Abstract
Background
Creation of human gene families was facilitated significantly by gene duplication and diversification. The (TG/CA)n repeats exhibit length variability, display genome-wide distribution, and are abundant in the human genome. Accumulation of evidences for their multiple functional roles including regulation of transcription and stimulation of recombination and splicing elect them as functional elements. Here, we report analysis of the distribution of (TG/CA)n repeats in human gene families.
Results
The 1,317 human gene families were classified into six functional classes. Distribution of (TG/CA)n repeats were analyzed both from a global perspective and from a stratified perspective based on their biological properties. The number of genes with repeats decreased with increasing repeat length and several genes (53%) had repeats of multiple types in various combinations. Repeats were positively associated with the class of Signaling and communication whereas, they were negatively associated with the classes of Immune and related functions and of Information. The proportion of genes with (TG/CA)n repeats in each class was proportional to the corresponding average gene length. The repeat distribution pattern in large gene families generally mirrored the global distribution pattern but differed particularly for Collagen gene family, which was rich in repeats. The position and flanking sequences of the repeats of Collagen genes showed high conservation in the Chimpanzee genome. However the majority of these repeats displayed length polymorphism.
Conclusion
Positive association of repeats with genes of Signaling and communication points to their role in modulation of transcription. Negative association of repeats in genes of Information relates to the smaller gene length, higher expression and fundamental role in cellular physiology. In genes of Immune and related functions negative association of repeats perhaps relates to the smaller gene length and the directional nature of the recombinogenic processes to generate immune diversity. Thus, multiple factors including gene length, function and directionality of recombinogenic processes steered the observed distribution of (TG/CA)n repeats. Furthermore, the distribution of repeat patterns is consistent with the current model that long repeats tend to contract more than expand whereas, the reverse dynamics operates in short repeats.
Background
The evolution of organisms with increasing complexity was significantly facilitated by duplication of genes and genomes followed by diversification [1,2]. Gene duplication per se produces two identical copies. Subsequently, one of the copies may either accumulate beneficial changes to give rise to a functionally diversified gene or accrue deleterious mutations to end up as a pseudogene, while the other copy retains its original function. The former mechanism leads to the creation of 'gene families' capable of carrying out diverse functions [2,3].
The classification of genes into gene families by Human Gene Nomenclature Committee (HGNC) on the basis of sequence similarity of the encoded proteins [4] and the availability of human genome sequence [5] allow us to carry out a comprehensive survey of a class of important functional element, namely the (TG/CA)n repeats. Analysis of the distribution of (TG/CA)nrepeats within genes in 'present day' gene families holds the potential to provide insights into the factors steering their abundance and selective distribution. Although the characteristic property of (TG/CA)n repeats exhibiting length polymorphism has been widely used in genetic mapping [6], a growing body of evidence accumulating over several years point to their multiple functional roles in various biological processes.
The (TG/CA)n repeats have a propensity to undergo structural transitions [7-10] and have been shown to modulate transcription in several genes including rat α-lactalbumin [9], rat prolactin [11], MMP-9 [12], IFN-γ [13], EGFR [14], HSD11B2 [15], tilipia prolactin1 [16] and human housekeeping genes [17]. Furthermore, the (TG)n tracts have been observed to act as stimulator in recombination and in mRNA splicing [18-22].
In the current study, the analysis of distribution of (TG/CA)n repeats in human gene families affords assessment of the distribution of these repeats by examining for positive association or negative association with respect to gene length and function.
Results
Characteristics of human gene families and their functional classification
Each of the 1,317 gene families included members with similar functional roles. The family sizes varied in a wide range between 2 to 223 members (Figure 1). The number of gene families was found to bear an inverse exponential relation to family size. About two-fifths of the gene families were duplex. Only three gene families had more than 100 members per family: Immunoglobulin heavy chain (162 genes), Zinc finger proteins (200 genes) and Solute carrier (223 genes).
Figure 1.
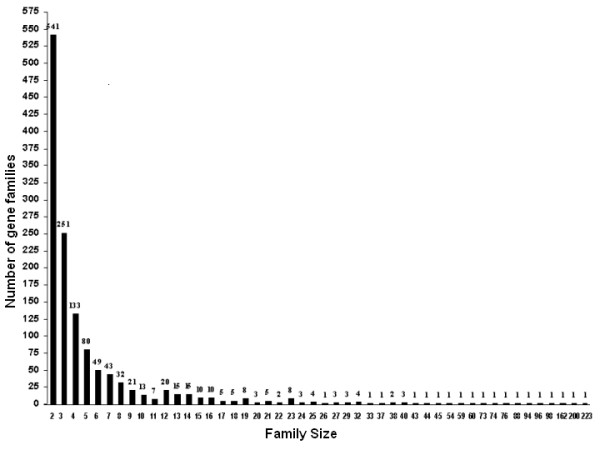
Distribution pattern of human gene families with respect to family sizes. X axis: family size (number of genes in each gene family). Y axis: number of gene families corresponding to various family sizes. Note the inverse exponential relationship.
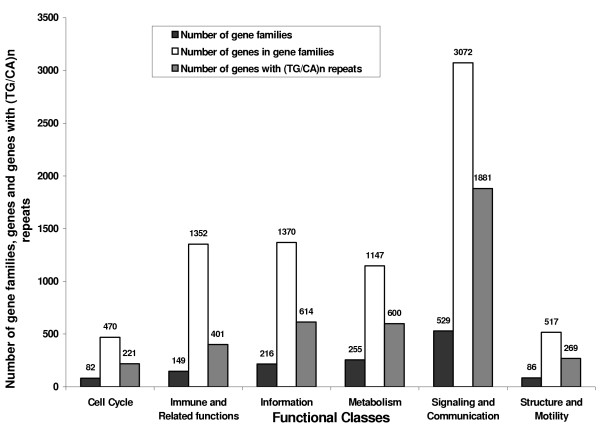
The functional classification of 1,317 gene families comprising 7,928 genes in the six functional classes unveiled that the Signaling and communication is largest with 529 families and 3,072 genes (Figure 2). The Cell cycle is the smallest with 82 families and 470 genes.
Figure 2.
Global distribution of gene families, genes and proportion of genes containing (TG/CA)n repeats classified into the six functional classes. The numbers correspond to the height of the vertical bars in each group.
Of the 1,317 gene families, 131 were entirely intrachromosomal. Chromosome 1 had the largest number with 17 families followed by chromosomes 19 and 11 with 13 and 12 families respectively. The remaining chromosomes had less than 10 intrachromosomal gene families per chromosome. The functional classification of these 131 intrachromosomal gene families revealed that the highest number (45) belonged to the class of 'Immune and related functions' closely followed by the class of Signaling and communication with 40 families. The remaining classes had the following distribution of gene families: Metabolism (24), Information (15), Structure and motility (5) and Cell cycle (2). These observations indicate that the creation of intrachromosomal human gene families was driven by large number of duplications followed by divergence in selected functional classes.
Global distribution of (TG/CA)n repeats (n ≥ 6 units) in gene families
Of the 1,317 gene families, 732 families had (TG/CA)n repeats in at least one of their members and 326 families had repeats in all their members. Of the 7,928 genes in 1,317 families, 3,986 genes had intragenic (TG/CA)n repeats of length greater than or equal to 6 units. All 3,986 genes had repeats in their introns. Only 277 genes had (TG/CA)n repeats in exons indicating that these repeats are mainly present in introns.
The distribution of genes with (TG/CA)n repeats in the six functional classes is displayed in Figure 2. It is apparent that the class of Signaling and communication had the highest number of genes with (TG/CA)n repeats. Comparison of the proportion of genes with repeats in each class with the global proportion showed that the class of Signaling and communication had significantly higher than the expected proportion (p < 0.0001, Binomial test). In contrast, the classes of Immune and related functions and Information had significantly lower than the expected proportion of genes with repeats (p < 0.0001 and p < 0.0002 respectively). The proportion of genes with repeats was not significantly different from the global proportion in the Cell cycle, Metabolism and Structure and motility classes. These observations show that the (TG/CA)n repeats exhibit positive association with the genes belonging to Signaling and communication whereas, they are negatively associated with the genes belonging to Immune and related functions and Information.
It has been shown that the human genome has an isochore structure that varies in GC content [5]. This variation raises the possibility that the observed selective distribution of (TG/CA)n repeats might have arisen due to fluctuations in the local %(G+C) content of the genomic region as opposed to function. We examined this by comparing the average %(G+C) content of the genes in the six functional classes with the corresponding proportions of genes with repeats. The average %(G+C) content was observed to be in the narrow range (47–49%) in the six functional classes whereas, the proportion of genes with repeats varies widely in the range 29.6–61%. These observations indicate that the proportion of genes with repeats is significantly determined by function instead of small fluctuations in %(G+C) content.
Correlation between gene length, function and global distribution of (TG/CA)n repeats
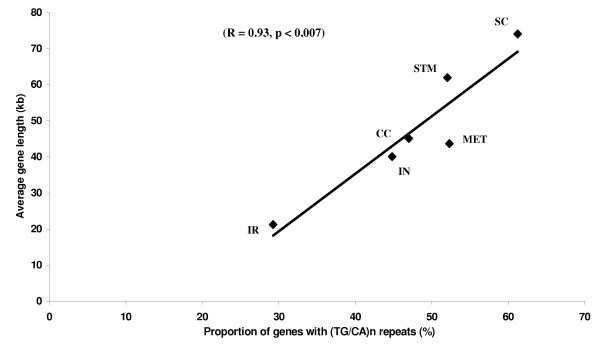
Comparison of the proportion of genes containing (TG/CA)n repeats with the average lengths of genes in each of the six functional classes revealed a linear relationship (Figure 3, correlation coefficient R = 0.93, p < 0.007). The signaling and communication class had the longest average gene length (74.07 kb) along with the highest proportion of genes with (TG/CA)n repeats (61.23%). The class of Immune and related functions had the shortest average gene length (21.26 kb) with the lowest proportion of genes with (TG/CA)n repeats (29.65%). These observations show that the proportion of genes with (TG/CA)n repeats bears a linear relationship to the length of genes.
Figure 3.
Relationship between proportion of genes with (TG/CA)n repeats in each functional class and the average gene length in the corresponding functional classes. X axis: Proportion of genes with (TG/CA)n repeats (%); Y axis: Average gene length (kb) (CC: Cell cycle; IN: Information; IR: Immune and related functions; MET: Metabolism; SC: Signaling and communication; STM: Structure and motility)
Trinity of (TG/CA)n repeats in gene families
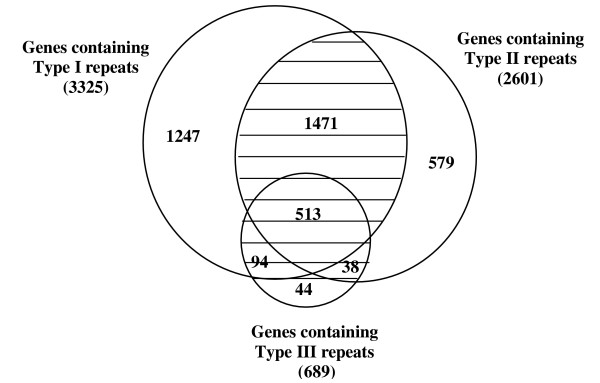
In order to examine the characteristics of distribution of (TG/CA)n repeats with respect to multiple functional roles principally governed by their length, we analysed the repeats stratified into three categories: type I (6 ≤ n < 12), type II (12 ≤ n < 23) and type III (n ≥ 23). The results are displayed in Figure 4. The number of genes containing (TG/CA)n repeats decreases with increasing repeat length. It is also apparent that several genes (53% of the total) have multiple types of repeats in various combinations.
Figure 4.
A Venn diagram of the genes with trinity of intragenic (TG/CA)n repeats (type I, II and III). Note that several genes (shaded area) have multiple types of repeats in various combinations.
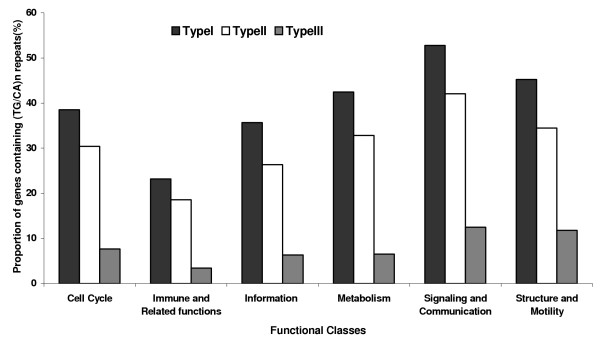
Classification of the distribution of genes with (TG/CA)n repeats stratified into three categories into six functional classes is shown in Figure 5. It is evident that the proportion of genes containing repeats decreases in the order I > II > III in all classes. The proportion of genes containing (TG/CA)n repeats of Signaling and communication were significantly higher than the expected proportion in all three categories of repeats (p < 0.0001, type I, II and III). On the other hand, the proportion of genes with (TG/CA)n repeats of Immune and related functions and Information were significantly lower than expected proportion in all three categories: Immune and related functions (p < 0.0001, type I, II and III), Information (p < 0.0001, type I and II, p < 0.004, type III). The proportion of genes with type III repeats was marginally lower than the expected proportion in Metabolism class (p < 0.01) and marginally higher than the expected proportion in Structure and motility class (p < 0.02). The proportion of genes with repeats in the three categories was not significantly different from the expected value in the class of Cell cycle. These observations show that repeats of all types are positively associated with the genes of Signaling and communication whereas they are negatively associated with the genes of Immune and related functions and Information.
Figure 5.
Distribution of proportion of genes with three types of (TG/CA)n repeats in the six functional classes.
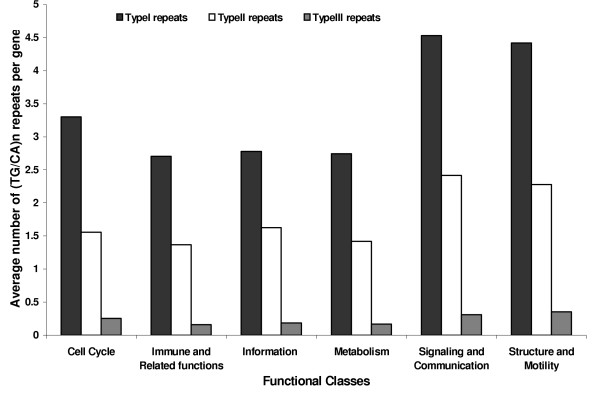
The distribution of average number of (TG/CA)n repeats per gene in the three categories in the six functional classes is displayed in Figure 6. Comparison of the average number of repeats per gene in the three categories with the global distribution pattern revealed that in most cases the observed number was significantly lower than the expected value, except for the genes belonging to Signaling and communication and Structure and motility, which had significantly higher average number of repeats per gene than the expected value (p < 0.0004 in all three categories, both classes). The average number of type III repeats per gene in the class of Cell cycle was not significantly different from the expected value. These observations show that the repeat densities were higher in the genes belonging to Signaling and communication and Structure and motility classes whereas, the genes belonging to other classes had lower repeat densities.
Figure 6.
Distribution of the densities of three types of (TG/CA)n repeats in the genes of six functional classes.
Large gene families
As a special case of this study, we examined the distribution of (TG/CA)n repeats in the top 2% large families (27). The family sizes of this category varied widely from 32 to 223 members. Functional classification of these large families revealed the following distribution: Immune and related functions (9), Signaling and communication (8), Information (6), Metabolism (2), Structure and motility (1) and Cell cycle (1).
The proportion of genes with (TG/CA)n repeats in large families is displayed in Table 1. Comparison with the global distribution showed that the proportion of genes with repeats was significantly higher than expected value in the Signaling and communication and Structure and motility classes (p < 0.0001, Binomial test). There was no significant difference between the observed and the expected proportion of genes with repeats in the class of Metabolism. In the remaining classes, the proportion of genes with repeats was significantly lower than the expected value (p < 0.0001, Binomial test). As observed with all gene families, a linear relationship was observed between gene lengths and proportion of genes with (TG/CA)n repeats (correlation coefficient R = 0.79, p < 0.0001).
Table 1.
Distribution of (TG/CA)n repeats in large gene families
| Functional class and gene families | Chromosomal Distributiona | Average Gene Length (kb)b | Genes in the family | Proportion of Genes with (TG/CA)n repeats | Number of Genes with (TG/CA)n repeats in three categories | Average number of repeats per gene in three categories | ||||
| Type I | Type II | Type III | Type I | Type II | Type III | |||||
| Cell cycle classc(1) | ||||||||||
| Histone proteins family | Dispersed | 3.55 | 76 | 7.9 | 4 | 4 | 0 | 1.7 | 0.7 | 0 |
| Immune and related functions class (9) | ||||||||||
| Interleukins | Dispersed | 14.67 | 43 | 32.6 | 9 | 6 | 2 | 1.3 | 0.9 | 0.1 |
| Serine (or cysteine) proteinase inhibitor family | Dispersed | 17.87 | 32 | 50 | 12 | 9 | 2 | 1.4 | 0.8 | 0.2 |
| Tumor necrosis factor (ligand) superfamily | Dispersed | 23.49 | 38 | 63.2 | 21 | 14 | 1 | 1.9 | 0.9 | 0.1 |
| CD antigens | Dispersed | 26.96 | 54 | 46.3 | 20 | 11 | 3 | 1.9 | 1.3 | 0.3 |
| Immunoglobulin heavy chains | Intrachromosomal | 0.38 | 162 | 0.6 | 1 | 0 | 0 | 1 | 0 | 0 |
| Immunoglobulin kappa chains | Intrachromosomal | 0.55 | 73 | 5.5 | 1 | 3 | 0 | 0.3 | 3 | 0 |
| Immunoglobulin lambda chains | Intrachromosomal | 0.35 | 88 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Interleukin receptors family | Dispersed | 29.77 | 32 | 59.4 | 17 | 10 | 0 | 3.4 | 0.9 | 0 |
| T cell receptor beta chains | 84 Intrachromosomal, 9 Dispersed | 0.42 | 94 | 9.6 | 5 | 3 | 1 | 0.8 | 0.3 | 0.1 |
| Information class (6) | ||||||||||
| Homeo box | Dispersed | 5.48 | 40 | 25 | 6 | 6 | 1 | 1.2 | 0.9 | 0.2 |
| Eukaryotic translation initiation factor | Dispersed | 36.54 | 33 | 45.5 | 13 | 10 | 3 | 2.1 | 0.9 | 0.2 |
| Zinc finger protein family | Dispersed | 30.33 | 200 | 42.5 | 63 | 44 | 6 | 2.2 | 1.1 | 0.1 |
| DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptides | Dispersed | 43.44 | 32 | 62.5 | 17 | 8 | 2 | 1.5 | 0.6 | 0.1 |
| Ribosomal protein genes | Dispersed | 4.94 | 96 | 6.3 | 6 | 0 | 0 | 1 | 0 | 0 |
| Mitochondrial ribosomal protein genes | Dispersed | 16.92 | 74 | 23 | 14 | 12 | 1 | 1.8 | 0.9 | 0.1 |
| Metabolism class (2) | ||||||||||
| Cytochrome P450 superfamily | Dispersed | 31.34 | 45 | 46.7 | 15 | 11 | 2 | 1.9 | 1 | 0.1 |
| Proteasome subunit genes | Dispersed | 25.64 | 40 | 32.5 | 11 | 8 | 0 | 1.5 | 0.8 | 0 |
| Signaling and Communication class (8) | ||||||||||
| G protein-coupled receptor family | Dispersed | 24.71 | 98 | 33.7 | 26 | 19 | 6 | 2.3 | 1.5 | 0.2 |
| Tripartite motif-containing family | Dispersed | 29.29 | 40 | 60 | 19 | 12 | 2 | 1.6 | 0.8 | 0.1 |
| Solute carrier family | Dispersed | 59.19 | 223 | 62.8 | 134 | 87 | 22 | 2.9 | 1.5 | 0.2 |
| RAS oncogene family | Dispersed | 39.92 | 60 | 65 | 38 | 17 | 4 | 1.8 | 0.7 | 0.1 |
| ATP-binding cassette transporters gene family | Dispersed | 73.85 | 44 | 68.2 | 29 | 24 | 4 | 3.6 | 2.5 | 0.3 |
| Guanine nucleotide binding protein (G protein) polypeptide genes | Dispersed | 58.83 | 32 | 59.4 | 18 | 12 | 5 | 3.3 | 1.9 | 0.4 |
| Potassium voltage-gated channel genes | Dispersed | 104.95 | 38 | 57.9 | 17 | 16 | 6 | 8.6 | 4.4 | 0.5 |
| Protein phosphatase subunit genes | Dispersed | 65.62 | 59 | 57.6 | 27 | 22 | 7 | 2.9 | 1.8 | 0.3 |
| Structure and motility class (1) | ||||||||||
| Collagen family | Dispersed | 132.83 | 37 | 86.5 | 29 | 23 | 10 | 5.7 | 2.3 | 0.4 |
a: Chromosomal distribution of the members of gene families. 'Dispersed' indicates that members are distributed on different chromosomes.
b: average gene length (in kb) for each gene family.
c:Numbers in parentheses show the number of large sized gene families in each functional class.
The large Collagen gene family belonging to the class of Structure and motility had the highest proportion of genes containing repeats (86.5%). In order to analyze this further, we examined the sequence conservation of the region flanking 200 bases upstream and downstream in addition to the repeats by comparing the human sequence with the available genome sequence of Chimpanzee (Pan troglodytes), a nearest ancestor to human [46]. We observed, that of the 268 sequence segments including repeats from human Collagen genes, 244 were conserved with greater than 92% identity in the chimpanzee. Of these 244 repeats in human Collagen genes, 73 repeats were identical in length, 142 repeats displayed length polymorphism in the chimpanzee, 27 repeats had point mutations and in 2 cases there were no repeats in the corresponding segments in the chimpanzee. These observations show that both human and chimpanzee Collagen genes have high repeat content, high conservation of position and flanking sequences of the repeats. However, majority of repeats exhibited length polymorphisms, which is consistent with their characteristic property [6].
Discussion
The inverse relationship between the number of gene families and their corresponding sizes, resulting in a large number of small sized gene families, suggests that several duplicated copies may have been lost during the first round of genome duplication itself, considering the hypothesis of two rounds of genome duplication in vertebrate evolution [1,23,24]. The non-uniform distribution of the number of gene families across the six functional classes suggests that widespread gene duplication across gene families spanning a wide range of functions may have been less productive in attaining higher levels of complexity. An alternate course involving large amount of duplications followed by divergence producing a wide range of functions in selected classes might have been favorable. The support for the latter hypothesis emerges from the fact that large sized gene families, inherently low in number, mainly belong to Immune and related functions (required to tackle a wide range of infections), Signaling and communication (required to respond to diverse environmental stimuli) and Information class (required to implement complex molecular processes through supramolecular assemblies or organelles). A few members of large sized gene families of Metabolism class function in bioenergetics and xenobiotic metabolism and of Cell cycle class function in packaging of nuclear DNA. Similarly the large Collagen gene family of Structure and motility class offers a useful repertoire for the formation of multiple tissues [25].
It is apparent that short repeats are abundant in human genes and long repeats are rare. Our findings are consistent with the observations by Whittaker et al. (2003), who showed that longer repeats are more likely to contract than expand [47]. Accordingly, contraction of long repeats in time would result in accumulation of higher number of short repeats.
Of the six functional classes, the Signaling and communication class was the richest in repeats including the proportion of genes with repeats and repeat densities. Many of the genes belonging to this class function at the interface between the body and its environment that appears to be a distinct feature of eukaryotes [28] to confer species-specific advantages [24,41]. The positive association of (TG/CA)n repeats associated with genes of this class strongly argues for a positive temporal regulatory role that could provide for variations in gene expression to complement the enormous diversity characteristic of this class. Compared to this class, the anciently evolved gene families of Information and Cell cycle are poor in repeats. Considering the fact that these genes are highly conserved [28-30] and are involved in implementing the molecular processes acting at the core of cellular physiology, these observations suggest that repeats are negatively associated with these genes to avoid unpredictable consequences for the normal functioning of the cell.
Another argument in favor of these inferences stems from the linear relationship between the average gene length of gene families belonging to the respective functional classes and the proportion of genes with repeats in these classes. The average length of genes belonging to Information class was short and this factor aids in obtaining high levels of expression of these genes [29]. This requirement, however, generates a space constraint to accommodate additional elements. This situation contrasts to that of genes of Signaling and communication class with higher average gene length offering more space for accommodating other regulatory elements. The analysis of Collagen gene family belonging to large sized families presents itself as an interesting case. Most of the members of this family have (TG/CA)n repeats. Sequence comparisons of repeat containing regions of human Collagen genes with the nearest ancestor to humans, the Chimpanzee, revealed that although there is high conservation in terms of content and position of repeats, majority of repeats were polymorphic, which is consistent with their characteristic property [6]. Among repeats that displayed polymorphism between human and Chimpanzee, nearly equal proportions of human repeats were either contracted or expanded in Chimpanzee. These results are also consistent with the Whittaker's model [47].
Strikingly, the genes of Immune and related functions class are poor in (TG/CA)n repeats in general and in type III repeats in particular. A characteristic trend of this class is to have large sized families with their genes arranged juxtaposed on the same chromosomal locations. This arrangement increases the possibility of these gene families to display more uniform sequence characteristics [31]. Further, these genes have the smallest average gene length indicating a compact arrangement, which is likely to act as a space constraint in the accommodation of (TG/CA)n repeats. In addition, the negative association of type III (TG/CA)n repeats in these genes may have a directional role. The immunoglobulin genes use the 7 bp and 9 bp repeats for generation of variants through VDJ recombination [33]. Accommodation of type III (TG/CA)n repeats (n ≥ 23) might introduce variations in this process and could result in loss of directional recombination essential to generate diversity in immunoglobulins and T cell receptor chains in an ordered manner.
Conclusion
The (TG/CA)n repeat distribution pattern observed in human gene families is consistent with Whittaker's model of repeat expansion and contraction. It appears that multiple factors including gene length, function and directionality of recombination processes steered the observed selective patterns of distribution of (TG/CA)n repeats in human gene families.
Methods
Sequence retrieval and mapping of (TG/CA)n repeats
Sequences of 35,114 human genes (build number 33) were retrieved from LocusLink http://www.ncbi.nlm.nih.gov/LocusLink/[43] using a JavaScript program. A sum of 192 genes could not be retrieved because of either inaccessibility to the LocusLink page or absence of the link for retrieving the gene sequence. A gene in this analysis is considered as the nucleotide sequence from the start of first exon to the end of last exon. If alternate splicing was reported, the gene length considered was the start of first exon to the last known exon including all alternatively spliced products for that gene.
Perl scripts, 'SimRep' and 'RepGene' were written for the identification and mapping of perfect intragenic (TG/CA)n repeats of length n ≥ 6 units in genes [17]. Throughout this work we have used n ≥ 6 units as the minimum cut-off to identify (TG/CA)n repeats. All repeats were scored in the intragenic region (exons and introns only).
Categorization of (TG/CA)n repeats
We grouped (TG/CA)n repeats into three categories (types I, II and III), according to their length and biological properties. Type I (TG/CA)n repeats, in the range 6 ≤ n <12 units, are short repeats based on the observation that a repeat length of 8 units (n = 8) is minimum to be likely polymorphic [34,35]. Type II (TG/CA)n repeats comprise of 12 ≤ n < 23 units and is based on the observation that more than 93% of the (CA)n repeats of n ≥ 12 units are polymorphic [6]. Further, repeats of this length have also been shown to have preferential binding to nuclear factors compared to short repeats [36] and can also stimulate mRNA splicing [21,22]. Type III repeats consist of relatively long reiterations of (TG/CA)n (n ≥ 23 units) and have propensity to adopt structures such as Z DNA [8,9,37]. Other studies have shown that (TG/CA)n repeats of length greater than 22.5 units can stimulate recombination [18-20].
Clustering of genes into gene families
Functional roles of a large number of human genes are not well known. Presently, these genes are assigned hypothetical annotations. Genes labeled as 'LOC', 'DFKZP', 'FLJ', 'HSPC', 'HSU', 'HT', 'KIAA', 'ORF', 'hypothetical', 'PRO' and 'pseudogenes' without clear functional details were filtered out. A total of 22,688 genes were removed in this filtering exercise. Out of the remaining 12,426 genes, a total of 8,778 genes (25% of total) were clustered into gene families based on their gene root symbols as defined in the guidelines of Human Gene Nomenclature Committee (2002) [4]. The remaining 3,648 genes could not be clustered into gene families and are solitary.
The HGNC guidelines consider sequence and functional similarity of proteins encoded by genes while grouping them into gene families [4,38,39]. A root symbol signifies a gene family. The family members are designated by Arabic numerals placed immediately after the gene root symbol, for example GPR1, GPR2, GPR3 for genes of the G protein-coupled receptor family. A Perl script namely Clustergene was written to cluster 8,778 human genes into 1,556 gene families. The Perl script called ChromoCluster was written to report gene families located on the same chromosome. Subsequently these gene families were classified into the six functional classes as described below.
Functional Classification of gene families for comparative analysis
The gene families were classified into six functional classes namely, 'Information', 'Cell cycle', 'Metabolism', 'Signaling and communication', 'Immune and related functions' and 'Structure and motility' based on the scheme defined by Adams et al. [40]. We combined the functional classes of replication, transcription, RNA processing and translation into 'Information' class based on Andrade et al. [41].
'Cell cycle' includes cell cycle, apoptosis, chromosomal structure and DNA repair; 'Immune and related functions' includes immunology, homeostasis, carrier proteins/membrane transport and stress response; 'Information' includes protein synthesis, translation factors, ribosomal proteins, post-translational modification/targeting, protein degradation, tRNA synthesis/metabolism, RNA synthesis, transcription factors, RNA polymerase, RNA processing, RNA degradation, DNA synthesis/replication and DNA repair; 'Metabolism' includes amino acids, nucleotides, sugars, lipids, cofactors, protein modification, energy and carrier proteins/membrane transport; 'Signaling and communication' includes receptors, hormone/growth factors, intracellular transducers, effectors/modulators, metabolism, cell adhesion and channels/transport proteins; 'Structure and motility' includes cytoskeletal, microtubule-associated proteins/motors and extracellular matrix.
Assignment of gene families to each of the functional classes was carried out according to their annotations in Gene Ontology [42] and LocusLink [43] databases. Out of the total 1,556 gene families, 1,317 could be classified into any of the six functional classes. The remaining 239 families could not be classified unambiguously due to limited information on gene function. Subsequent analysis, with respect to functional classification and distribution of (TG/CA)n repeats, presented here is from 1,317 gene families comprising of 7,928 genes.
Alignment of human (TG/CA)n repeats and flanking sequences with Chimpanzee genome sequence
The repeats present in human Collagen genes were aligned with Chimpanzee (Pan troglodytes) genome by using 'BLAT' software available at UCSC Genome Bioinformatics Site http://www.genome.ucsc.edu/cgi-bin/hgBlat[49]. Nucleotide segments including the repeats and containing 200 nucleotides upstream of the start and 200 nucleotides downstream from the end of each of the (TG/CA)n repeat were extracted for human Collagen genes [48]. These segments were aligned with the Chimpanzee genome (Build 1, version 1, Nov 2003) using BLAT. Only those segments that showed more than 92% identity were noted as conserved.
Statistical methods
Significance of the differences between the proportions of genes containing repeats and repeats densities in the six functional classes compared with global distribution was tested using Binomial proportions test. The observed proportion in each class was tested against the expected proportion, which was computed assuming no preference with respect to function. Correlation coefficient (R) was computed to examine the relationship between average gene length of gene families belonging to a functional class and the proportion of genes with (TG/CA)n repeats in the corresponding functional classes. The 'Interactive Statistical Calculation Pages' website http://members.aol.com/johnp71/javastat.html was used to perform the statistical tests.
Authors' contributions
VKS conceived of the idea, developed algorithms in Perl, carried out the analysis and wrote the manuscript. SKB gave scientific suggestions for improving the quality of the work and participated in manuscript preparation. SR is the group leader, gave scientific suggestions, helped in the statistical analysis, critical examination, presentation, writing and manuscript preparation.
Acknowledgments
Acknowledgements
VKS is a recipient of Senior Research Fellowship from CSIR. We thank Pankaj Bhatnagar for help in writing programs and the anonymous reviewers for their insightful comments. SKB and SR thank CSIR for funding support in the form of a grant (CMM0017) Task Force on "In Silico Biology for Drug target development".
Contributor Information
Vineet K Sharma, Email: vsharma@igib.res.in.
Samir K Brahmachari, Email: skb@igib.res.in.
Srinivasan Ramachandran, Email: ramu@igib.res.in.
References
- Meyer A, Schart M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Current Opinion in Cell Biology. 1999;11:699–704. doi: 10.1016/S0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Wagner A. Selection and gene duplication: a view from the genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-reviews1012. reviews1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RC, Stansfield WD. A Dictionary of Genetics. Oxford University Press; 1990. [Google Scholar]
- Wain HM, Bruford EA, Lovering RC, Lush MJ, Wright MW, Povey S. Guidelines for human gene nomenclature. Genomics. 2002;79:464–470. doi: 10.1006/geno.2002.6748. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–4. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- Brahmachari SK, Meera G, Sarkar PS, Balagurumoorthy P, Tripathi J, Raghavan S, Shaligram U, Pataskar S. Simple repetitive sequences in the genome: structure and functional significance. Electrophoresis. 1995;16:1705–14. doi: 10.1002/elps.11501601283. [DOI] [PubMed] [Google Scholar]
- Nordheim A, Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci. 1983;80:1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meera G, Ramesh N, Brahmachari SK. Zintrons in rat α-lactalbuman gene. FEBS Lett. 1989;251:245–249. doi: 10.1016/0014-5793(89)81463-2. [DOI] [PubMed] [Google Scholar]
- Tripathi J, Brahmachari SK. Distribution of simple repetitive (TG/CA)n and (CT/AG)n sequences in human and rodent genomes. J Biomol Struct Dyn. 1991;9:387–97. doi: 10.1080/07391102.1991.10507919. [DOI] [PubMed] [Google Scholar]
- Naylor LH, Clark EM. d(TG)n.d(CA)n sequences upstream of the rat prolactin gene form Z-DNA and inhibit gene transcription. Nucleic Acids Res. 1990;18:1595–1601. doi: 10.1093/nar/18.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimajiri S, Arima N, Tanimoto A, Murata Y, Hamada T, Wang KY, Sasaguri Y. Shortened microsatellite d(CA)21 sequence down-regulates promoter activity of matrix metalloproteinase 9 gene. FEBS Lett. 1999;455:70–4. doi: 10.1016/S0014-5793(99)00863-7. [DOI] [PubMed] [Google Scholar]
- Pravica V, Asderakis A, Perrey C, Hajeer A, Sinnott PJ, Hutchinson IV. In vitro production of IFN-gamma correlates with CA repeat polymorphism in the human IFN-gamma gene. Eur J Immunogenet. 1999;26:1–3. doi: 10.1046/j.1365-2370.1999.00122.x. [DOI] [PubMed] [Google Scholar]
- Gebhardt F, Zanker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem. 1999;274:13176–13180. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- Agarwal AK, Giacchetti G, Lavery G, Nikkila H, Palermo M, Ricketts M, McTernan C, Bianchi G, Manunta P, Strazzullo P, Mantero F, White PC, Stewart PM. CA-Repeat polymorphism in intron 1 of HSD11B2: effects on gene expression and salt sensitivity. Hypertension. 2000;36:187–94. doi: 10.1161/01.hyp.36.2.187. [DOI] [PubMed] [Google Scholar]
- Streelman JT, Kocher TD. Microsatellite variation associated with prolactin expression and growth of salt-challenged tilapia. Physiol Genomics. 2002;9:1–4. doi: 10.1152/physiolgenomics.00105.2001. [DOI] [PubMed] [Google Scholar]
- Sharma VK, B-Rao C, Sharma A, Brahmachari SK, Ramachandran S. (TG/CA)n repeats in human housekeeping genes. J Biomol Struct Dyn. 2003;21:303–10. doi: 10.1080/07391102.2003.10506926. [DOI] [PubMed] [Google Scholar]
- Tracy RB, Baumohl JK, Kowalczykowski SC. The preference for GT-rich DNA by the yeast Rad51 protein defines a set of universal pairing sequences. Genes Dev. 1997;11:3423–31. doi: 10.1101/gad.11.24.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutreix M. (GT)n repetitive tracts affect several stages of RecA-promoted recombination. J Mol Biol. 1997;273:105–13. doi: 10.1006/jmbi.1997.1293. [DOI] [PubMed] [Google Scholar]
- Majewski J, Ott J. GT Repeats are associated with recombination on human chromosome 22. Genome Research. 2000;10:1108–14. doi: 10.1101/gr.10.8.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini N. A polymorphic GT repeat from the human cardiac Na + Ca2+ exchanger intron 2 activates splicing. Eur J Biochem. 2001;268:1076–83. doi: 10.1046/j.1432-1327.2001.01974.x. [DOI] [PubMed] [Google Scholar]
- Hui J, Stangl K, Lane WS, Bindereif A. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat Struct Biol. 2003;10:33–7. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- Hinegardner R. Evolution of cellular DNA content in teleost fishes. Am Nat. 1968;102:517–523. doi: 10.1086/282564. [DOI] [Google Scholar]
- Fyodor A, Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-research0008. RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelse K, Poschl E, Aigner T. Collagens – structure, function, and biosynthesis. Adv Drug Deliv Rev. 2003;55:1531–46. doi: 10.1016/j.addr.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Hardison RC. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/S0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Tagle DA, Koop BF, Goodman M, Slightom JL, Hess DL, Jones RT. Embryonic epsilon and gamma globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J Mol Biol. 1988;203:439–455. doi: 10.1016/0022-2836(88)90011-3. [DOI] [PubMed] [Google Scholar]
- Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci USA. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA. Selection for short introns in highly expressed genes. Nat Genet. 2002;31:415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- Dotan I, Ziv E, Dafni N, Beckman JS, McCann RO, Glover CV, Canaani D. Functional Conservation between the Human, Nematode, and Yeast CK2 Cell Cycle Genes. Biochem Biophys Res Commun. 2001;288:603–609. doi: 10.1006/bbrc.2001.5804. [DOI] [PubMed] [Google Scholar]
- Dover GA. Evolution of genetic redundancy for advanced players. Curr Opin Genet Dev. 1993;3:902–10. doi: 10.1016/0959-437X(93)90012-E. [DOI] [PubMed] [Google Scholar]
- Ramachandran GN, Sasisekharan V. Structure of collagen. Nature. 1961;190:1004–5. doi: 10.1038/1901004a0. [DOI] [PubMed] [Google Scholar]
- Alt FW, Oltz EM, Young F, Gorman J, Taccioli G, Chen J. VDJ recombination. Immunol Today. 1992;13:306–14. doi: 10.1016/0167-5699(92)90043-7. [DOI] [PubMed] [Google Scholar]
- Fondon JW, 3rd, Mele GM, Brezinschek RI, Cummings D, Pande A, Wren J, O'Brien KM, Kupfer KC, Wei MH, Lerman M, Minna JD, Garner HR. Computerized polymorphic marker identification: experimental validation and a predicted human polymorphism catalog. Proc Natl Acad Sci. 1998;95:7514–7519. doi: 10.1073/pnas.95.13.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Wray GA. Abundant raw material for Cis – regulatory evolution in humans. Mol Biol Evol. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- Epplen C, Santos EJ, Maueler W, van Helden P, Epplen JT. On simple repetitive DNA sequences and complex diseases. Electrophoresis. 1997;18:1577–85. doi: 10.1002/elps.1150180916. [DOI] [PubMed] [Google Scholar]
- Yagil G. Paranemic structures of DNA and their role in DNA unwinding. Crit Rev Biochem Mol Biol. 1991;26:475–559. doi: 10.3109/10409239109086791. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Owens IS, Burchell B, Bock KW, Bairoch A, Belanger A, Fournel-Gigleux S, Green M, Hum DW, Iyanagi T, Lancet D, Louisot P, Magdalou J, Chowdhury JR, Ritter JK, Schachter H, Tephly TR, Tipton KF, Nebert DW. The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics. 1997;7:255–69. doi: 10.1097/00008571-199708000-00001. [DOI] [PubMed] [Google Scholar]
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O, Sutton G, Blake JA, Brandon RC, Chiu M, Clayton RA, Cline RT, Cotton MD, Earle-Hughes J, Fine LD, FitzGerald LM, FitzHugh WM, Fritchman JL, Geoghagen NSM, Glodek A, Gnehm CL, Hanna MC, Hedblom E, Hinkle PS, Jr, Kelley JM, Klimek KM, Kelley JC, Liu L, Marmaros SM, Merrick JM, Moreno-Palanques RF, McDonald LA, Nguyen DT, Pellegrino SM, Phillips CA, Ryder SE, Scott JL, Saudek DM, Shirley R, Small KV, Spriggs TA, Utterbach TR, Weidman JF, Li Y, Barthlow R, Bednarik DP, Cao L, Cepeda MA, Coleman TA, Collins E, Dimke D, Feng P, Ferrie A, Fischer C, Hastings GA, He W, Hu J, Huddleston KA, Greene JM, Gruber J, Hudson P, Kim A, Kozak DL, Kunsch C, Ji H, Li H, Meissner PS, Olsen H, Raymond L, Wei Y, Wing J, Xu C, Yu G, Ruben SM, Dillon PJ, Fannon MR, Rosen CA, Haseltine WA, Fields C, Fraser CM, Venter JC. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377:3–174. [PubMed] [Google Scholar]
- Andrade MA, Ouzounis C, Sander C, Tamames J, Valencia A. Functional classes in the three domains of life. J Mol Evol. 1999;49:551–557. doi: 10.1007/pl00006576. [DOI] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R, Gene Ontology Consortium The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004:D258–61. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Edgar R, Federhen S, Helmberg W, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Suzek TO, Tatusova TA, Wagner L. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2004;32:D35–40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Rifkin SA, White KP, Li Wen-Hsiung. Duplicate genes increase gene expression diversity within and between species. Nature Genetics. 2004;36:577–579. doi: 10.1038/ng1355. [DOI] [PubMed] [Google Scholar]
- Thornton JW, DeSalle R. Gene family evolution and homology: Genomics Meets Phylogenetics. Annu Rev Genomics Hum Genet. 2000;1:41–73. doi: 10.1146/annurev.genom.1.1.41. [DOI] [PubMed] [Google Scholar]
- Check E. Geneticists study chimp-human divergence. Nature. 2004;428:242. doi: 10.1038/428242a. [DOI] [PubMed] [Google Scholar]
- Whittaker JC, Harbord RM, Boxall N, Mackay I, Dawson G, Sibly RM. Likelihood-based estimation of microsatellite mutation rates. Genetics. 2003;164:781–787. doi: 10.1093/genetics/164.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainudiin R, Durrett RT, Aquadro CF, Nielsen R. Microsatellite mutation models: insights from a comparison of humans and chimpanzees. Genetics. 2004;168:383–95. doi: 10.1534/genetics.103.022665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ, University of California Santa Cruz. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–4. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]