Abstract
A facile synthetic route for the 4′-thioribonucleoside building block 4′SN (N = U, C, A and G) with the ribose O4′ replaced by sulfur is presented. Conversion of l-lyxose to 1,5-di-O-acetyl-2,3-di-O-benzoyl-4-thio-d-ribofuranose was achieved via an efficient four-step synthesis with high yield. Conversion of the thiosugar into the four ribonucleoside phosphoramidite building blocks was accomplished with additional four steps in each case. Incorporation of 4′-thiocytidines into oligoribonucleotides improved the thermal stability of the corresponding duplexes by ∼1°C per modification, irrespective of whether the strand contained a single modification or a consecutive stretch of 4′SC residues. The gain in thermodynamic stability is comparable to that observed with oligoribonucleotides containing 2′-O-methylated residues. To establish potential conformational changes in RNA as a result of the 4′-thio modification and to better understand the origins of the observed stability changes, the crystal structure of the oligonucleotide 5′-r(CC4′SCCGGGG) was determined and analyzed using the previously solved structure of the native RNA octamer as a reference. The two 4′-thioriboses adopt conformations that are very similar to the C3′-endo pucker observed for the corresponding sugars in the native duplex. Subtle changes in the local geometry of the modified duplex are mostly due to the larger radius of sulfur compared to oxygen or appear to be lattice-induced. The significantly increased RNA affinity of 4′-thio-modified RNA relative to RNA, and the relatively minor conformational changes caused by the modification render this nucleic acid analog an interesting candidate for in vitro and in vivo applications, including use in RNA interference (RNAi), antisense, ribozyme, decoy and aptamer technologies.
INTRODUCTION
RNA affinity is a key criterion for assessing the potential utility of a particular nucleic acid modification for a variety of nucleic acid based applications, including use in RNA interference (RNAi), antisense, ribozyme, decoy and aptamer technologies. Enhancement of thermal stability of a duplex between a modified strand and its RNA complement relative to the corresponding unmodified duplex is desirable (1). Over the past years, numerous modifications were introduced, and for ∼200 of them, structural features were correlated with changes in the thermal stability of modified duplexes (2). The majority of modifications had a destabilizing effect and only few actually led to enhanced stability. Oligonucleotides that will serve as effective research reagents and therapeutic agents also require chemical modification in order to protect them against nuclease digestion. The origins of the enhanced resistance against nuclease degradation afforded by the first-generation phosphorothioate DNA (PS-DNA) and second-generation 2′-O-(3-amino-propyl) RNA (AP-RNA) have been studied using three-dimensional structures of exonuclease–oligonucleotide complexes [ref. (3) and (4), respectively]. Incorporation of chemically modified nucleosides at particular sites in hammerhead-type ribozymes can also lead to improved activity of the catalytic RNA (5,6). Moreover, in vitro and in vivo use of RNAi will most probably benefit significantly from the incorporation of chemically modified nucleosides into small interfering RNA (siRNA) (7,8).
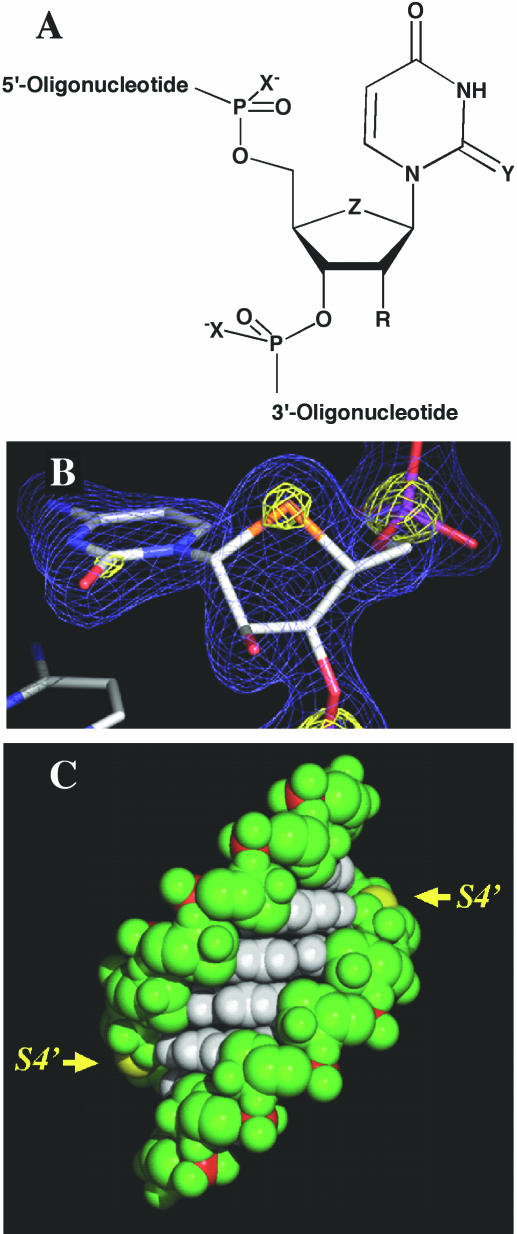
Substitution of particular oxygen atoms by sulfur is a common way to chemically modify both DNA and RNA (Figure 1A). Thus, PS-DNA, in which one of the non-bridging oxygen atoms in the DNA backbone is replaced by sulfur, constitutes the most widely used antisense modification (9,10). PS-DNA:RNA duplexes exhibit reduced thermal stability relative to the corresponding natural DNA–RNA hybrids (∼−0.7°C per PS modification). However, the increased nuclease resistance of PS-DNA relative to DNA rendered it an attractive antisense analog both for in vitro applications and as a therapeutic agent in the clinic (11,12). Replacement of the exocyclic keto oxygen 2 by sulfur in the thymine base was also reported (13). Incorporation of 2-thio-T into DNA strands increased the Tm of the modified DNA–RNA duplexes by 1.4°C on average per modification (2).
Figure 1.
(A) Selected sulfur analogs of DNA (R = H) and RNA (R = OH). X = S and R = H: PS-DNA; Y = S: 2-thio-U; and Z = S: 4′-thio-nucleic acid. (B) Final Fourier sum (2Fo-Fc) electron density around residue 4′SC3, drawn at the 1.3σ (blue) and 3.9σ levels (yellow). The sulfur atom is highlighted in yellow. (C) Van der Waals representation of the [r(CC4′SCGGGG)]2 duplex. Bases are colored white, the sugar-phosphate backbone is green and phosphorus atoms are red. The sulfur atoms of the 4′-thioribocytidines in the RNA duplex are colored yellow and their positions in the minor groove are highlighted by arrows.
The effects on duplex stability resulting from replacement of the sugar 4′-oxygen with sulfur were investigated for both DNA and RNA. Incorporation of 4′-thio-2′-deoxyribonucleosides into DNA strands reduces the thermal stability of the corresponding DNA–RNA hybrids by ∼1°C (14–16) and provides improved protection against endo- but not exo-nucleolytic degradation (14). Interestingly, the 4′-thio-DNA modification renders the RNA strand in modified DNA–RNA duplexes much more resistant to degradation by RNase H (16). DNA containing 4′-thio-2′-deoxy-C was found to inhibit methylation by a methyltransferase (17). An X-ray crystal structure of a B-DNA dodecamer duplex with two incorporated 4′-thio-Ts revealed changes in the conformation of the sugar-phosphate backbone at and adjacent to the modification sites (18). In addition, all four 4′-thio-2′-deoxyribose sugars adopted a C3′-exo conformation instead of C1′-exo, the one observed in the structure of the native dodecamer.
Incorporation of 4′-thio-U residues into RNA strands leads to enhanced thermal stability of duplexes between modified strands and RNA relative to the native counterparts (19,20). 4′-Thio-RNA binds more tightly to RNA than to DNA (19,20), consistent with the previous notion that RNA duplexes are more stable than RNA–DNA hybrids (21,22). The presence of sulfur at the 4′-position can be expected to change the pseudorotational equilibrium in the ribose moiety. For example, the gauche effect for [S-C-C-O] fragments is most probably weaker compared with the 4′-oxo counterparts because sulfur is less electronegative than oxygen (23). The S4′-C1′-N9/1 anomeric effect is weaker in pyrimidines than in purines (23), whereas the situation is reversed in the case of 4′-oxonucleosides (24). Overall, the S4′-C1′-N9/1 anomeric effect is weaker than the O4′-C1′-N9/N1 one. Similar to the effect of the 2′-hydroxyl group on the sugar conformation in 4′-oxonucleosides, where an electronegative 2′-substituent stabilizes a C3′-endo (N) type pucker, the N-type rotamer is preferred by 4′-thioribonucleosides compared with 4′-thio-2′-deoxyribonucleosides. For example, solution NMR experiments revealed a ΔΔH° of 3.8 kJ mol−1 between 4′-thio-C and 4′-thio-2′-deoxyribo-C in favor of the modified ribonucleoside (23). More recently, the relative overall order of thermal stabilities of duplexes was reported to be 4′-thio-RNA:4′-thio-RNA ≫ 4′-thio-RNA:RNA > RNA:RNA > RNA:DNA > 4′-thio-RNA:DNA (25). In addition to the changes observed at the thermodynamic level, 4′-thio-oligo-U and 4′-thio-modified RNA strands with mixed sequences were found to exhibit improved resistance to a variety of exo- and endonucleases (19,25,26). However, no details of the changes induced by the 4′-thio RNA modification at the structural level beyond Circular Dichroism experiments have been reported thus far.
The synthesis of 4′-thio nucleosides has attracted the attention of many groups' efforts over the past three decades, resulting in a wide array of synthetic approaches. The first synthetic approach for this class of compounds was by Reist et al. (27). Recent developments in the synthesis of 4′-thioribonucleosides have focused upon the synthesis of peracylated 4-thio-d-ribofuranose as a thiosugar precursor to glycosylation (20). This approach takes advantage of acyloxonium ion formation at C1 to give higher selectivity in generating the desired nucleoside β anomer product (28). However, overall synthetic yields have remained low. Very recently, the synthesis of all four 4′-thioribonucleoside phosphoramidite building blocks based on a new synthetic protocol using the Pummerer reaction was reported (25,29).
We have developed a synthetic scheme that offers considerable advantage in the overall yield for both the thiosugar and nucleoside analogs. In this contribution, we present a novel synthetic route for the generation of all four 4′-thioribonucleoside building blocks, which is a modification of Reist's approach (27). Based on this approach, oligoribonucleotides containing single and multiple 4′-thioribocytosines (4′SC) were prepared. We have measured the thermodynamic stability of such RNA oligonucleotides. In addition, an RNA octamer duplex carrying a single 4′SC residue per strand was crystallized and its structure determined at 1.93 Å resolution. The thermodynamic stability data are being analyzed on the basis of the insights gained from the crystal structure of the RNA duplex with incorporated 4′SC residues.
MATERIALS AND METHODS
General methods
NMR spectra were recorded on a Varian Gemini 400 spectrometer operating at 400.075 MHz for proton and 161.947 MHz for phosphorus. Chemical shifts in p.p.m. refer to trimethyl silane and H3PO4, respectively and are reported in parts per million (δ), and signals are expressed as s (singlet), d (doublet), dd (doublet of doublet), t (triplet), q (quartet) or m (multiplet). Analytical thin-layer chromatography (TLC) was performed with Whatman MK6F silica gel 60 Å F254 plates and column chromatography using Merck 0.040–0.063 mm silica gel 60.
Synthesis of the 4-thio-d-ribofuranose building block
Methyl 2,3-O-isopropylidine-α-l-lyxopyranoside (1)
A solution of l-lyxose (50 g, 0.33 mol) in anhydrous 0.5% methanolic hydrogen chloride (900 ml) was refluxed for 6 h. The reaction mixture was then cooled to room temperature, neutralized by addition of silver carbonate (48 g) and filtered through a Celite pad. Evaporation of the filtrate afforded crude methyl α-l-lyxopyranoside as a colorless syrup. To this syrup was added 2,2-dimethoxypropane (205 ml, 1.66 mol) followed by p-toluenesulfonic acid (1.25 g, 6.66 mmol). The suspension upon sonication gave a homogeneous solution that was stirred at room temperature for 2 h. The reaction mixture was then diluted with four volumes of dichloromethane and washed with saturated NaHCO3. The dichloromethane layer was dried (Na2SO4) and concentrated in vacuo. The residue was purified by silica gel column chromatography, eluted with 10–50% EtOAc in hexane to give 1 (54 g, 80% as an oil). 1H NMR (DMSO-d6): 5.16 (d, J4, OH = 5.4, 1H, 4-OH), 4.58 (s, 1H, H1), 3.91 (m, 2H, H2, H3), 3.56 (m, 1H, H4), 3.44 (dd, J5,4 = 4.3, J5,5′ = 11.6, 1H, H5), 3.36 (dd, J5′,4 = 7.8, J5′,5 = 11.6, 1H, H5′), 3.29 (s, 3H, OCH3), 1.40 (s, 3H, CH3), 1.27 (s, 3H, CH3).
Methyl 2,3-O-isopropylidine-4-S-benzoyl-β-d-ribopyranoside (2)
To a solution of 1 (52 g, 0.25 mol) in dichloromethane (1 l) was added 2,6-di-tert-butyl-4-methylpyridine (170 g, 0.8 mol) and stirred under argon. The resulting pale yellow solution was cooled to −10°C with an ethanol/ice bath and triflic anhydride (126 ml, 0.75 mol) was added dropwise via addition funnel. After 10 min of stirring, a white precipitate formed and stirring of the resulting slurry was continued at −10°C for an additional 45 min. The reaction mixture was diluted with three volumes of anhydrous toluene and filtered over a Celite bed. After washing the filter bed with anhydrous toluene, the resulting pink filtrate was evaporated in vacuo, co-evaporated three times with anhydrous toluene to afford a pink glass. The crude triflate material was resuspended in anhydrous DMF (1 l) and a solution of potassium thiobenzoate (135 g, 0.83 mol) in anhydrous DMF (500 ml) was added dropwise and stirred under argon. After stirring at room temperature for 2 h, the clear red reaction mixture was diluted with two volumes of dichloromethane and then partitioned with water. The dichloromethane layer was dried (Na2SO4) and concentrated in vacuo. The residue was purified by silica gel column chromatography, eluted with 3–5% EtOAc in hexane to yield 2 (43.0 g, 56% as a clear yellowish oil). 1H NMR (CDCl3): 7.99–7.43 (m, 5H, benzoyl), 4.53 (dd, J3,2 = 5.2, J3,4 = 6.0, 1H, H3), 4.46 (d, J1,2 = 5.8, 1H, H1), 4.39 (m, 1H, H4), 4.08 (dd, J2,1 = 5.8, J2,3 = 5.2, 1H, H2), 3.90 (dd, J5,4 = 5.6, J5,5′ = 10.8, 1H, H5), 3.73 (dd, J5′,4 = 11.8, J5′,5 = 10.8, 1H, H5′), 3.53 (s, 3H, OCH3), 1.56 (s, 3H, CH3), 1.39 (s, 3H, CH3).
Methyl 2,3-di-O-benzoyl-4-S-benzoyl-β-d-ribopyranoside (3)
A −20°C solution of trifluoroacetic acid, dioxane and water (2:1:1, 500 ml) was added to 2 (43.0 g, 0.139 mol) at −10°C with stirring. The yellow reaction mixture was maintained at 4°C for 48 h, diluted with dichloromethane (1 l) and neutralized with saturated aqueous NaHCO3 followed by solid NaHCO3 (200 g). The dichloromethane layer was dried (Na2SO4) and concentrated in vacuo, and was co-evaporated three times with anhydrous pyridine. The residue was resuspended in anhydrous pyridine (800 ml) and cooled to 0°C with stirring under argon. Benzoyl chloride (35.5 ml, 0.31 mol) was added dropwise using an addition funnel to the stirred reaction mixture, the reaction was warmed to room temperature and after 1 h, quenched with anhydrous EtOH (20 ml). Pyridine was removed in vacuo and the residue partitioned between dichloromethane and water. The dichloromethane layer was dried (Na2SO4) and concentrated in vacuo and the residue was purified by silica gel column chromatography, eluted with 3–5% EtOAc in hexane to yield 3 (62.3 g, 94% as an off-white foam). 1H NMR (CDCl3): 8.18–7.34 (m, 15H, benzoyl), 5.98 (dd, J3,2 = 3.2, J3,4 = 4.1, 1H, H3), 5.44 (dd, J2,1 = 3.9, J2,3 = 3.2, 1H, H2), 4.93 (d, J1,2 = 3.9, 1H, H1), 4.53 (dd, J4,3 = 4.1, J4,5 = 3.3, J4,5′ = 5.0, 1H, H4), 4.35 (dd, J5,4 = 3.3, J5,5′ = 12.5, 1H, H5), 3.96 (dd, J5′,4 = 5.0, J5′,5 = 12.5, 1H, H5′), 3.51 (s, 3H, OCH3).
1,5-Di-O-acetyl-2,3-di-O-benzoyl-4-thio-d-ribofuranose (4)
An ice-cold solution of AcOH, Ac2O and H2SO4 (400, 400 and 40 ml, respectively) was added to 3 (62.0 g, 0.130 mol) at −10°C. The light yellow solution was left at 4°C for 18 h. The acid in the reaction mixture was decomposed by adding sodium acetate (120 g). The reaction mixture was reduced to ∼50% volume in vacuo and was subsequently co-evaporated with toluene three times to ∼50% volume. After diluting with dichloromethane (1.5 l), the solution was treated with saturated aqueous NaHCO3, followed by solid NaHCO3 until neutral. The dichloromethane layer was dried (Na2SO4) and concentrated in vacuo, and the residue was purified by silica gel column chromatography, eluted with 5–40% EtOAc in hexane to afford 4 (51.0 g, quantitative as a pale yellow glass). 1H NMR (CDCl3): 8.03–7.36 (m, 10H, aromatic), 6.04 (d, J1,2 = 1.7, 1H, H1), 5.94 (dd, J2,3 = 3.5, J2,1 = 1.7, 1H, H2), 5.78 (dd, J3,4 = 8.2, J3,2 = 3.5, 1H, H3), 4.46 (dd, J5,4 = 6.0, J5,5′ = 11.0, 1H, H5), 4.28 (dd, J5′,4 = 6.6, J5′,5 = 11.0, 1H, H5′), 4.09 (m, J4,3 = 8.2, J4,5 = 6.0, J4,5′ = 6.6, 1H, H4), 2.16 (s, 3H, OAc), 1.98 (s, 3H, OAc).
Synthesis of 4′-thioribonucleoside phosphoramidite units
1-[2,3-Di-O-benzoyl-5-O-acetyl-4-thio-β-d-ribofuranosyl]uracil (5)
To a suspension of uracil (0.69 g, 6.12 mmol) in anhydrous acetonitrile (50 ml), N,O-Bis(trimethylsilyl)acetamide (BSA) (5.04 ml, 20.4 mmol) was added, and the reaction mixture was refluxed for 1 h. After cooling to room temperature, a solution of 4 (2.0 g, 5.10 mmol) in anhydrous acetonitrile (10 ml) was added, followed by addition of SnCl4 (0.9 ml, 7.65 mmol). The reaction mixture was heated to 40°C for 5 h. An additional portion of SnCl4 (0.3 ml, 2.55 mmol) was added and the reaction continued at 40°C for an additional 5 h. The reaction mixture was diluted with two volumes of dichloromethane and washed with saturated aqueous NaHCO3. The dichloromethane layer was dried (Na2SO4) and concentrated in vacuo, and the residue was purified by silica gel column chromatography, eluted with 30–60% EtOAc in hexane to give 5 (1.75 g, 75% as a white foam). 1H NMR (CDCl3): 8.0 (br s, 1H, NH), 7.88 (d, J6,5 = 8.2, 1H, H6), 8.06–7.36 (m, 10H, benzoyl), 6.68 (d, J1′,2′ = 7.1, 1H, H1′), 5.89 (dd, J5,6 = 8.3, J5,NH = 1.8, 1H, H5), 5.82 (m, 2H, H2′, H3′), 4.55 (dd, J5′,4′ = 5.5, J5′,5′ = 11.0, 1H, H5′), 4.37 (dd, J5′′,4′ = 5.8, J5′,5′′ = 11.0, 1H, H5′′), 3.92 (m, 1H, H4′), 2.20 (s, 3H, OAc).
1-[5-O-Dimethoxytrityl-4-thio-β-d-ribofuranosyl]uracil (6)
To a −10°C solution of 5 (3.3 g, 7.17 mmol) in pyridine (44 ml) and methanol (9 ml) was added a pre-cooled solution of 2 M NaOH (13 ml) with stirring. The reaction mixture was stirred at −10°C for 20 min before being neutralized to pH 7 with Dowex 50W×4 pyr+ resin. The resin was filtered off and the filter bed washed with pyridine/water and the filtrate evaporated in vacuo to afford an off-white foam which was co-evaporated repeatedly with anhydrous pyridine. The crude foam was resuspended in anhydrous pyridine (100 ml) under an argon atmosphere followed by the addition of dimethoxytrityl chloride (2.88 g, 8.52 mmol). The reaction mixture was stirred at room temperature for 48 h before being quenched with anhydrous EtOH (10 ml). Pyridine was removed in vacuo and the residue partitioned between dichloromethane and saturated aqueous NaHCO3. The dichloromethane layer was dried (Na2SO4) and concentrated in vacuo and the residue was purified by silica gel column chromatography, eluted with 3–10% EtOH in dichloromethane to yield 6 (2.0 g, 54.4% as an off-white foam). 1H NMR (DMSO-d6): 10.93 (s, 1H, NH), 7.69 (d, J6,5 = 8.2, 1H, H6), 7.39–6.88 (m, 15H, trityl), 5.82 (d, J1′,2′ = 5.6, 1H, H1′), 5.54 (d, JOH,2′ = 5.0, 1H, 2′OH), 5.50 (d, J5,6 = 8.2, 1H, H5), 5.27 (d, JOH,3′ = 4.5, 1H, 3′OH), 3.99 (m, 2H, H2′, H3′), 3.72 (s, 6H, 2×OMe), 3.37–3.22 (m, 3H, H4′, H5′, H5′′).
1-[2-O-tert-Butyldimethylsilyl-5-O-dimethoxytrityl-4-thio-β-d-ribofuranosyl]uracil (7)
To a solution of 6 (2.0 g, 3.9 mmol) in anhydrous THF (20 ml) at room temperature was added silver nitrate (1.72 g, 10.14 mmol) with stirring under argon. After 15 min, pyridine (0.63 ml, 7.8 mmol) was added followed by tert-butyldimethylsilyl chloride (TBDMSCl) (0.77 g, 5.07 mmol) and the reaction mixture was stirred in the dark for 20 h. Silver salts were removed by filtration over a Celite pad and the filter bed was washed with THF. The filtrate was then evaporated in vacuo and the resulting foam partitioned between dichloromethane and saturated aqueous NaHCO3. The dichloromethane layer was dried (Na2SO4) and concentrated in vacuo and the residue was purified by silica gel column chromatography, eluted with 30–70% EtOAc in hexane to give 7 (0.93 g, 38% as a white foam) in addition to the 3-O-TBDMS derivative (0.51 g, 20.8%). 2-O-TBDMS 1H NMR (DMSO-d6): 11.37 (s, 1H, NH), 7.82 (d, J6,5 = 8.1, 1H, H6), 7.42–6.91 (m, 15H, trityl), 5.85 (d, J1′,2′ = 5.2, 1H, H1′), 5.52 (d, J5,6 = 8.1, 1H, H5), 5.26 (d, JOH,3′ = 5.0, 1H, 3′OH), 4.11 (dd, J2′,1′ = 5.2, J2′,3′ = 4.0, 1H, H2′), 3.96 (dd, J3′,2′ = 4.0, J3′,4′ = 4.4, 1H, H3′), 3.75 (s, 6H, 2×OMe), 3.39–3.24 (m, 3H, H4′, H5′, H5′′) 0.83 (s, 9H, tBu), 0.03 (s, 3H, CH3), −0.01 (s, 3H, CH3).
1-{2-O-tert-Butyldimethylsilyl-3-O-[(2-cyanoethoxy) (N,N-diisopropylamino)phosphino]-5-O-dimethoxytrityl-4-thio-β-d-ribofuranosyl}uracil (8)
Compound 7 (0.89 g, 1.27 mmol) was dissolved in anhydrous dichloromethane (8 ml) under an argon atmosphere and cooled to 0°C in an ice bath. N,N-Diisopropylethylamine (2.8 eq., 0.62 ml) was added slowly to the stirred solution and after 15 min, 2-cyanoethyl N,N-diisopropylchlorophosphoramidite (1.4 eq., 0.4 ml) was added dropwise followed by 1-methylimidazole (1.0 eq., 0.1 ml). The resulting clear solution was allowed to equilibrate to room temperature. After 5 h of stirring at room temperature, the reaction was complete (TLC) and was subsequently quenched with anhydrous EtOH (0.2 ml). The reaction mixture was diluted with hexane and applied directly to a flash silica gel column and eluted with a gradient of 10–50% EtOAc in hexane to give 8 (1.1 g, 98% as a white foam). 31P NMR (CDCl3): 148.01 (s).
Synthesis of C, G and A 4′-thioribo-phosphoramidites (compounds 9–20)
See Supplementary Material.
Oligonucleotide synthesis and purification
Oligonucleotides were synthesized at Sirna Therapeutics Inc. on an ABI 394 synthesizer using 4′-thio-C-phosphoramidite 12, and commercially available ribonucleoside phosphoramidite units on a 1.0 μmol scale, S-ethyltetrazole (0.25 M) as the activator, and following synthesis and deprotection protocols as previously described (30). For a typical synthesis, each of phosphoramidite units was used at a concentration of 0.1 M in dry acetonitrile and the coupling time was increased to 10 min for each step. After completion of the synthesis, the CPG support was treated with NH4OH/EtOH (3:1) at 65°C for 5 h, followed by desilylation using TBAF at room temperature for 24 h. After desalting, the oligonucleotides were HPLC-purified on an RP-C18 column (Rainin) at pH 7.0 (50 mM TEAA buffer) and eluted with an acetonitrile gradient (flow rate 0.75 ml/min.). The oligonucleotides were desalted on a Sepak column and after lyophilization, the stock concentration was adjusted to 8 mM based on measurements of UV absorption.
Crystallization and data collection
Crystals of the purified octamer r(CC4′SCCGGGG) were grown in hanging drops over a period of 6 months from a solution containing 1 mM RNA (single-strand concentration), 50 mM Na HEPES pH 7.5, 1% v/v PEG 400 and 1 M ammonium sulfate, equilibrated against 100 mM Na HEPES pH 7.5, 2% v/v PEG 400 and 2 M ammonium sulfate (Condition 39, Crystal Screen I, Hampton Research, Aliso Viejo, CA). Crystals of the octamers r(C4′SCCCGGGG) and r(CCC4′SCGGGG) could also be grown, but were of lower quality and only diffracted to limited resolution.
An r(CC4′SCCGGGG) crystal of approximate dimensions 0.2 × 0.2 × 0.2 mm was picked up from the droplet with a nylon loop, swiped through a 30% glycerol solution of the above reservoir for cryoprotection and then directly transferred into the −170°C cold N2 stream. X-ray diffraction data were collected on a Rigaku R-AXIS IIc image plate system, mounted on a Rigaku rotating anode X-ray generator. The detector-crystal distance was 120 mm and 100 frames were measured with a 2° oscillation angle and 10 min of exposure time. Data were processed with the DENZO/SCALEPACK program package (31) and yielded 3077 unique reflections above the 1σ level between 25 and 1.93 Å resolution (Rmerge = 6.7%; the shell between 1.93 and 2.02 Å resolution is 50% complete). Selected crystal data are listed in Table 1.
Table 1.
Selected crystal data and refinement parameters
| r(CC4′SCCGGGG)2a | |
|---|---|
| Space group | R32 |
| Crystal system | rhombohedral |
| Unit cell dimensions (hexagonal setting) | a = b = 42.44 Å |
| c = 127.66 Å | |
| α = β = 90° | |
| γ = 120° | |
| Strands per asymm. Unit | 2 |
| Vasym (Å3) | 11 063 |
| V/base pair (Å3) | 1383 |
| Temperature | −170°C |
| Unique data | 2962 > 2σ(Fo) |
| Resolution | 10.0–1.93 Å |
| R-merge | 6.7% |
| Completeness | 86% |
| Non-hydrogen atoms | 338 |
| Solvent molecules | 69 |
| r.m.s. deviations | 0.03 Å (bonds) |
| 3.8°(angles) | |
| R-factor | 20.2% |
a4′SC = 4′-thioribo-C.
Structure determination and refinement
The structure of the 4′-thio-modified octamer duplex was determined with the Molecular Replacement method using the program AMoRe (32). The coordinates of the RNA duplex from the room temperature r(C4G4) structure served as the starting model (33). Rotational and translational searches confirmed an orientation of the duplex identical to the one in the r(C4G4) structure. The initial model was refined with the program CNS (34) using updated dictionary files (35). To calculate the R-free, 5% of the reflections were set aside in a random fashion (36). Solvent molecules were included in the model in groups of five and conventional crystallographic refinement converged at 20.6% for 2962 reflections with intensities above the 2σ level in the resolution range between 10.0 and 1.93 Å. After reaching an R-free of 25%, all reflections were included in the final rounds of refinement. Selected crystallographic refinement parameters are listed in Table 1 and an example of the quality of the final electron density is depicted in Figure 1B. Structure factors and final coordinates have been deposited in the Brookhaven Protein Data Bank (PDB ID 2A0P).
Thermal denaturation studies
Absorbance versus temperature profiles were recorded at 260 nm on a Cary Bio-1 spectrophotometer equipped with a Peltier temperature control device. The measurements were conducted with five oligonucleotides, the native octamer r(C4G4) and the four modified strands r(C4′SCCCGGGG), r(CC4′SCCGGGG), r(CCC4′SCGGGG) and r(C4′SC4′SC4′SCGGGG). The samples were prepared under sterile conditions in 0.01 M Tris–HCl buffer (pH 7.4) containing 0.15 M NaCl solution and were subsequently degassed. The measurements were carried out at 0.75, 1.5, 2.5, 4.8 and 9.2 μM concentrations of the duplexes. Prior to the measurements, each sample was briefly heated to 80°C and a layer of silicon oil solution was placed on the surface of the sample solutions in the quartz cuvettes. The melting curves were obtained with both a cooling and a heating ramp of 0.3°C/min. The melting temperatures Tm were calculated by differentiation of the melting curves and the thermodynamic data were obtained according to Marky and Breslauer (37).
RESULTS AND DISCUSSION
Synthesis of 4′-thioribonucleoside phosphoramidites
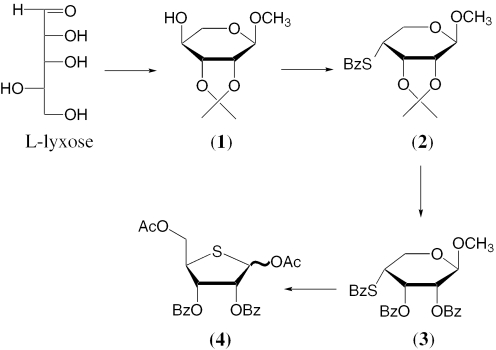
The synthesis of 1,5-di-O-acetyl-2,3-di-O-benzoyl-4-thio-d-ribofuranose 4 is based on modification of the procedure developed by Reist and coworkers for the synthesis of 1,2,3,5-tetra-O-acetyl-4-thio-d-ribofuranose (27). Modifications include the ‘one pot’ triflate/thiobenzoate SN2 displacement at position 4 of the sugar, instead of the reported tosylate/S-benzoate by Reist et al. (27). Conversion of l-lyxose to methyl-2,3-O-isopropylidine-α-l-lyxopyranoside takes place in ‘one pot’ by treatment with methanolic HCl to form methyl-α-l-lyxopyranoside which is then exposed to 2,2-dimethoxypropane in the presence of p-toluenesulfonic acid (Scheme 1). Flash chromatography gives the pure product 1 in 80% yield. A sulfur-containing nucleophile is then introduced at C4, using triflation followed by displacement with potassium thiobenzoate. The use of the highly hindered base, 2,6-di-tert-butyl-4-methylpyridine during triflation gives the desired product in the highest yield. Initial attempts to generate the 4-S-benzoyl ribopyranose derivative under Mitsunobu conditions failed to give the product in satisfactory yield. The ‘one pot’ triflation/displacement provides methyl-2,3-O-isopropylidine-4-S-benzoyl-β-d-ribopyranoside 2 in 56% yield after flash chromatography.
Scheme 1.
Synthesis of 1,5-di-O-acetyl-2,3-di-O-benzoyl-4-thio-d-ribofuranose.
Subsequent cleavage of isopropylidine protection takes place by treatment with trifluoroacetic acid. The crude deprotection mixture is then neutralized with sodium bicarbonate and treated with benzoyl chloride in pyridine to afford methyl-2,3-di-O-benzoyl-4-S-benzoyl-β-d-ribopyranoside 3 in 94% yield after chromatography. Exposure of the pyranoside to acetolysis conditions results in the formation of a labile anomeric acetate which acts as a good leaving group for the thiobenzoyl nucleophile. Spontaneous ring contraction then results in the formation of 1,5-di-O-acetyl-2,3-di-O-benzoyl-4-thio-d-ribofuranose 4 in quantitative yield (43% overall yield in 4 steps). This synthesis is amenable to scale up as demonstrated by our conversion of 50 g of l-lyxose into 51 g of 1,5-di-O-acetyl-2,3-di-O-benzoyl-4-thio-d-ribofuranose 4.
Conversion of the thiosugar 4 into the corresponding 4′-thioribonucleosides was accomplished by silylation of the desired base using BSA followed by glycosylation in the presence of tin tetrachloride (Scheme 2). Acyl groups were then cleaved selectively from the sugar using sodium hydroxide in dioxane. Tritylation of the 5′-hydroxyl groups took place under standard conditions upon treatment with dimethoxytrityl chloride in pyridine. Tritylation proved to be slow in all cases, future attempts should include silver triflate for activation. In the case of the uracil derivative, protection of the 2′-hydroxyl with TBDMSCl proceeded in the presence of silver nitrate with good selectivity and without cleavage of the carbon–sulfur bond. However, when the same conditions were applied to the cytosine analog, carbon–sulfur bond cleavage was observed. Subsequent introduction of TBDMS protection was therefore limited to the use of TBDMSCl, imidazole/DMF. Phosphitylations were carried out using standard conditions: N,N-diisopropylethylamine, 2-cyanoethyl N,N-diisopropylchlorophosphoramidite, 1-methylimidazole/dichloromethane. Phosphitylations carried out in the case of U, C and A went smoothly, however, in the case of G, high equivalents of the reagent was required for satisfactory conversion (see Supplementary Material for the C, G and A building blocks).
Scheme 2.
Synthesis of the 4′-thioribo-U, -C, -G and -A phosphoramidites.
Overall conformation of the chemically modified duplex and geometry of 4′-thio-cytidines
The RNA duplex bearing 4′-thio modifications at residues C3 and C11 (nucleotides in the two strands are numbered 1–8 and 9–16, respectively) adopts a regular A-form geometry. The sulfur atoms in residues 3 and 11 are located at the border of the minor groove (Figure 1C). Selected helical parameters are listed in Tables 2 and 3. The average inclination of base pairs relative to the overall helix axis is 10° and the average values for helical rise and twist are 2.8 Å and 32.3°, respectively. The structures of the chemically modified and native [r(CCCCGGGG)]2 duplexes are isomorphous. Two structures of the native octamer duplex had been determined previously. The initial model was based on diffraction data extending to 1.80 Å resolution (38). Subsequently, the resolution of the structure was improved to 1.45 Å and the hydration of the RNA duplex was analyzed in detail (33). To determine if 4′-thio modification alters the conformation of RNA locally or globally, we compared the structure of the RNA duplex with chemical modifications to that of the native duplex obtained at a comparable resolution (38) (Nucleic Acid Database (39) entry ARH064). Since the resolutions of the two structures are similar (1.93 versus 1.80 Å), possible differences between their geometries can be expected to be a consequence of chemical modification.
Table 2.
Average global helical parameters
| x-Displacement (dx) | −4.7 (0.1) |
| y-Displacement (dy) | 0.0 (0.2) |
| Inclination η (°) | 10.0 (1.1) |
| Tip Θ (°) | −0.3 (1.8) |
| Shear Sx (Å) | 0.1 (0.2) |
| Stretch Sy (Å) | 0.2 (0.1) |
| Stagger Sz (Å) | −0.2 (0.1) |
| Shift Dx (Å) | 0.3 (0.3) |
| Slide Dy (Å) | −0.1 (0.3) |
| Intra-strand P–P distance (Å) | 5.8 (0.3) |
Table 3.
| Base pair | Tilt (°) | Roll (°) | Twist (°) | Buckle (°) | Prop. (°) | Open. (°) | Rise (Å) |
|---|---|---|---|---|---|---|---|
| C(1)–G(16) | −3.0 (−2.6) | 1.2 (−2.8) | 30.9 (31.6) | 10.2 (8.4) | −15.3 (−11.6) | 1.2 (−2.1) | 2.8 (2.8) |
| C(2)–G(15) | −2.6 (−2.3) | −5.0 (−1.6) | 33.0 (35.0) | 10.5 (12.0) | −25.2 (−19.1) | −1.6 (−0.2) | 3.3 (2.9) |
| 4′SC(3)–G(14) | −0.l (0.5) | 7.7 (0.9) | 34.0 (31.8) | 7.8 (8.5) | −17.0 (−13.4) | 8.9 (2.2) | 2.5 (2.8) |
| C(4)–G(13) | −0.8 (−1.1) | 4.6 (4.3) | 30.9 (32.6) | −0.7 (3.8) | −10.3 (−12.0) | 1.6 (2.0) | 2.8 (2.7) |
| G(5)–C(12) | 0.1 (0.0) | 5.0 (4.2) | 27.2 (30.5) | −5.0 (−3.3) | −11.1 (−7.1) | 1.6 (1.1) | 2.5 (2.6) |
| G(6)–4′SC(11) | 3.2 (−0.6) | 4.6 (1.3) | 36.1 (33.0) | −3.0 (−3.3) | −12.4 (−10.8) | 9.3 (4.6) | 2.8 (2.9) |
| G(7)–C(10) | 2.5 (4.3) | −2.5 (−1.3) | 33.9 (33.2) | −11.1 (−9.0) | −18.2 (−12.6) | −2.9 (1.7) | 3.2 (3.0) |
| −12.1 (−10.9) | −23.4 (−20.9) | 2.8 (0.9) | |||||
| Average | −0.1 | 2.2 | 32.3 | −0.4 | −16.6 | 2.6 | 2.8 |
| SD | 2.5 | 4.6 | 2.9 | 9.1 | 5.5 | 4.4 | 0.3 |
There appear to be no drastic changes among either the global or local helical parameters as a result of the introduction of the two 4′-thio-modified residues (Table 3). At the C2p4′SC3 step, the helical rise in the modified duplex is increased by 0.4° relative to the corresponding step in the native RNA. At the adjacent 4′SC3pC4 step in the modified duplex, the rise is reduced by 0.3 Å compared to the native duplex. However, in both cases the observed changes are within the standard deviations. A further subtle change is observed in the helical twists between base pairs involving chemically modified nucleotides. Compared to the native duplex, the helical twist between base pairs C2–G15 and 4′SC3–G14 is 2° lower in the modified duplex (Table 3). However, the twist between 4′SC3–G14 and C4–G13 is increased by 2.2° in the modified duplex. Similarly, the helical twists between base pairs G5–C12 and G6–4′SC11 and between G6–4′SC11 and G7–C10 are reduced (by 3.3°, Table 3) and increased (by 3.1°), respectively.
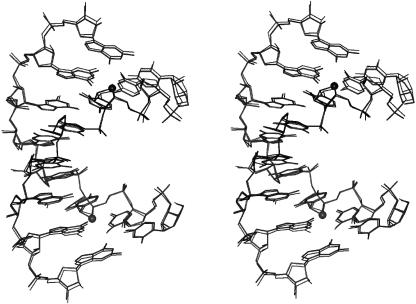
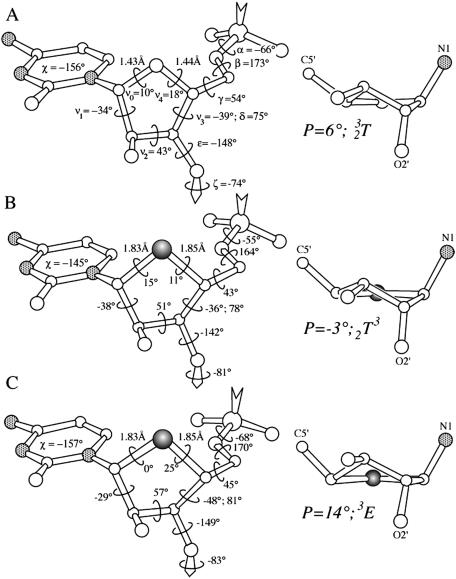
Superposition of the native and [r(CC4′SCCGGGG)]2 duplexes reveals three regions of notable geometric deviations (Figure 2). These comprise the two modified sugars of residues C3 and C11 and the backbone of residue G13. Deviations between the sugar portions of nucleotides C3 and C11 in the modified and reference duplexes are mainly due to the longer C1′–S4′ and C4′–S4′ bonds in the former compared with the corresponding bonds in the 4′-oxo sugars (Figure 3). All sugars in the two duplexes adopt ‘Northern’ type puckers. However, the puckers of the two 4′-thio-modified nucleotides differ subtly. In residue 4′SC3, the sugar pucker is C2′-exo and in residue 4′SC11 it is C3′-endo. By comparison, the pucker of both C3 and C11 in the native duplex is C3′-endo (only C3 shown in Figure 3). However, in the native duplex, the sugar of residue G13 also exhibits a C2′-exo conformation. The altered sugar pucker is accompanied by an extended backbone variant, characterized by anti-periplanar conformations of the α and γ backbone torsion angles (Figure 2; see Figure 3 for torsion angle definition). This feature gives rise to the third region of significant conformational deviation between the native and chemically modified RNA octamer duplexes.
Figure 2.
Stereo overlay plot of the [r(CC4′SCGGG)]2 (this work, thick lines) and [r(C4G4)]2 duplexes [thin lines (38)]. Phosphate groups of the two duplexes were superimposed, resulting in an overall root mean square (r.m.s.) deviation between all atoms of 0.3 Å. 4′-Sulfur atoms of modified riboses are drawn as filled circles.
Figure 3.
Geometry of ribose and 4′-thioribose sugars in the [r(C4G4]2 and [r(CC4′SCGGGG)]2 duplexes, respectively. (A) Left-hand side: Residue C3 from the r(C4G4) structure (32). The backbone torsion angles are defined as O3′n−1-P-α-O5′-β-C5′-γ-C4′-δ-C3′-ε-O3′-ζ-P-O5′n+1, sugar torsion angles are C1′-ν1-C2′-ν2-C3′-ν3-C4′-ν4-O4′-ν0-C1′, χ is the glycosydic torsion angle, and the C1′–O4′ and O4′–C4′ bond lengths are indicated. Right-hand side: Sugar conformation of residue C3. Pseudorotation phase angle P and pucker type are listed, and nitrogen atoms are stippled in grey. (B) Geometry and sugar conformation of 4′SC3 in the [r(CC4′SCCGGGG)]2 duplex. (C) Geometry and sugar conformation of 4′SC11 in the [r(CC4′SCCGGGG)]2 duplex. 4′-Sulfur atoms are highlighted as filled circles.
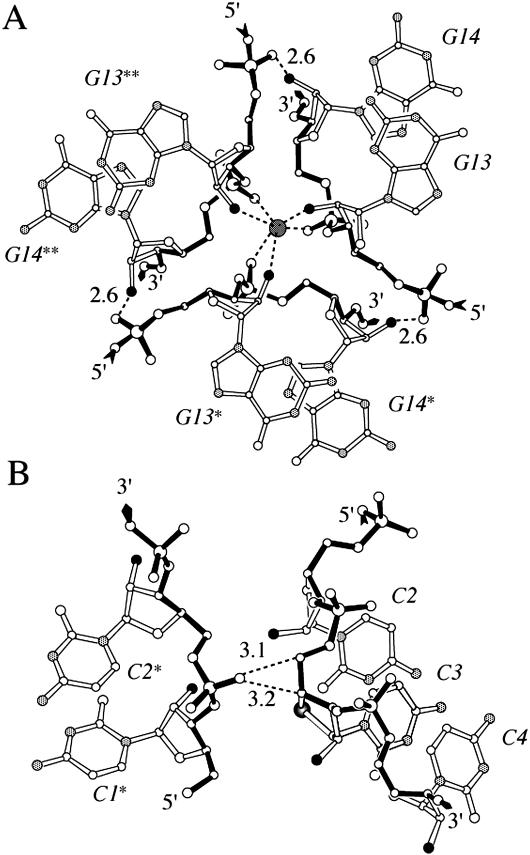
Lattice interactions
In the rhombohedral lattice, infinite columns of stacked helices are arranged around 3-fold rotation axes (Figure 4). Lateral contacts include hydrogen bonds between 2′-hydroxyl and phosphate groups from neighboring helices. Adjacent symmetry mates are bridged by water molecules, the latter located on 3-fold rotation axes. Each of these water molecules forms a total of six hydrogen bonds to 2′-hydroxyl and phosphate groups from three duplexes (Figure 4A). Although the electron density peaks on the rotation axes may constitute sodium ions, they were refined as water molecules. At the present resolution of the structure of ∼2 Å, it is impossible to determine the precise nature of these peaks. Additional lateral contacts between neighboring duplexes are mediated by phosphate groups and sugar carbon atoms (Figure 4B). For example, a non-bridging phosphate oxygen acts as acceptor in C–H⋯O type hydrogen bonds to the C4′ and C5′ atoms of residue 4′SC3 (Figure 4B). In the structure of the modified duplex, the distances between C4′ (4′SC3) and O1P(C2*) (an asterisk denotes a symmetry mate) and C5′ (4′SC3) and O1P(C2*) are 3.2 and 3.1 Å, respectively. In the lattice of the native RNA duplex, these contacts are loosened and the distances between corresponding atom pairs are 3.6 and 3.3 Å, respectively. Interestingly, the sugar pucker observed for residue C3 in case of the native RNA duplex is C3′-endo while it is C2′-exo for 4′SC3 in the modified duplex (vide supra). It is likely that the pucker of the latter is affected by the above C–H⋯O type lattice interactions. Therefore, we may assume that the preferred conformation of 4′SC is the one exhibited by 4′SC11 (C3′-endo, Figure 3), since this residue is not involved in any inter-duplex contacts.
Figure 4.
Interactions between symmetry-related octamer strands in the lattice of [r(CC4′SCCGGGG)]2. (A) Three-way junction around a crystallographic 3-fold rotation axis, under formation of hydrogen bonds between O2′ (black circles) and phosphate oxygen atoms and additional coordination of a putative sodium ion (dark gray circle). The putative sodium ion interacts with O2′ and phosphate oxygen atoms from three adjacent duplex molecules forming a distorted octahedron (dashed lines). (B) C–H⋯O type hydrogen bonds between two symmetry-related duplexes, involving a phosphate oxygen atom from one molecule and hydrogen atoms of C4′ and C5′ from the other. Hydrogen bonds are dashed and their lengths are given in Å.
Hydration of the 4′-thio-cytidines
Based on the high-resolution crystal structure of [r(CCCCGGGG)]2, the ribose 4′-oxygens were observed to be engaged in the lowest number of hydrogen bonds among all potential hydrogen bond acceptor and donor atoms in RNA (33). Moreover, hydrogen bonds of the 3′→5′ intra-strand type between O2′ and O4′ were found to be weak and typically longer than the standard cutoff length of 3.3 Å. The average length of such hydrogen bonds was 3.68 Å, with a minimum of 3.37 Å and a maximum of 3.89 Å. It was concluded that water molecules are better acceptors of hydrogen bonds from 2′-hydroxyl groups than ribose 4′-oxygen atoms. Auffinger and Westhof later provided convincing evidence that the primary interaction between adjacent intra-strand riboses is a C2′–H2′⋯O4′ hydrogen bond (40). For all these reasons, replacement of the 4′-oxygen by sulfur would not be expected to affect hydration of the RNA backbone and the minor groove to a significant extent. Indeed, in the 1.8 Å crystal structure of the native [r(CCCCGGGG)]2 duplex, only a single hydrogen bond with a length ∼3.3 Å was observed (to O4′ of residue C3) (38). The 4′-sulfur of the equivalent residue in the structure of the modified duplex at 1.93 Å resolution does not exhibit any close contacts to water molecules. However, the 4′-sulfur of 4′SC11 on the opposite strand forms a contact to a water molecule with a distance of 3.45 Å. In addition, O4′ of G13 is engaged in a hydrogen bond to a water molecule (distance 3.33 Å). As expected, presence of the larger sulfur atom results in a slightly wider spacing between adjacent riboses. This is apparent from the longer C2′ (3′)⋯S4′ (5′) distances at modified steps relative to the corresponding C2′⋯O4′ distances. The distance between S4′ of 4′SC3 and C2′ of C2 is 3.86 Å and the distance between S4′ of 4′SC11 and C2′ of C10 is 3.77 Å. The corresponding distances in the native duplex are 3.73 and 3.52 Å. The average distance between O4′ atoms and adjacent C2′ atoms in the modified duplex is 3.52 Å.
Stability of RNA duplexes with incorporated 4′SC residues
The UV melting temperatures Tm and thermodynamic stabilities of four duplexes were measured and compared with the stability parameters of the native RNA duplex. The modified octamers comprise the octamers r(C4′SCCCGGGG), r(CC4′SCCGGGG), r(CCC4′SCGGGG) and r(C4′SC4′SC4′SCGGGG) (Table 4). The stabilities of the four 4′S-modified oligoribonucleotides are enhanced relative to the native octamer. The melting temperatures of the three oligonucleotides carrying single 4′-thiocytidines are increased by ∼1°C. The thermodynamic parameters indicate that the gain in stability is enthalpy based. In all three cases, the entropies are unfavorable compared with the native duplex. Interestingly, the entropy term becomes less favorable as the modified nucleoside is moved toward the center of the duplex (Table 4). Thus, r(CCC4′SCGGGG) exhibits the highest gain in enthalpy among the five analyzed octamers. At the same time, its entropy term is the least favorable one. Upon duplex formation of r(CCC4′SCGGGG), modified nucleosides are arranged on opposite strands within a dinucleotide step. In the case of the octamer featuring three consecutive 4′-thiocytidines, the enthalpy contribution does not increase further (Table 4). Rather, it is decreased relative to r(CCC4′SCGGGG) and comparable to the enthalpy seen with r(CC4′SCCGGGG). Similarly, the entropy term for the octamer carrying three modifications is slightly more favorable than in the case of r(CCC4′SCGGGG), but still considerably less favorable compared with the native octamer.
Table 4.
UV melting temperaturesa and thermodynamic datab of duplex formation for r(C4G4) and four isosequential 4′-thio-modified RNA octamers
| Oligonucleotide | ΔH° (kcal/mol) | ΔS° (cal/mol) | ΔG°298 (kcal/mol) | Tm (°C) |
|---|---|---|---|---|
| R(CCCCGGGG) | −70.2 | −179.5 | −16.7 | 74.1 |
| r(C4′SCCCGGGG) | −84.6 | −220.5 | −18.9 | 74.8 |
| r(CC4′SCCGGGG) | −90.2 | −236.7 | −19.7 | 74.7 |
| r(CCC4′SCGGGG) | −111.8 | −298.5 | −22.9 | 75.1 |
| r(C4′SC4′SC4′SCGGG) | −96.3 | −251.6 | −21.3 | 78.0 |
a9.2 μM oligonucleotide in 10 mM Tris–HCl, pH 7.4 and 0.15 M NaCl.
bFrom concentration-dependent transition temperatures.
The interpretation of the thermodynamic parameters is not straightforward, but the contributions to stability clearly vary between RNAs carrying a single nucleoside analog and those carrying multiple ones. From the small number of chemically modified oligonucleotides studied here, we may conclude that 4′-thio modification of RNA is stabilizing. Although the enthalpic and entropic contributions vary significantly depending on whether a single 4′-thio-modified C or multiple ones are incorporated, it appears that the gain in stability is additive and amounts to ∼1.0°C per modification (Table 4). This is in-line with recently reported stability gains for all-modified 4′-thio RNAs that amounted to ∼2.5°C per base pair relative to RNA (25). It is very likely that the increased stabilities are due in part to the altered gauche and anomeric effects in 4′-thio-modified nucleosides compared to the native 4′-oxo species.
CONCLUSIONS
We have presented a facile route for the preparation of the 4′-thio-U, -C, -G and -A phosphoramidites. The protected and activated 4′-thio-d-ribofuranose precursor was obtained in just four steps and with high yield and is converted into the phosphoramidite building block in a further four steps. Our protocol offers considerable advantages in terms of ease of synthesis and overall yield compared to existing procedures (19,25,26,29). Incorporation of isolated and consecutive stretches of 4′-thio-C residues, into an RNA octamer, all significantly stabilized the RNA duplex. The gains were somewhat more moderate than those reported for fully modified 4′-thio RNAs (25), but they serve to reiterate that 4′-thio modification boosts RNA affinity. The structural changes induced by replacement of the 4′-oxygen with sulfur are only minimal as demonstrated by the crystal structure at medium resolution of an RNA octamer duplex with incorporated 4′-thio-C residues. In particular, the sugar pucker is virtually unaffected by the O→S replacement. The observed structural differences between RNA and 4′-thio RNA are limited to geometrical changes as a result of the somewhat bulkier sulfur in the sugar. Combined with the excellent protection against endo- and exo-nuclease degradation afforded by the modification (25), these properties render 4′-thio RNA a promising RNA analog for in vitro and in vivo applications of RNAi. For example, recent studies indicate that selective stabilization at the 3′ end of the antisense strand of an siRNA duplex with concomitant destabilization at the 5′ end of the siRNA duplex can afford improved RNAi selectivity and activity (41–43). In initial gene silencing experiments, the modification appeared to be better tolerated in the sense siRNA than in the antisense siRNA and its effectiveness was higher when placed at the termini of strands rather than in the center [reviewed in (7)]. Although our crystal structure shows that the conformational consequences of the 4′-thio modification in RNA are relatively minor, in agreement with the results from CD experiments in solution (25), a more detailed understanding of the effects on stability and hydration by the 4′-thio modification will require a high-resolution crystal structure of an RNA duplex containing a consecutive stretch of 4′-thioribonucleosides.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We are grateful to Dr Leonid Beigelman for his important contributions to the design of the synthetic route, and Dr Nassim Usman for his enthusiastic support. We would like to thank Drs Xiaolin Wu and Stefan Pitsch for help with acquisition of the thermodynamic data. This work was supported by the National Institutes of Health (grant GM55237 to M.E.). Funding to pay the Open Access publication charges for this article was provided by NIH GMS grant 2 R01 GM55237.
Conflict of interest statement. None declared.
REFERENCES
- 1.De Mesmaeker A., Häner R., Martin P., Moser H.E. Antisense Oligonucleotides. Acc. Chem. Res. 1995;28:366–374. [Google Scholar]
- 2.Freier S.M., Altmann K.H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1996;24:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brautigam C.A., Steitz T.A. Structural principles for the inhibition of the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J. Mol. Biol. 1998;277:363–377. doi: 10.1006/jmbi.1997.1586. [DOI] [PubMed] [Google Scholar]
- 4.Teplova M., Wallace S.T., Minasov G., Tereshko V., Symons A., Cook P.D., Manoharan M., Egli M. Structural origins of the exonuclease resistance of a zwitterionic RNA. Proc. Natl Acad. Sci. USA. 1999;96:14240–14245. doi: 10.1073/pnas.96.25.14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baidya N., Ammons G.E., Matulic-Adamic J., Karpeisky A., Beigelman L., Uhlenbeck O.C. Functional groups on the cleavage site pyrimidine nucleotide are required for stabilization of the hammerhead transition state. RNA. 1997;3:1135–1142. [PMC free article] [PubMed] [Google Scholar]
- 6.Burgin A.B., Jr, Gonzalez C., Matulic-Adamic J., Karpeisky A.M., Usman N., McSwiggen J.A., Beigelman L. Chemically modified hammerhead ribozymes with improved catalytic rates. Biochemistry. 1996;35:14090–14097. doi: 10.1021/bi961264u. [DOI] [PubMed] [Google Scholar]
- 7.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 9.Eckstein F. Nucleoside phosphorothioates. Ann. Rev. Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- 10.Zon G., Stec W. Phosphorothioate oligonucleotides. In: Eckstein F., editor. Oligonucleotides Analogues. Oxford: IRL Press; 1991. pp. 87–108. [Google Scholar]
- 11.Akhtar S., Agrawal S. In vivo studies with antisense oligonucleotides. Trends Pharmacol. Sci. 1997;18:12–18. doi: 10.1016/s0165-6147(96)01002-4. [DOI] [PubMed] [Google Scholar]
- 12.Cook P.D. Second generation antisense oligonucleotides: 2′-modifications. Ann. Rep. Med. Chem. 1998;33:313–325. [Google Scholar]
- 13.Rajeev K.G., Prakash T.P., Manoharan M. 2′-Modified-2-thiothymidine oligonucleotides. Org. Lett. 2003;5:3005–3008. doi: 10.1021/ol0348607. [DOI] [PubMed] [Google Scholar]
- 14.Jones G.D., Lesnik E.A., Owens S.R., Risen L.M., Walker R.T. Investigation of some properties of oligodeoxy-nucleotides containing 4′-thio-2′-deoxynucleotides: duplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1996;24:4117–4122. doi: 10.1093/nar/24.21.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones G.D., Altmann K.H., Hüsken D., Walker R.T. Duplex- and triplex-forming properties of 4′-thio-modified oligodeoxynucleotides. Bioorg. Med. Chem. Lett. 1997;7:1275–1278. [Google Scholar]
- 16.Walker R.T. 4′-Thio-2′-deoxyribonucleosides, their chemistry and biological properties–a review. R. Soc. Chem. Spec. Publ. (Anti-Infectives) 1997;198:203–237. [Google Scholar]
- 17.Kumar S., Horton J.R., Jones G.D., Walker R.T., Roberts R.J., Cheng X. DNA containing 4′-thio-2′-deoxycytidine inhibits methylation by HhaI methyltransferase. Nucleic Acids Res. 1997;25:2773–2783. doi: 10.1093/nar/25.14.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boggon T.J., Hancox E.L., McAuley-Hecht K.E., Connolly B.A., Hunter W.N., Brown T., Walker R.T., Leonard G.A. The crystal structure analysis of d(CGCGAASSCGCG)2, a synthetic DNA dodecamer duplex containing four 4′-thio-2′-deoxythymidine nucleotides. Nucleic Acids Res. 1996;24:951–961. doi: 10.1093/nar/24.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leydier C., Bellon L., Barascut J.L, Morvan F., Rayner B., Imbach J.L. 4′-Thio-RNA: synthesis of mixed 4′-thio-oligoribonucleotides, nuclease resistance and base pairing properties with complementary single and double strand. Antisense Res. Dev. 1995;5:167–174. doi: 10.1089/ard.1995.5.167. [DOI] [PubMed] [Google Scholar]
- 20.Leydier C., Bellon L., Barascut J.L., Imbach J.L. 4′-Thio-β-d-oligoribonucleotides: nuclease resistance and hydrogen bonding properties. Nucleosides Nucleotides. 1995;14:1027–1030. [Google Scholar]
- 21.Hall K.B., McLaughlin L.W. Thermodynamic and structural properties of pentamer DNA•DNA, RNA•RNA, and DNA•RNA duplexes of identical sequence. Biochemistry. 1991;30:10606–10613. doi: 10.1021/bi00108a002. [DOI] [PubMed] [Google Scholar]
- 22.Lesnik E.A., Freier S.M. Relative thermodynamic stability of DNA, RNA and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry. 1995;34:10807–10815. doi: 10.1021/bi00034a013. [DOI] [PubMed] [Google Scholar]
- 23.Crnugelj M., Dukhan D., Barascut J.-L., Imbach J.-L., Plavec J. How S-C-N anomeric effect and energetic preference across [S-C-C-O] fragments steer conformational equilibria in 4′-thionucleosides. 1H NMR and ab initio MO study. J. Chem. Soc. Perkin Trans. 2000;2:255–262. [Google Scholar]
- 24.Thibaudeau C., Chattopadyaya J. Stereoelectronic Effects in Nucleosides and Nucleotides and their Structural Implications. Uppsala, Sweden: Uppsala University Press; 1999. [Google Scholar]
- 25.Hoshika S., Minakawa N., Matsuda A. Synthesis and physical and physiological properties of 4′-thioRNA: application to post-modification of RNA aptamer toward NF-κB. Nucleic Acids Res. 2004;32:3815–3825. doi: 10.1093/nar/gkh705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellon L., Barascut J.L., Maury G., Divita G., Goody R., Imbach J.L. 4′-Thio-oligo-β-d-ribonucleotides: synthesis of β-4′-thio-oligouridylates, nuclease resistance, base pairing properties, and interaction with HIV-1 reverse transcriptase. Nucleic Acids Res. 1993;21:1587–1593. doi: 10.1093/nar/21.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reist E.J., Gueffroy D.E., Goodman L. Synthesis of 4′-thio-d- and -l-ribofuranose and the corresponding adenine nucleosides. J. Am. Chem. Soc. 1964;86:5658–5663. [Google Scholar]
- 28.Vorbrüggen H., Bennua-Skalmowski B. New simplified nucleoside synthesis. Tetrahedron Lett. 1978;19:1339–1342. [Google Scholar]
- 29.Naka T., Minakawa N., Abe H., Kaga D., Matsuda A. The stereoselective synthesis of 4′-β-thioribonucleosides via the Pummerer reaction. J. Am. Chem. Soc. 2000;122:7233–7243. [Google Scholar]
- 30.Wincott F., DiRenzo A., Shaffer C., Grimm S., Tracz D., Workman C., Sweedle D., Gonzalez C., Scaringe S., Usman N. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A. 1994;50:157–163. [Google Scholar]
- 33.Egli M., Portmann S., Usman N. RNA hydration: a detailed look. Biochemistry. 1996;35:8489–8494. doi: 10.1021/bi9607214. [DOI] [PubMed] [Google Scholar]
- 34.Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., Read R.J., Rice L.M., Simonson T., Warren G.L. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Parkinson G., Vojtechovsky J., Clowney L., Brünger A.T., Berman H.M. New parameters for the refinement of nucleic acid-containing structures. Acta Crystallogr. D. 1996;52:57–64. doi: 10.1107/S0907444995011115. [DOI] [PubMed] [Google Scholar]
- 36.Brünger A.T. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 37.Marky L.A., Breslauer K.J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 38.Portmann S., Usman N., Egli M. The crystal structure of r(CCCCGGGG) in two distinct lattices. Biochemistry. 1995;34:7569–7575. doi: 10.1021/bi00023a002. [DOI] [PubMed] [Google Scholar]
- 39.Berman H.M., Olson W.K., Beveridge D.L., Westbrook J., Gelbin A., Demeny T., Hsieh S.-H., Srinivasan A.R., Schneider B. The nucleic acid database: a comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auffinger P., Westhof E. Rules governing the orientation of the 2′-hydroxyl group in RNA. J Mol. Biol. 1997;274:54–63. doi: 10.1006/jmbi.1997.1370. [DOI] [PubMed] [Google Scholar]
- 41.Ui-Tei K., Naito Y., Takahashi F., Haraguchi T., Ohki-Hamazaki H., Juni A., Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin F., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 43.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 44.Lavery R., Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.