various protein concentrations, ranging from 250 to 3000 nM, were perfused on the chips and signals obtained at the equilibrium (Req) were plotted as a function of protein concentration. Apparent KD values were deduced from these plots, as the concentration corresponding to Req(max)/2.
Anti-syntenin-2 antibodies
Two rabbits were immunized (Eurogentec, Belgium) with purified recombinant human syntenin-2 produced in the impact T7 system according to the instructions of the manufacturer (New England Biolabs, UK). Anti-syntenin-2 polyclonal antibodies were isolated from the sera from these animals by affinity purification on a syntenin-2-Sepharose 4B column (Amersham Biosciences, UK). Characterization of these antibodies indicated that they do not crossreact with syntenin-1 and recognize epitopes in the N-terminal, PDZ-1, PDZ-2 and C-terminal domains of syntenin-2, in Western blot and in immunocytochemistry. However, they fail to recognize the nuclear speckles-associated form of syntenin-2.
Western blotting
Cells were lysed in DIP buffer (1% Triton X-100, 0.1% SDS and 1% NP-40 in PBS). Western blotting was performed as described before (Zimmermann et al, 2001). The blots were incubated with anti-syntenin-2 (1 μg/ml) or anti-calnexin (StressGen Biotech. Corp., Canada) antibodies and appropriate secondary antibodies coupled to horseradish peroxidase (Amersham Biosciences, UK). Signals were visualized by enhanced chemiluminescence.
Cell biology and microscopy
Cell cultures, transfections and stainings were performed as described before (Zimmermann et al, 2001). For microscopical analysis of fixed samples, cells were plated on uncoated eight-well chamber slides (Nalgene Nunc International, USA). For live cell imaging, cells were plated on two-well chamber borosilicate coverglass (Nalgene Nunc International, USA). For plasma membrane PIP2 breakdown experiments, cells were washed in Krebs–Ringer buffer (Varnai and Balla, 1998) before incubation with 10 μg/μl of ionomycin (Sigma, USA) in the same buffer. For inhibition of transcription, MCF-7 cells were treated with 5 μg/ml of α-amanitin (Roche, UK) for 6 h. For PLC activation assays, IGF-I was purchased from Invitrogen (USA) and m-3M3 FBS (Bae et al, 2003) from Calbiochem EMD Biosciences (USA). Anti-syntenin-2 antibodies were used at 10 μg/ml to detect endogenous syntenin-2 and at 1 μg/ml to detect overexpressed syntenin-2. Anti-PIP2 antibodies (2C11) were applied as described elsewhere (Osborne et al, 2001). Antinucleolar antigen antibody was purchased from Biomeda (Foster City, CA). RNase A (Roche UK; 0.5 mg/ml, 5 min) and DNase I (Roche UK; 100 μg/ml, 1 h) treatments were performed as described elsewhere (Osborne et al, 2001). Incorporation of BrdU was detected with anti-BrdU antibody (Becton Dickinson, San Jose, CA) in cells that were pulsed for 15 min with 20 μM BrdU (Calbiochem, EMD Biosciences, USA). Images were obtained by digital imaging fluorescence microscopy using a cooled charge-coupled device camera (Photometrics, Tucson, AZ) or by confocal microscopy (MRC-1024 Laser Scanning Confocal Imaging System, Bio-Rad, CA). Live cell imaging experiments were performed with a Leica AS MDW workstation (Leica Microsystems, Wetzlar, Germany).
siRNA
siRNAs, specifically targeting syntenin-2, were purchased from Dharmacon Inc. (USA). They consist of four RNA duplexes (sigenome-on-target™ duplexes for human-SDCBP2). Duplex 2 was proven to be the best to downregulate endogenous syntenin-2 and was used in all our assays. NT (siSTABLE #1, Dharmacon Inc.) was used as a control. Cells were transfected at 40–50% density with 100 nM siRNA or NT together with an expression vector for farnesylated eGFP to recognize targeted cells and were analyzed after 48 h. Transfection was achieved with oligofectamine (Invitrogen, USA), according to the recommendations of the manufacturer. For live experiments, cells were grown in two-well chamber borosilicate coverglass to allow a parallel study of siRNA- (well 1) or NT- (well 2) treated cells. Cell fate was recorded every 3 min by fluorescence and DIC time-lapse microscopy.
Supplementary Material
Supplementary Information 1
Supplementary Information 2
Supplementary Information 3
Supplementary Information 4
Supplementary Information 5
Acknowledgments
We thank Dr G Schiavo for providing 2C11 antibodies, Dr JP Borg for providing GST-PSD95 PDZ1-3 and GST-Mint3 PDZ1-2 expression vectors, Dr K Schuh for providing the GST-nNOS PDZ1 expression vector, Dr J Franklin and Dr R Coffey for providing the GST-Magi-3 FL expression vector, Dr M Bhat for providing the GST-Dlt FL expression vector, Dr Wodarz for the Bazooka cDNA and Dr F Wolf and Dr S Geley for providing the U-2 OS cell line stably expressing RFP-histone 2B. We also thank Dr S Dove for supplying materials and for helpful discussions. This work was supported by grants from the Fund for Scientific Research-Flanders (FWO), by the Flanders Interuniversity Institute for Biotechnology, by the Belgian Federation against Cancer, by the Concerted Actions Program of the KU Leuven, by the Interuniversity Attraction Poles of the Prime Ministers' Services and by FB insurance Fortis. PZ is a Postdoctoral Fellow of the FWO. Request for materials should be addressed to PZ.
References
- Bae YS, Lee TG, Park JC, Hur JH, Kim Y, Heo K, Kwak JY, Suh PG, Ryu SH (2003) Identification of a compound that directly stimulates phospholipase C activity. Mol Pharmacol 63: 1043–1050 [DOI] [PubMed] [Google Scholar]
- Bilder D, Schober M, Perrimon N (2003) Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat Cell Biol 5: 53–58 [DOI] [PubMed] [Google Scholar]
- Boronenkov IV, Loijens JC, Umeda M, Anderson RA (1998) Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol Biol Cell 9: 3547–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell-Pages M, Fernandez-Larrea J, Borroto A, Rojo F, Baselga J, Arribas J (2000) The carboxy-terminal cysteine of the tetraspanin L6 antigen is required for its interaction with SITAC, a novel PDZ protein. Mol Biol Cell 11: 4217–4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco L, Martelli AM, Gilmour RS, Ognibene A, Manzoli FA, Irvine RF (1988) Rapid changes in phospholipid metabolism in the nuclei of Swiss 3T3 cells induced by treatment of the cells with insulin-like growth factor I. Biochem Biophys Res Commun 154: 1266–1272 [DOI] [PubMed] [Google Scholar]
- Czech MP (2003) Dynamics of phosphoinositides in membrane retrieval and insertion. Annu Rev Physiol 65: 791–815 [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Godi A (2004) PI-loting membrane traffic. Nat Cell Biol 6: 487–492 [DOI] [PubMed] [Google Scholar]
- Divecha N, Banfic H, Irvine RF (1991) The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-I) in the plasma membrane, and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase C to the nucleus. EMBO J 10: 3207–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divecha N, Banfic H, Irvine RF (1993) Inositides and the nucleus and inositides in the nucleus. Cell 74: 405–407 [DOI] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J (2003) The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114: 99–111 [DOI] [PubMed] [Google Scholar]
- Grootjans JJ, Reekmans G, Ceulemans H, David G (2000) Syntenin–syndecan binding requires syndecan-synteny and the co-operation of both PDZ domains of syntenin. J Biol Chem 275: 19933–19941 [DOI] [PubMed] [Google Scholar]
- Grootjans JJ, Zimmermann P, Reekmans G, Smets A, Degeest G, Durr J, David G (1997) Syntenin, a PDZ protein that binds syndecan cytoplasmic domains. Proc Natl Acad Sci USA 94: 13683–13688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Ward DC (1996) Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp Cell Res 224: 163–173 [DOI] [PubMed] [Google Scholar]
- Haas M, Jost E (1993) Functional analysis of phosphorylation sites in human lamin A controlling lamin disassembly, nuclear transport and assembly. Eur J Cell Biol 62: 237–247 [PubMed] [Google Scholar]
- Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman MA, Kanaho Y (1999) Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 99: 521–532 [DOI] [PubMed] [Google Scholar]
- Hung AY, Sheng M (2002) PDZ domains: structural modules for protein complex assembly. J Biol Chem 277: 5699–5702 [DOI] [PubMed] [Google Scholar]
- Irvine RF (2003) Nuclear lipid signalling. Nat Rev Mol Cell Biol 4: 349–360 [DOI] [PubMed] [Google Scholar]
- Jones DR, Divecha N (2004) Linking lipids to chromatin. Curr Opin Genet Dev 14: 196–202 [DOI] [PubMed] [Google Scholar]
- Kachel N, Erdmann KS, Kremer W, Wolff P, Gronwald W, Heumann R, Kalbitzer HR (2003) Structure determination and ligand interactions of the PDZ2b domain of PTP-Bas (hPTP1E): splicing-induced modulation of ligand specificity. J Mol Biol 334: 143–155 [DOI] [PubMed] [Google Scholar]
- Koroll M, Rathjen FG, Volkmer H (2001) The neural cell recognition molecule neurofascin interacts with syntenin-1 but not with syntenin-2, both of which reveal self-associating activity. J Biol Chem 276: 10646–10654 [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL (2003) Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol 4: 605–612 [DOI] [PubMed] [Google Scholar]
- Lemmon MA (2003) Phosphoinositide recognition domains. Traffic 4: 201–213 [DOI] [PubMed] [Google Scholar]
- Maraldi NM, Zini N, Santi S, Manzoli FA (1999) Topology of inositol lipid signal transduction in the nucleus. J Cell Physiol 181: 203–217 [DOI] [PubMed] [Google Scholar]
- Martelli AM, Manzoli L, Cocco L (2004) Nuclear inositides: facts and perspectives. Pharmacol Ther 101: 47–64 [DOI] [PubMed] [Google Scholar]
- Nourry C, Grant SG, Borg JP (2003) PDZ domain proteins: plug and play!. Sci STKE 2003: RE7. [DOI] [PubMed] [Google Scholar]
- Okada M, Fujii M, Yamaga M, Sugimoto H, Sadano H, Osumi T, Kamata H, Hirata H, Yagisawa H (2002) Carboxyl-terminal basic amino acids in the X domain are essential for the nuclear import of phospholipase C delta1. Genes Cells 7: 985–996 [DOI] [PubMed] [Google Scholar]
- Osborne SL, Thomas CL, Gschmeissner S, Schiavo G (2001) Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J Cell Sci 114: 2501–2511 [DOI] [PubMed] [Google Scholar]
- Roh MH, Margolis B (2003) Composition and function of PDZ protein complexes during cell polarization. Am J Physiol Renal Physiol 285: F377–F387 [DOI] [PubMed] [Google Scholar]
- Sheng M, Sala C (2001) PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci 24: 1–29 [DOI] [PubMed] [Google Scholar]
- Skare P, Kreivi JP, Bergstrom A, Karlsson R (2003) Profilin I colocalizes with speckles and Cajal bodies: a possible role in pre-mRNA splicing. Exp Cell Res 286: 12–21 [DOI] [PubMed] [Google Scholar]
- Tang Y, Tang J, Chen Z, Trost C, Flockerzi V, Li M, Ramesh V, Zhu MX (2000) Association of mammalian trp4 and phospholipase C isozymes with a PDZ domain-containing protein, NHERF. J Biol Chem 275: 37559–37564 [DOI] [PubMed] [Google Scholar]
- Toker A (2002) Phosphoinositides and signal transduction. Cell Mol Life Sci 59: 761–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Balla T (1998) Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol 143: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Lin X, Lee SB, Tuymetova G, Bondeva T, Spat A, Rhee SG, Hajnoczky G, Balla T (2002) Inositol lipid binding and membrane localization of isolated pleckstrin homology (PH) domains. Studies on the PH domains of phospholipase C delta 1 and p130. J Biol Chem 277: 27412–27422 [DOI] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR (1999) A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285: 96–100 [DOI] [PubMed] [Google Scholar]
- Yu H, Fukami K, Watanabe Y, Ozaki C, Takenawa T (1998) Phosphatidylinositol 4,5-bisphosphate reverses the inhibition of RNA transcription caused by histone H1. Eur J Biochem 251: 281–287 [DOI] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA (2004) Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell 13: 677–688 [DOI] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR (1998) Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95: 625–636 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Meerschaert K, Reekmans G, Leenaerts I, Small JV, Vandekerckhove J, David G, Gettemans J (2002) PIP(2)-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol Cell 9: 1215–1225 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Tomatis D, Rosas M, Grootjans J, Leenaerts I, Degeest G, Reekmans G, Coomans C, David G (2001) Characterization of syntenin, a syndecan-binding PDZ protein, as a component of cell adhesion sites and microfilaments. Mol Biol Cell 12: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information 1
Supplementary Information 2
Supplementary Information 3
Supplementary Information 4
Supplementary Information 5


