Abstract
When telomerase is absent and/or telomeres become critically short, cells undergo a progressive decline in viability termed senescence. The telomere checkpoint model predicts that cells will respond to a damaged or critically short telomere by transiently arresting and activating repair of the telomere. We examined the senescence of telomerase-deficient Saccharomyces cerevisiae at the cellular level to ask if the loss of telomerase activity triggers a checkpoint response. As telomerase-deficient mutants were serially subcultured, cells exhibited a progressive decline in average growth rate and an increase in the number of cells delayed in the G2/M stage of the cell cycle. MEC3, MEC1, and DDC2, genes important for the DNA damage checkpoint response, were required for the cell cycle delay in telomerase-deficient cells. In contrast, TEL1, RAD9, and RAD53, genes also required for the DNA damage checkpoint response, were not required for the G2/M delay in telomerase-deficient cells. We propose that the telomere checkpoint is distinct from the DNA damage checkpoint and requires a specific set of gene products to delay the cell cycle and presumably to activate telomerase and/or other telomere repair activities.
INTRODUCTION
Telomeres, the nucleotide-protein structures at the ends of linear chromosomes, serve as a cap to protect the ends of chromosomes (reviewed in Blackburn, 2000). The function of this cap must strike a balance between facilitating and limiting access to the telomere. The cap must allow access to telomeric DNA for DNA replication enzymes, replication forks, and telomerase. In contrast, access to other factors that degrade and/or modify DNA ends must be limited. Thus, the cap must be dynamic, coordinating access to telomere DNA with other cellular events such as DNA replication or mitosis.
Telomere DNA is normally replicated by telomerase, a specialized reverse transcriptase that utilizes an RNA template that is an integral component of the enzyme. Telomerase is activated late in S phase, around the time when telomeres are replicated (Wellinger et al., 1993a, 1993b). Cells that are telomerase-deficient due to mutations in the catalytic component of the enzyme, the template RNA, or other required factors undergo senescence, a progressive loss of viability that is dependent on the number of divisions after loss of telomerase (Lundblad and Szostak, 1989; McEachern and Blackburn, 1996; Nakamura et al., 1997). During senescence, telomeres become progressively shorter as population viability declines (Singer and Gottschling, 1994; Lendvay et al., 1996; McEachern and Blackburn, 1996; Lingner et al., 1997). In yeasts, senescence can be detected as reduced numbers of colony-forming units and decreased colony size on solid media or as a decline in the average culture growth rate in liquid cultures (Singer and Gottschling, 1994; Lendvay et al., 1996; McEachern and Blackburn, 1996; Lingner et al., 1997).
Changes in telomere structure can cause cellular abnormalities. In human cells, overexpression of a dominant-negative form of TRF2, a telomere-regulating protein, causes frequent chromosome rearrangements and apoptosis (Karlseder et al., 1999). Dominant-negative telomerase mutations, which inhibit telomerase activity, also trigger apoptosis (Zhang et al., 1999). In Tetrahymena, a mutation in the telomerase template RNA leads to abnormal mitosis in the micronucleus and abnormally large cells (Kirk et al., 1997). In the yeast Kluyveromyces lactis, a mutation in the telomerase template RNA causes slow growth, abnormal karyotypes, and aberrant nuclear divisions (Smith and Blackburn, 1999). These data argue that the loss of telomere cap function leads to abnormal growth and loss of cell cycle coordination (reviewed in Blackburn, 2001).
Cell cycle checkpoints coordinate many processes in which one event must be completed before another is initiated. The DNA damage checkpoint is triggered by single stranded DNA (ssDNA) or broken DNA ends (reviewed in Longhese et al., 1998; Zhou and Elledge, 2000), which trigger a cell cycle delay and the activation of damage repair processes. Failure of this checkpoint results in cells that continue to divide a damaged genome, eventually leading to cell death. The DNA damage checkpoint is mediated by several interdependent pathways that require the products of MEC3, MEC1, DDC2, TEL1, RAD53, and RAD9 as well as other genes (Usui et al., 2001, reviewed in Longhese et al., 1998; Zhou and Elledge, 2000).
Normal telomeres terminate with a 3′ ssDNA overhang (Blackburn, 2000), yet they do not appear to activate a DNA damage checkpoint. Furthermore, loss of telomerase does not immediately lead to cell death; rather, senescence occurs (Singer and Gottschling, 1994; Lendvay et al., 1996; McEachern and Blackburn, 1996; Lingner et al., 1997). This suggests that the mechanism that monitors replication or genome integrity functions differently at telomeres than it does at other regions of the genome. Thus, the DNA ends at telomeres are either masked from DNA damage checkpoint detection pathways or they actively signal to the DNA damage checkpoint that telomere structure and/or function is normal.
Despite the fact that intact telomeres do not appear to be recognized as double strand breaks (DSBs), telomeres require several checkpoint genes for their replication and maintenance. In Saccharomyces cerevisiae, cells lacking the two ATM-like kinases, TEL1 and MEC1, do not maintain telomere length (Craven and Petes, 1999; Ritchie et al., 1999) and undergo senescence similar to that observed in telomerase-deficient cells, despite having functional telomerase components (Chan et al., 2001). Furthermore, telomerase activation requires the ATM kinase functions of either TEL1 or MEC1 in humans, S. cerevisiae, and Schizosaccharomyces pombe (Vaziri, 1997; Naito et al., 1998; Ritchie et al., 1999; Mallory and Petes, 2000; Chan et al., 2001). This implies that ATM kinases positively regulate telomerase activity either directly or indirectly.
The telomere checkpoint model (Blackburn, 2000) posits the existence of a telomere-specific checkpoint that arrests cells and activates telomere synthesis in response to the loss of cap function. This loss of cap function is proposed to occur when telomeres become critically short because of lack of telomerase activity or because alteration of telomere components renders the telomere inaccessible to telomerase. In this report, we tested the telomere checkpoint model by analyzing telomerase-deficient cells undergoing senescence.
MATERIALS AND METHODS
Yeast Strains and Culture Conditions
All strains used in this study were isogenic with W303 and are listed in Table 1. The TLC1 disruption was made by a one step gene replacement in diploid YJB334 using plasmid pBlue61::LEU2 (Singer and Gottschling, 1994). tlc1 isolates from early passages were used in standard crosses. The mec3::TRP1, mec1::HIS3, ddc2::KanMX4, and sml1::Kanx4 alleles were obtained from M.P. Longhese (Milan, Italy) in strains DMP2145/16C, DMP2952/2B, and DM2995/1B, respectively (Paciotti et al., 1998, 2000). The rad53-K227A kinase domain allele was obtained from M. Foiani (Milan, Italy) in yeast strain CY2034 (Pellicioli et al., 1999). The rad9::URA3 allele was obtained from O. Tsuchiya (Higashi-Hiroshima, Japan) in yeast strain W-DR9a (Mizunuma et al., 1998). The sml1::HIS3 and tel1::URA3 alleles were obtained from T. Petes (University of North Carolina) in yeast strain JMY303 and SPY40 (Ritchie et al., 1999). The GFP-TUB1 allele was obtained from K. Blumer (Washington University) in yeast strain KBY215 (Holly and Blumer, 1999). All of these alleles were introduced into our W303 strain background by standard crosses and multiple backcrosses. Sporulation and tetrad dissection were performed according to standard methods (Sherman and Hicks, 1991). Strains were maintained in standard yeast media (Sherman, 1991).
Table 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| YJB195 | MATaura3-1 ade2-1 his3-11 leu2-3, 112 can1-100 trp1-1 |
| YJB209 | MATα ura3-1 ade2-1 his3-11 leu2-3, 112 can1-100 trp1-1 |
| YJB334 | YJB195 × YJB209 |
| YJB2768 | YJB334 +nmd2::HIS3/NMD2 VR-ADE2-TEL/VR-TEL tlc1Δ::LEU2/TLC1 |
| YJB3867 | YJB334 +VR-ADE2-TEL/VR-TEL tlc1Δ::LEU2/TLC1 rad9Δ::URA3/RAD9 |
| YJB4565 | YJB334 +tlc1Δ::LEU2/TLC1 mec3Δ::TRP1/MEC3 |
| YJB6361 | YJB334 +VR-ADE2-TEL/VR-TEL tlc1Δ::LEU2/TLC1 sml1Δ::HIS3/SML1 ddc2Δ::KanMX4/DDC2 |
| YJB6689 | YJB334 +VR-ADE2-TEL/VR-TEL tlc1Δ::LEU2/TLC1 sml1Δ::HIS3/SML1 mec1Δ::HIS3/MEC1 VIIL::UR3-TEL/VIIL-TEL |
| YJB6690 | YJB334 +tlc1Δ::LEU2/TLC1 rad53K227A::KanMX4/RAD53 |
| YJB6741 | YJB334 +tlc1Δ::LEU2/TLC1 GFP-TUB1::HIS3/his3-11 |
| YJB6744 | YJB334 +tlc1Δ::LEU2/TLC1 mec3Δ::TRP1/MEC3 GFP-TUB1::HIS3/his3-11 ADE2::URA3/ura3-1 |
| YJB7448 | YJB334 +tlc1Δ::LEU2/TLC1 tel1Δ::URA3/TEL1 VR-ADE2-TEL/VR-TEL |
Serial Plate Passages of Senescing Cultures
Telomerase-deficient cells were obtained by sporulation and dissection of heterozygous diploids (YJB2768, 3867, 4565, 6361, 6689, 6690, 6741, 6744, and 7448). At least six independently derived spores of each genotype were restreaked from the tetrad dissection plate onto fresh solid medium and grown for 24 h at 30°C. The senescing cultures were serially restreaked from the thickest region of the plate for up to 10 passages. Images of each passage were captured on a Nikon Cool Pix 900 digital camera mounted on a Zeiss stereoscope Stemi DRC. To examine the spindle structure of wild-type and tlc1Δ senescing cultures, strains containing GFP-TUB1 were obtained by sporulation and dissection of heterozygous diploids and were serially passaged by successive restreaking on YPAD plates. Fluorescence microscopy of >400 live cells per passage, (mounted in 15% glycerol), were scored for spindle length and bud size.
Quantitative Measurements of Colony Sizes from Senescing Serial Liquid Cultures
To calibrate the assay, the number of cells in six independent wild-type colonies was determined. We found that estimates of cell number based on colony forming units in early passages was variable and often resulted in significant underestimates (3- to 10-fold) of actual cell number. We established the relationship between measured colony area and cell number by measuring colonies of different sizes for colony area and then manually dissecting and counting the total number of cells in each colony. Measurements of the colony area were reproducible and readily distinguished twofold differences in cell number in the wild-type strain.
Colony areas were measured for senescing cultures after limiting passages in liquid media and outgrowth on solid media. Telomerase-deficient cells were obtained by sporulation and dissection of the relevant heterozygous diploid strains. Spore colonies were grown for 2 d after dissection on solid medium at 30°C. These colonies were suspended in 15% glycerol, and ∼105 cells were inoculated into 1 ml of YPAD. The liquid cultures were grown for 24 h at 30°C, at which point they had typically completed 10 population doublings and had reached stationary phase. The 24-h liquid cultures (passage 1) were diluted 1:1000 into fresh YPAD liquid and grown again for 24 h at 30°C (passage 2). Single cells from each passage were spotted onto solid YPAD media and grown for 24 h at 30°C to quantitate and visualize individual cell growth. Six serial cultures were generated by successive dilution (1:1000) of the previous 24-h culture. Images of colonies from each passage were captured with a Nikon Cool Pix 900 digital camera (Melville, NY) mounted on a Zeiss stereoscope Stemi DRC (Sterling Heights, MI).
Cells that remained from the serial liquid cultures were prepared for DAPI staining by dilution into fresh YPAD medium and growth at 30°C for 6 h. Cells were fixed with 75% ethanol for 30 min, washed one time with water, and stained with 1 ng/ml DAPI for 24 h at 4°C before visualization of nuclear DNA by fluorescence microscopy. For each strain and passage, >400 cells from each independent isolate were scored for nuclear position and bud morphology. Chi square values for each data set were calculated (Snedecor and Cocharan, 1980) to test the null hypothesis that each data set would contain a single mean (i.e., that passaging the cells would not alter their cell cycle distribution).
RESULTS
Progressive Decrease in Colony Size During Senescence Is Due to a Decreased Rate of Cell Division
Yeast strains that incur DNA damage or are deficient in proteins required for DNA replication undergo an abrupt cell cycle arrest (Zhou and Elledge, 2000). Telomerase-deficient yeast, however, undergo replicative senescence and a progressive decline in viability (Lundblad and Szostak, 1989; McEachern and Blackburn, 1996; Nakamura et al., 1997). Senescence may be due to a reduction in the rate of cell division or to an increase in the rate of cell death events. To examine the process of senescence in telomerase-deficient yeast, we isolated mutant cells lacking TLC1, the RNA component of telomerase, by sporulating a diploid tlc1Δ/TLC1 strain (Singer and Gottschling, 1994).
Six independent tlc1Δ isolates were serially subcultured by restreaking them on plates every 24 h for 10 consecutive days. We refer to each serial subculture as a “plate passage” to distinguish these experiments from the later assays utilizing liquid cultures. For reference, wild-type cells undergo about six doublings during a single plate passage. As expected, all tlc1Δ spores senesced, despite some variation in the timing of senescence. This variation was a property associated with each individual spore because early or late senescence of specific spore progeny was reproducible. Cultures were plated at low density and examined after 24 h to determine if cells in the senescent population were either failing to divide or were dividing at a reduced rate. We followed one representative spore in detail.
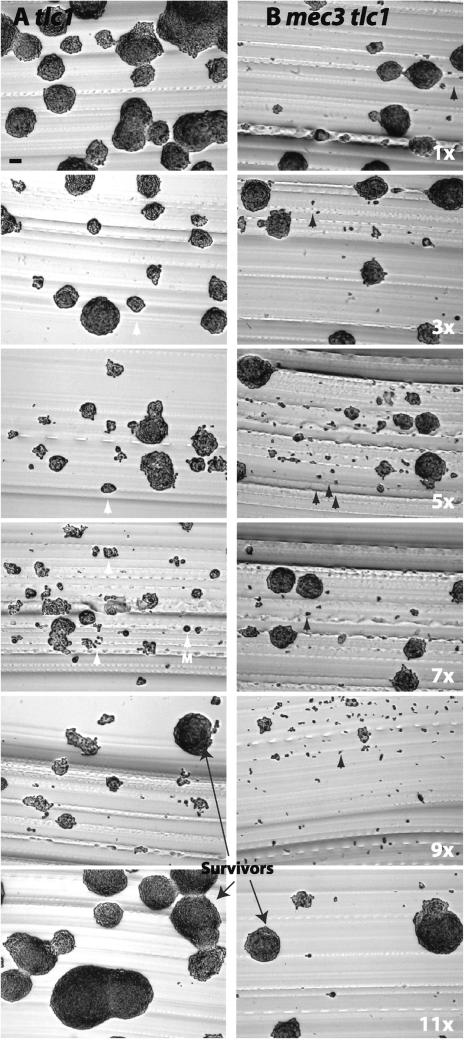
tlc1Δ cells streaked directly from the tetrad dissection plate (founder cells) formed uniform sized colonies that were nearly wild type in colony size (Figure 1A). However, cells from serial plate passages 1–4 formed colonies that were progressively smaller with each passage. Importantly, the smaller colonies (white arrows) were not accompanied by the presence of significant numbers of dead cells, which could have accounted for the colony size decrease (as seen with mec3Δ tlc1Δ, Figure 1B). This implies that cell death is not a frequent event during early plate passages in telomerase-deficient cells.
Figure 1.
Senescence of tlc1Δ and mec3Δ tlc1Δ strains during serial plate passages. (A) Diploid strain YJB 2768 was sporulated and tlc1Δ progeny were isolated. Photomicrographs were taken of successive passages on YPAD plates restreaked onto a fresh plate and grown at 30°C for 24 h. White arrow heads, small senescing colonies; M, monster cells. (B) Diploid strain YJB4565 was sporulated, mec3Δ tlc1Δ progeny were isolated and grown as in A. Black arrowheads, single cells that did not divide during the course of the plate outgrowth. Note the absence of monster cells in mec3Δ tlc1Δ cultures. Bar, 0.2 mm. Thin arrows, survivor colonies that form in later plate passages. ×, number of plate passages.
Plate passages 4–5 exhibited the lowest levels of viability, as determined by the growth of the population. On solid medium, a mixture of small colonies and large colonies was evident. One striking feature of the small colonies was that they contained cells that had irregular shapes and very large sizes (Figure 1A, arrow marked M). Some of these individual conspicuous cells were about five times larger than individual wild-type cells, implying that the cells increased in cell size in the absence of cell division. We conclude that the primary reason for senescence during the early passages of telomerase-deficient cells is not due to an increase in cell death but rather appears to be caused by a reduced rate of cell division in the population.
In addition to the small colonies composed of irregular, large cells that began to appear in plate passages 4–5, colonies with wild-type-like size appeared in later plate passages. Eventually, either the culture became inviable or wild-type-like colonies became predominant in the culture. The wild-type-like colonies were “survivors” (Lundblad and Blackburn, 1993) that had undergone recombination events at their telomeres (our unpublished results). We focused our attention on the senescence events that occurred before the appearance of these survivors.
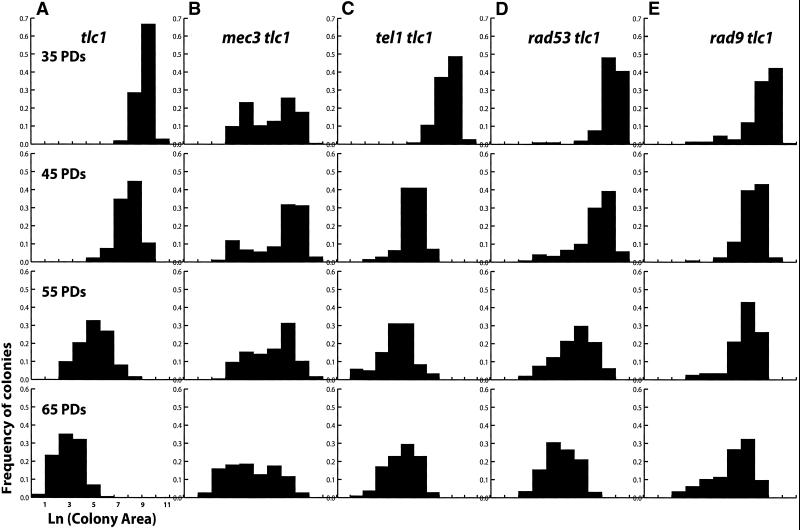
The telomere checkpoint model suggests that a cell cycle delay is triggered when one or more telomeres become critically short. It follows that an increased number of short telomeres may trigger a longer cell cycle delay. Late passage cells lacking telomerase should have more critically short telomeres than from earlier passages. To test this model, we measured and compared the rate of cell division in telomerase-deficient cells from different passages. Colony area was used as a quantitative indicator of the number of divisions the tlc1Δ cells were able to complete (i.e., growth rate). Analysis of wild-type colony sizes indicated a good relationship between colony area and the number of cells per colony (see MATERIALS AND METHODS). Small colony areas indicate that the senescing tlc1Δ cells underwent fewer divisions and therefore had a slower growth rate. For these experiments, we serially cultured tlc1Δ cells in liquid media so that they were limited to a maximum of ∼10 population doublings per passage (“liquid passage”). Cells from these cultures were then plated on solid medium and grown for 24 h, and the colony area was measured. These measurements were plotted as a histogram on a semilog scale (Figure 2A).
Figure 2.
Quantitative measurement of colony area during senescence in liquid passages. Cells were advanced by 10 population doublings in liquid culture and then plated onto YPAD plates. PDs, the average number of population doublings in each culture. Colony area was measured and converted to a natural log scale, expressed as arbitrary units of colony area. Wild-type colonies are typically 4.9 U at 8 h, 5.3 U at 12 h, and 6.8 U at 15.5 h. (A) Colony area after 24 h of growth at 30°C was performed on tlc1Δ isolates from diploid strain YJB2768. Note that the tlc1Δ median colony area decreases progressively with a Gaussian distribution. (B) Colony area for mec3Δ tlc1Δ isolates from diploid strain YJB4565. Note the bimodal distribution of colony areas. (C) Colony area for tel1Δtlc1Δ isolates from diploid strain YJB7448. (D) Colony area for rad53-K227A tlc1Δ isolates from diploid strain YJB6690. (E) Colony area for rad9Δ tlc1Δ isolates from diploid strain YJB3867.
For tlc1Δ progeny in the first liquid passage, the colony area after the log transformation resembled a Gaussian distribution with a single peak. The shape of the curve and the narrow range of colony areas suggest that members of the population behave in a similar manner. In subsequent liquid passages, the colony size distribution curve of the tlc1Δ spore progeny continued to exhibit a Gaussian distribution with a single peak. However, the mean colony size decreased with each successive liquid passage (Figure 2A). The mean colony size as well as the sizes of the largest and smallest colonies decreased in a gradual, progressive manner with each liquid passage. No abrupt transition from the wild-type growth rate to the reduced division rate was evident. These results indicate that (1) the division rate of the entire population of cells was decreasing and (2) the division rate continued to decrease with increasing numbers of passages.
From these experiments we conclude that senescence is primarily a progressive reduction in cell division rate that correlates with the number of population doublings that have occurred in the absence of telomerase. The progressive nature of the reduction in average cell division rate is consistent with the idea that critically short telomeres signal a need for cell cycle delay and that, as cells continue to divide in the absence of telomerase, more telomeres per cell reach this critically short length. In this case, the signal becomes stronger, resulting in a progressively longer cell cycle delay. It is also possible that the number of cells in the population that undergo a cell cycle delay increases as the number of cell divisions after loss of telomerase increases.
The Decreased Rate of Cell Division in tlc1Δ Cells Is Due to a G2/M Delay
To determine if the tlc1Δ cells exhibit a slower average division rate because of a specific cell cycle arrest event, we analyzed the cell cycle distribution of tlc1Δ cells during senescence. Unbudded cells, cells with a small bud and a single nucleus, and attached cells with two nuclei (that have completed mitosis) are considered to be in G1 or S phase. G2/M cells are those that contain a large bud with a single nucleus near or spanning the mother-bud neck. Several spores were selected for each strain and followed over serial passages (as indicated below). Greater than 400 tlc1Δ cells per passage, from each individual isolate, were stained with DAPI and observed by microscopy.
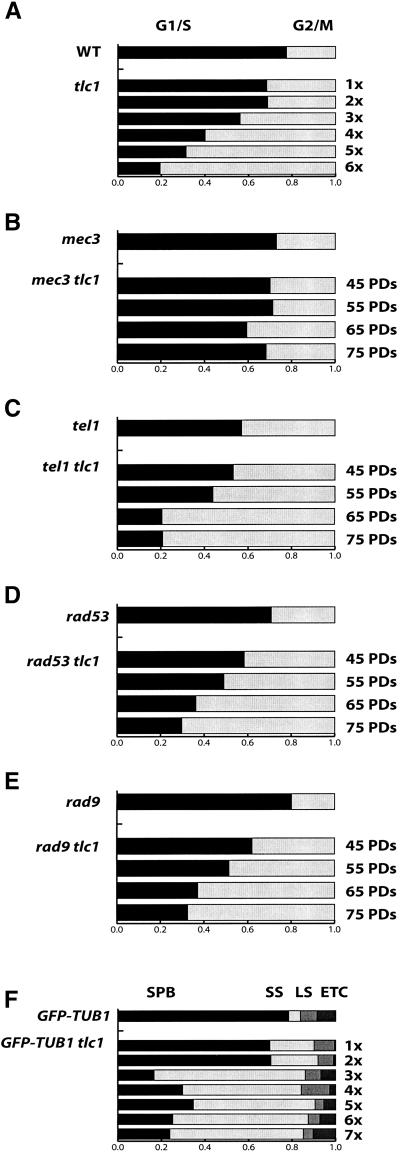
During plate passages 1–3, the cell cycle distribution of tlc1Δ cells was similar to that of wild-type cells (Figure 3A). As cell division slowed in plate passages 4–6, the proportion of tlc1Δ cells in G2/M phase increased progressively. By plate passage 6, monster cells made up 10% of the population (Figure 1A and our unpublished results). They often contained multiple large buds and fragmented, condensed DNA. Monster cells were excluded from the cell cycle distribution analysis because they do not have normal cell cycle landmarks. Monster cells are likely the result of cells increasing in volume while dividing only rarely because they are held in a cell cycle-delayed state (Reed, 1980; Hadwiger et al., 1989; Richardson et al., 1989). The population of tlc1Δ cells in G2/M increased progressively, and by plate passage 6, 80% of tlc1Δ cells (excluding monsters) were in G2/M.
Figure 3.
Senescing cultures exhibit an increase in the proportion of cells in the G2/M phase of the cell cycle. ×, the number of plate passages. PDs, the number of culture population doublings in liquid passages. Each bar graph represents the nuclear morphology scored on at least 400 DAPI stained cells from a single passage. Serially passaged cultures were derived from a single spore isolate. (A–E) Proportion of wild-type and tlc1Δ cells with a small bud and single nucleus (G1/S, black bar) or a large bud and a single nucleus near or stretched through the mother-bud neck (G2/M, gray bar). (A) Both wild-type and tlc1Δ spores were isolated from YJB2768. (B) mec3Δ and mec3Δ tlc1Δ cells isolated from YJB4565. (C) tel1Δ and tel1Δtlc1Δ cells isolated from YJB7448. (D) rad9Δ and rad9Δ tlc1Δ cells isolated from YJB3867. (E) rad53K227A and rad53K227A tlc1Δ cells isolated from YJB6690. (F) wild-type and tlc1Δ strains expressing GFP-tubulin isolated from YJB 6741. Unduplicated or duplicated spindle pole bodies (SPB, black), short preanaphase spindles (SS, light gray), long anaphase spindles (LS, medium gray), and aberrant spindle forms (ETC, dark gray). Chi square analysis was performed to determine if serially passaged cells altered their cell cycle distribution. tlc1Δ, rad53K227A tlc1Δ, and rad9Δ tlc1Δ mutants underwent a significant change in cell cycle distribution during senescence (p < 0.005). The mec3Δ tlc1Δ strain did not exhibit significant change in cell cycle distribution (p > 0.1) until after 55 PDs.
To determine if cells with the nucleus at, or spanning the neck, had initiated anaphase, we used fluorescently tagged tubulin (GFP-Tub1p) to examine spindle morphology in tlc1Δ mutant cells. Approximately 60% of cells from passages 3–6 exhibited “partially elongated spindles,” spindles with a length intermediate between the typical short (S/G2) and typical long (M phase) spindles of wild-type cells (Figure 3F). These partially elongated spindles suggest that the tlc1Δ mutants delay before the metaphase-to-anaphase transition. Monster cells often contained partially elongated spindles as well (our unpublished results).
The decreased colony expansion rate, the accumulation of cells with nuclei near or spanning the mother-bud neck, and the prevalence of partially elongated spindles in tlc1Δ cells indicates that telomerase-deficient cells exhibit a significant delay in the G2/M stage of the cell cycle that increases progressively with the number of passages after loss of TLC1. This observation is consistent with the idea that a checkpoint triggers a cell cycle arrest in response to telomere defects. Furthermore, this checkpoint functions in the absence of TLC1, the telomerase RNA. We observed similar results with est1, est2, est3, and cdc13–2 cells (our unpublished results). Thus, neither the components of the telomerase enzyme, nor its regulators, are required to activate this telomere checkpoint.
The DNA Damage Checkpoint Protein, MEC3, Is Required for the G2/M Delay of Senescence
Strains lacking a specific checkpoint component fail to arrest the cell cycle in response to the damage signal that normally activates that checkpoint. Checkpoint-deficient cells die as a consequence of the damage (Weinert and Hartwell, 1988). Accordingly, a strain lacking both telomerase and a component of the telomere checkpoint should fail to arrest the cell cycle and should die as a consequence of the lack of telomerase. Thus, a double mutant lacking both TLC1 and a telomere checkpoint component should not exhibit the reduced division rate, the G2/M delay, or the prevalence of monster cells seen in tlc1Δ cells. Rather, the primary mode of senescence seen in the checkpoint-deficient tlc1Δ mutants is expected to be an increase in cell death due to cell division in the presence of critically short telomeres.
Mec3p is a checkpoint protein required for arrest in the G2/M phase of the cell cycle in response to DNA damage. It participates with Ddc1p, Rad17p, and Rad24p to prevent cells from completing cellular division when nuclear DNA has been damaged (Weinert, 1992). To ask if Mec3p is involved in a telomere checkpoint in response to critically short telomeres, we isolated mec3Δ tlc1Δ strains from a diploid parent heterozygous at both loci. Seven independent mec3Δ tlc1Δ spores were cultured by serial plate passages (Figure 1B). All mec3Δ tlc1Δ cultures eventually became inviable or gave rise to survivors after multiple passages in both liquid and solid media, indicating that mec3Δ tlc1Δ cells, like tlc1Δ cells, undergo senescence. Because mec3Δ TLC1 strains are viable, senescence and death are presumably due to the lack of telomerase in mec3Δ tlc1Δ cells.
We then followed a representative isolate in detail. When observed at the cellular level, senescing mec3Δ tlc1Δ cultures were clearly different from tlc1Δ cells. Immediately after the loss of telomerase, the mec3Δ tlc1Δ founder cells formed colonies that were indistinguishable in size from the mec3Δ, tlc1Δ and wild-type sibling progeny (our unpublished results). However, as early as plate passage 1, mec3Δ tlc1Δ cells formed two distinct types of colonies: large colonies (>1000) cells and microcolonies (<10 cells; Figure 1B). The frequency of microcolonies within the population increased with successive plate passages (Figure 1B, black arrows). This suggests that mec3Δ tlc1Δ cells were not undergoing the uniform, progressive reduction in division rate that was seen in tlc1Δ cells (Figure 1A). Rather, a subpopulation of the mec3Δ tlc1Δ cells grew at a normal rate, forming colonies that were larger than the corresponding colonies in the tlc1Δ cultures, whereas the other subpopulation of cells exhibited a high death rate (as early as plate passage 1). Thus, MEC3 is required for the high levels of viability in early plate passages of tlc1Δ cells.
Quantitative analysis of the colony area distribution of serial liquid passages of a single isolate also revealed differences in the mec3Δ tlc1Δ colonies relative to tlc1Δ cultures. In contrast to the Gaussian distribution of MEC3 tlc1Δ colony sizes, the mec3Δ tlc1Δ colonies exhibited a bimodal distribution, even in early liquid passages (Figure 2B). A subset of the population continued to produce colonies with a near-wild-type expansion rate (right side of the distribution curve, Figure 2B). Even in early liquid passages, a large population of the mec3Δ tlc1Δ population exhibited reduced colony size, eventually producing primarily cells that never divided (left end of the distribution curve). Thus, rather than exhibiting a progressively decreasing division rate, early liquid passage mec3Δ tlc1Δ cells grew either at a wild-type rate or failed to divide. Furthermore, the population of dead cells in mec3Δ tlc1Δ cultures (measured by vital staining of YJB4565 spore progeny, our unpublished results) increased with successive passages after the loss of telomerase activity.
The cell cycle distribution of mec3Δ tlc1Δ cells during senescence was determined from cell shape and nuclear distribution measurements (Figure 3B) as performed on the tlc1Δ cells. In contrast to tlc1Δ mutants, no dramatic increase in G2/M cells was observed. The mec3Δ tlc1Δ liquid passages contain ∼30% of cells classified as G2/M, and mec3Δ tlc1Δ cells exhibited similar distributions of cell cycle stages throughout most of the senescence process. We do observe a small but reproducible increase in G2/M cells at ∼65 PDs after loss of telomerase, which may indicate that a Mec3p-independent response occurs at a later stage in the senescence process. In plate passage 4 (Figure 1B), the largest cells in mec3Δ tlc1Δ cultures had a diameter twice that of wild-type cells, whereas tlc1 monster cells had up to a fivefold increase in diameter, indicating that mec3Δ tlc1Δ cells do not accumulate monster cells and suggesting that mec3Δ tlc1Δ do not exhibit the major G2/M delay seen in tlc1Δ cells. This is consistent with the idea that mec3Δ tlc1Δ cells either divided and formed large colonies, died as individual cells, or died after several divisions, forming microcolonies. Taken together with the colony analysis, these data indicate that MEC3 is required for the progressively reduced colony size, monster cell formation, and the G2/M delay observed during senescence in cells that lack telomerase. Therefore, MEC3 is a candidate for a component of the telomere checkpoint pathway.
MEC1 and DDC2 Are Also Required for the Cell Cycle Arrest of Telomerase-deficient Cells
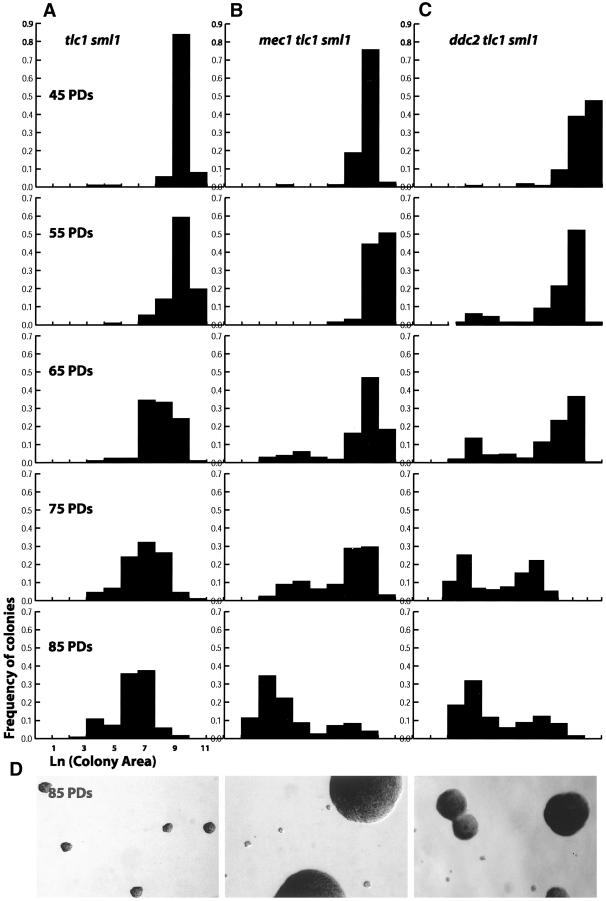
MEC1 and DDC2/LCD1/PIE1 are two essential genes that have a central role in the DNA damage response (Paciotti et al., 2000; Rouse and Jackson, 2000; Wakayama et al., 2001). Mec1p is an ATM-like kinase required for the DNA damage checkpoint response. Ddc2p forms a physical complex with Mec1p and regulates the Mec1p kinase activity (Paciotti et al., 2000; Rouse and Jackson, 2000; Wakayama et al., 2001). When Sml1p, which regulates nucleotide pool levels, is absent, mec1Δ sml1Δ and ddc2Δ sml1Δ strains are viable (Zhao et al., 1998). We first asked if Sml1p affects the process of senescence in tlc1Δ mutants by measuring colony area after successive passages in liquid medium. Like tlc1Δ strains, tlc1Δ sml 1Δ strains exhibited a normal distribution of colony sizes that progressively decreased in area with successive liquid passages (Figure 4A). Consistent with previous reports (Ritchie et al., 1999), loss of Sml1p caused a delay in the senescence relative to tlc1Δ (our unpublished results). Yet, tlc1Δ sml1Δ cells, like tlc1Δ cells, clearly exhibited a progressive increase in the population of G2/M cells with successive passages (Figure 5A, from 30 to 60% G2/M) when nuclear morphology was examined by DAPI staining. Furthermore, monster cells were evident in passages 5–8 (our unpublished results). Thus, tlc1Δ sml1Δ mutants, like tlc1Δ strains, exhibit a significant G2/M delay during the senescence process, and the sml1Δ-mediated delay in the onset of senescence did not affect the process of senescence.
Figure 4.
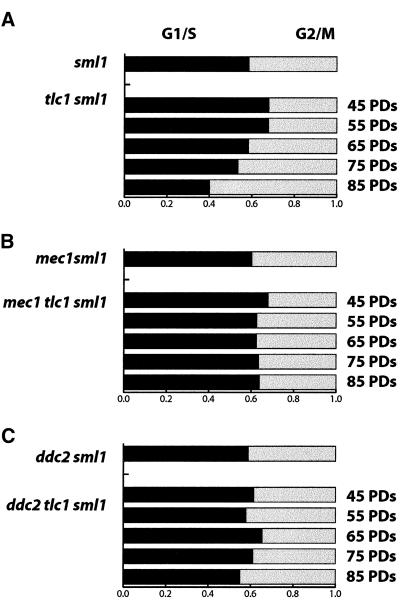
MEC1 and DDC2 are required for the cell cycle delay in senescing populations of telomerase-deficient cells. Quantitative measurements of colony areas were determined as described in the legend to Figure 2. (A) tlc1Δ sml1Δ isolates were spore progeny from strain YJB6689. (B) mec1Δ tlc1Δ sml1Δ isolates were from diploid strain YJB6689. (C) ddc2Δ tlc1Δ sml1Δ isolates were from strain YJB6361. (D) Representative colony populations of tlc1Δ sml1Δ, mec1Δ tlc1Δ sml1Δ, and ddc2Δ tlc1Δ sml1Δ cultures measured at 85 PDs in the histograms directly above the photomicrographs.
Figure 5.
MEC1 and DDC2 are required for the G2/M cell cycle delay in telomerase-deficient cells during senescence. The population of cells in G1/S (black bar) or G2/M (gray bar) were determined as described in the legend to Figure 3. (A) tlc1Δ and tlc1Δ sml1Δ isolates were spore progeny from YJB6689. Nuclear morphology was scored on at least 400 DAPI-stained cells for each passage and genotype. (B) mec1Δ sml1Δ and mec1Δ tlc1Δ sml1Δ isolates were from YJB6689. (C) ddc2Δ sml1Δ and ddc2Δ tlc1Δ sml1Δ isolates were from YJB6361. Chi square analysis was performed as described in the legend to Figure 3. tlc1Δ sml1Δ demonstrated a significant (p < 0.005) change during serial passages, whereas no significant change (p > 0.1) was detected for the mec1Δ tlc1Δ sml1Δ and ddc2Δ tlc1Δ sml1Δ passages.
To determine if MEC1 and DDC2 contribute to the telomere checkpoint pathway that causes a cell cycle delay in telomerase-deficient cells, we compared the senescence of mec1Δ tlc1Δ sml1Δ and ddc2Δ tlc1Δ sml1Δ cells with tlc1Δ sml1Δ cultures. We examined seven independent spores of each genotype by successive plate passages (our unpublished results) and followed one representative spore of each genotype in detail by serial liquid passages and measurements of colony area. The mec1Δ tlc1Δ sml1Δ and ddc2Δ tlc1Δ sml1Δ mutants senesced more like the mec3Δ tlc1Δ mutants described above (Figure 1B), with microcolonies and dead cells appearing in early passages and increasing in frequency with serial passage of the cultures (Figure 4, B and C). Some cells continued dividing and formed large colonies throughout the senescence process, indicating that a subpopulation of the mec1Δ tlc1Δ sml1Δ and ddc2Δ tlc1Δ sml1Δ mutants grew at a normal rate (Figure 4, B and C). In addition, neither the mec1Δ tlc1Δ sml1Δ nor the ddc2Δ tlc1Δ sml1Δ cultures exhibited an obvious accumulation of cells in the G2/M phase of the cell cycle (Figure 5, B and C) or the appearance of monster cells. Taken together the reduced colony expansion rate and G2/M accumulation data, these results indicate that MEC1 and DDC2 are required during senescence in telomerase-deficient cells for cell cycle arrest and monster cell formation.
TEL1, an ATM Kinase, Is not a Telomere Checkpoint Component
Tel1p is an IP3 kinase that is most similar to MEC1 and to the human ATM kinase. TEL1, along with MEC1, is required for maintenance of telomere length: double mutants have extremely short telomeres and undergo senescence despite having functional telomerase (Ritchie et al., 1999; Craven and Petes, 2000). Recent experiments have implicated TEL1 as a telomere adapter for the MRX complex that helps recruit telomerase to the telomere (Diede and Gottschling, 2001; Tsukamoto et al., 2001). In addition, TEL1 can act as a DNA damage sensor that activates RAD9 and RAD53 independent of either MEC3 or MEC1 (D'Amours and Jackson, 2001; Grenon et al., 2001; Usui et al., 2001). Therefore, TEL1 appeared to be a good candidate for a telomere checkpoint sensor.
We constructed tel1Δ tlc1Δ heterozygous diploids and examined 10 independent progeny after sporulation. After plate passages, each of the progeny formed colonies that, like the tlc1Δ mutant, were progressively smaller with each passage (our unpublished results). One segregant was examined in detail, and colony area was measured after advance in serial liquid cultures. The resulting histogram of colony area (Figure 2C) contained a single peak that, as in tlc1Δ cells, moved progressively toward the left side of the distribution (Figure 2A). The cell cycle distribution of this segregant was also measured. The tel1Δ tlc1Δ cells demonstrated a gradual increase in the proportion of cells in G2/M, with 47% in the earliest passage and 79% in the last passage, compared with 42% in the tel1Δ single mutant (Figure 3C). In addition, later passages of tel1Δ tlc1Δ cells contained monster cells (our unpublished results). Taken together, these data indicate that, in contrast to MEC1, TEL1 is not required for the cell cycle arrest in response to shortening telomeres.
RAD53 and RAD9 Are not Required for the Telomere Checkpoint
Both the telomere checkpoint and DNA damage checkpoint utilize Mec3p, Mec1p, and Ddc2p. To ask if the cell cycle arrest caused by a lack of telomerase is a specific “telomere checkpoint” response or a general DNA damage response that occurs after the telomeres become uncapped (Blackburn, 2001) and the ends become detected as double-strand breaks or single-stranded DNA, we examined the role of the DNA damage checkpoint genes, RAD53 and RAD9, in response to telomere damage caused by a lack of telomerase.
Rad53p is an essential protein kinase that is the central signal transducer in the DNA damage response pathway (Longhese et al., 1998; Zhou and Elledge, 2000). Rad53p activates the transcriptional response to damage and is also required for cell cycle arrest at G2/M, presumably through its kinase activity. The hypomorphic rad53-K227A allele contains a point mutation in the protein kinase domain that eliminates the Rad53p-dependent DNA damage response while not eliminating the essential functions of Rad53p (Fay et al., 1997). After sporulation of the appropriate heterozygous diploid strain, tlc1Δ rad53K227A progeny were passaged on solid (our unpublished results) and in liquid media as described for tlc1Δ mutants. Like tlc1Δ colonies, the rad53K227A tlc1Δ progeny exhibited a progressive decline in colony area that resembled the dynamics of colony area reduction seen in the tlc1Δ strains (Figure 2D). In addition, dead cells appeared in passage 2 and increased in numbers with successive passages (Figure 2D). The increased numbers of dead cells may occur because the kinase activity of Rad53p is required to keep the tlc1Δ cells alive during the arrest in response to loss of telomerase activity. Nonetheless, the viable tlc1Δ rad53-K227A cells gave rise to progressively smaller colonies with successive liquid passages, suggesting that the decline in division rate is similar to that seen in tlc1Δ cultures (Figure 2A). Like tlc1Δ mutants, the tlc1Δ rad53-K227A strains also accumulated a large proportion of G2/M cells in successive passages (Figure 3D, 70% at 75 PDs), indicating that Rad53p kinase activity is not required for the G2/M cell cycle delay in telomerase-deficient cells.
The RAD9 signaling response is independent of the MEC3 pathway, although both pathways activate RAD53 (Zhou and Elledge, 2000). Previous work demonstrated that RAD9 was required to arrest cells with an engineered double-strand break located adjacent to a telomere (Sandell and Zakian, 1993). We asked if RAD9 was also required for the cell cycle arrest that occurs during senescence in telomerase-deficient cells. Like tlc1Δ and rad53-K227A tlc1Δ strains, rad9Δ tlc1Δ strains initially formed large colonies that progressively decreased in average size with successive liquid passages (Figure 2E), indicating that rad9Δ tlc1Δ strains experience a reduction in cell division rate during senescence. We note that average colony size did not decrease as rapidly in rad9Δ tlc1Δ strains as it did in the tlc1Δ strains. Nonetheless, rad9Δ tlc1Δ cells accumulated in G2/M with successive passages (Figure 3E, 68% at 75 PDs). Thus Rad9p is not required for the G2/M delay observed during senescence in telomerase-deficient cells. In the tlc1Δ rad9Δ strains, dead cells were present at levels much higher than in otherwise wild-type tlc1Δ cultures. Thus, Rad9p, like the Rad53 kinase activity, appears to be important for maintaining cell viability during senescence. However, neither Rad9p, nor the kinase activity of Rad53p, are required for the cell cycle delay observed in viable telomerase-deficient cells, indicating that the telomere checkpoint is distinct from the general DNA damage checkpoint response.
DISCUSSION
In the absence of telomerase, cells undergo senescence, a process primarily caused by a reduced rate of cell cycle progression during the first several passages of growth. A telomere checkpoint causes a cell cycle delay with the majority of cells exhibiting partially elongated spindles, indicative of a late G2 or early M phase arrest. This delay or arrest is dependent on Mec3p, Mec1p, and Ddc2p, which are also components of the DNA damage checkpoint response. Loss of any one of these checkpoint genes eliminates the progressive increase in cells delayed in G2/M such that cells either continue to divide in the absence of telomerase activity or die, presumably because their telomeres have eroded.
In telomerase-deficient cultures, colonies become progressively smaller and the extent of the G2/M delay in the population increases. Very large monster cells, which continue to increase in size while dividing or budding only rarely, become conspicuous. These observations imply that telomerase-deficient cells spend an increasing amount of time in G2/M as a function of increasing numbers of population doublings after the loss of telomerase. One model to explain this is that a single defective (or critically short) telomere is sufficient to activate a checkpoint-mediated G2/M delay signal and that as more telomeres become defective, the signal becomes proportionally stronger, resulting in a longer G2/M delay. In telomerase-deficient cells, critically short telomeres cannot be repaired. Thus the level of irreparable damage (the numbers of critically short telomeres) increases with increasing passages.
Interestingly, the G2/M arrest in telomerase-deficient cells does not require RAD9 or RAD53. This is surprising because a double-strand break introduced near the telomere triggers a RAD9-dependent arrest (Sandell and Zakian, 1993). The rate at which rad9Δ tlc1Δ colonies become smaller is not as dramatic as the rate of tlc1Δ colony size reduction. This could be because cell cycle delays (in addition the G2/M delay that occurs in both strains) occur in tlc1Δ but not in rad9Δ tlc1Δ cells. Furthermore, rad53-K227A strains were able to activate the telomere checkpoint (Figure 3C), yet they do not activate the DNA damage checkpoint (Fay et al., 1997). We observed similar results with another rad53 allele (sad1-1, our unpublished results). Thus, the genetic requirements for the senescence response to a loss of telomerase, or to loss of critical telomere components (e.g., Est1p and Est3p) are distinct from the genetic requirements for the response to other forms of DNA damage, including the extensive single-stranded DNA generated in cdc13-1 mutants (Gardner et al., 1999). This suggests that a critically short telomere is not perceived as a double-strand DNA break (DSB) and that it elicits a response different than the response elicited by a DSB induced only six base pairs away from the telomere (Sandell and Zakian, 1993). The differences between telomere ends and DSBs is likely due to the constellation of telomere-associated proteins, including Ku70/Ku80, the MRX complex, and the Est proteins that ensure that a normal telomere is not a substrate of DNA repair activities (Dubrana et al., 2001).
Because TEL1 plays a dual role in both telomere maintenance and the DNA damage checkpoint pathway, it might be expected to play a central role in a telomere checkpoint. Surprisingly, TEL1 is not required for the cell cycle arrest at G2/M in response to eroding telomeres. The MRX complex (MRE11, RAD50, XRS2) acts with TEL1 to maintain telomere length and an intact DNA damage checkpoint (Ritchie and Petes, 2000; D'Amours and Jackson, 2001; Grenon et al., 2001; Tsukamoto et al., 2001; Usui et al., 2001). We also observed that rad50Δ tlc1Δ mutants, like tlc1Δ cultures, formed progressively smaller colonies during senescence (our unpublished results). This indicates that the MRX complex also is not required for the telomere checkpoint. The MRX complex is thought to activate cell cycle arrest in response to DNA damage by sending signals that feed into the RAD53-RAD9 pathway (Grenon et al., 2001; Usui et al., 2001). Our results indicate that the telomere checkpoint does not initially utilize TEL1, RAD53, or RAD9. We do not, however, discount the involvement of these proteins in the overall response, because there are differences in colony size distributions between tlc1Δ and tel1Δ tlc1Δ, rad53Δ tlc1Δ, or rad9Δ tlc1Δ (Figures 2 and 4). We suggest that initial telomere erosion activates the MEC3-MEC1-DDC2--dependent telomere checkpoint, resulting in a G2/M delay. As telomere erosion continues into later passages, a secondary event likely triggers the more conventional DSB damage response.
Our results can be explained by the telomere checkpoint model (Figure 6), which posits that, in wild-type cells, shorter telomeres elicit a checkpoint response that targets them for elongation by telomerase. When telomeres become critically short, the telomere checkpoint delays cell cycle progression, presumably to activate and/or recruit telomerase to the short telomeres. In contrast, the DNA damage checkpoint, mediated by Tel1p, Rad9p, and Rad53p, activates DNA repair activities (Usui et al., 2001, reviewed in Longhese et al., 1998; Zhou and Elledge, 2000). Consistent with the telomere checkpoint model, when a telomere is short, the kinetics of telomere elongation is initially fast, but as telomere length approaches the average wild-type size, the rate of telomere elongation is reduced (Marcand et al., 1999; Ray and Runge, 1999). The telomere checkpoint model may be conserved through evolution because telomere elongation in mouse cells is specifically targeted to the shortest telomeres (Hemann et al., 2001).
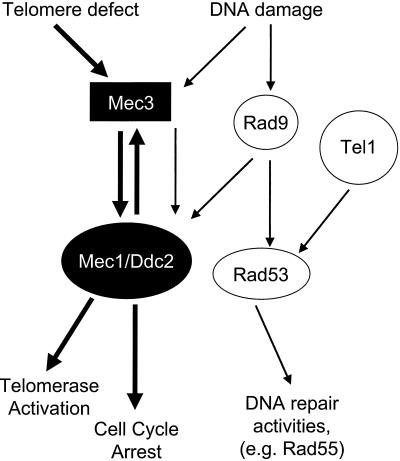
Figure 6.
The Telomere Checkpoint model. A defect in the telomere, such as a critically short telomere, triggers the checkpoint pathway (thick black arrows), which requires the activity of Mec3p, Mec1p, and Ddc2p. This leads to cell cycle arrest followed by activation and/or recruitment of telomerase to the short telomeres. The DNA damage pathway (thin black arrows) signals to Mec3p, Mec1p, and Ddc2p and a parallel pathway involving Tel1p is activated. The two DNA damage pathways converge at Rad9p and Rad53p. Activation of the DNA damage checkpoint leads to cell cycle arrest and activation of DNA repair activities but does not activate telomerase.
Recent work on checkpoints indicates that cell cycle arrest is coordinated with the activation of appropriate cellular responses such as DNA repair pathways (Zhou and Elledge, 2000). For example, Ddc1p and Ddc2p are recruited to sites of double-strand breaks (Kondo et al., 2001; Melo et al., 2001) where they presumably recruit repair activities. For example, in response to DNA damage, the Mec1p-Ddc2p and the Mecp3-Rad17p-Ddc1p complexes are recruited independently of each other, and yet both complexes are found at a DNA lesion (Melo et al., 2001).
Similarly, the telomere checkpoint must have at least two roles at telomeres. First, as demonstrated in this work, it arrests cells at G2/M. Second, we propose that the telomere checkpoint recruits telomerase to chromosome ends, especially those that are critically short, by a mechanism that involves Mec1p, Ddc2p, and Mec3p. Support for the latter role comes from studies of Mec1p, which is required for telomere elongation and the recruitment and/or activation of telomerase in the absence of Tel1p (Ritchie et al., 1999; Tsukamoto et al., 2001), although it does not affect the levels of soluble telomerase activity in the cells (Chan et al., 2001). An intriguing question is how proteins such as Mec1p, Mec3p, and Ddc2p, which are components of both the DNA damage checkpoint and the telomere checkpoint, distinguish between lesions such as DSBs, where DNA repair activities are deployed, and critically short telomeres, where telomerase is recruited and/or activated.
In telomerase-deficient cells, recombination events that occur in later passages (after 85 PDs) eventually lead to the formation of survivors. Because survivors grow faster than senescing cells, this implies that activities that induce survivor formation are late events that are induced only after failed attempts at telomerase recruitment in the early passages. Furthermore, the induction of survivors, which requires Rad52p as well as Rad51p and/or Mre11p/Rad50p/Xrs2p (Le et al., 1999; Teng et al., 2000; Chen et al., 2001), does not require the telomere checkpoint genes: survivors arose in the later passages of all strains studied in this work (Figure 1). Thus, cells induce these recombination activities, which give rise to survivors, only as a last resort mechanism to maintain viability.
ACKNOWLEDGMENTS
We thank M.P. Longhese, M. Foiani, O. Tsuchyia, T. Petes, and K. Blumer for yeast strains; Duncan Clarke and members of the Berman laboratory for many helpful discussions; and Kyle Kilburn for the initial observations of senescing cultures. This work was supported by National Institutes of Health grants GM 38626 (J.B.) and F32 GM 63352 (L.G.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–02–0012. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–02–0012.
REFERENCES
- Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Telomere states and cell fates. Nature. 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- Chan SW, Chang J, Prescott J, Blackburn EH. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr Biol. 2001;11:1240–1250. doi: 10.1016/s0960-9822(01)00391-8. [DOI] [PubMed] [Google Scholar]
- Chen Q, Ijpma A, Greider CW. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol. 2001;21:1819–1827. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RJ, Petes TD. Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics. 1999;152:1531–1541. doi: 10.1093/genetics/152.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RJ, Petes TD. Involvement of the checkpoint protein Mec1p in silencing of gene expression at telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:2378–2384. doi: 10.1128/mcb.20.7.2378-2384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 2001;15:2238–2249. doi: 10.1101/gad.208701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- Dubrana K, Perrod S, Gasser SM. Turning telomeres off and on. Curr Opin Cell Biol. 2001;13:281–289. doi: 10.1016/s0955-0674(00)00210-6. [DOI] [PubMed] [Google Scholar]
- Fay DS, Sun Z, Stern DF. Mutations in SPK1/RAD53 that specifically abolish checkpoint but not growth-related functions. Curr Genet. 1997;31:97–105. doi: 10.1007/s002940050181. [DOI] [PubMed] [Google Scholar]
- Gardner R, Putnam CW, Weinert T. RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J. 1999;18:3173–3185. doi: 10.1093/emboj/18.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenon M, Gilbert C, Lowndes NF. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat Cell Biol. 2001;3:844–847. doi: 10.1038/ncb0901-844. [DOI] [PubMed] [Google Scholar]
- Hadwiger JA, Wittenberg C, Richardson HE, de Barros Lopes M, Reed SI. A family of cyclin homologs that control the G1 phase in yeast. Proc Natl Acad Sci USA. 1989;86:6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- Holly SP, Blumer KJ. PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J Cell Biol. 1999;147:845–856. doi: 10.1083/jcb.147.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- Kirk KE, Harmon BP, Reichardt IK, Sedat JW, Blackburn EH. Block in anaphase chromosome separation caused by a telomerase template mutation. Science. 1997;275:1478–1481. doi: 10.1126/science.275.5305.1478. [DOI] [PubMed] [Google Scholar]
- Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001;294:867–870. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- Le S, Moore JK, Haber JE, Greider CW. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Mallory JC, Petes TD. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc Natl Acad Sci USA. 2000;97:13749–13754. doi: 10.1073/pnas.250475697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Gilson E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18:3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunuma M, Hirata D, Miyahara K, Tsuchiya E, Miyakawa T. Role of calcineurin and Mpk 1 in regulating the onset of mitosis in budding yeast. Nature. 1998;392:303–306. doi: 10.1038/32695. [DOI] [PubMed] [Google Scholar]
- Naito T, Matsuura A, Ishikawa F. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet. 1998;20:203–206. doi: 10.1038/2517. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Paciotti V, Clerici M, Lucchini G, Longhese MP. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- Paciotti V, Lucchini G, Plevani P, Longhese MP. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Runge KW. Varying the number of telomere-bound proteins does not alter telomere length in tel1Δ cells. Proc Natl Acad Sci USA. 1999;96:15044–15049. doi: 10.1073/pnas.96.26.15044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SI. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics. 1980;95:561–577. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson HE, Wittenberg C, Cross F, Reed SI. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- Ritchie KB, Mallory JC, Petes TD. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC 1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KB, Petes TD. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics. 2000;155:475–479. doi: 10.1093/genetics/155.1.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signaling pathway in Saccharomyces cerevisiae. EMBO J. 2000;19:5801–5812. doi: 10.1093/emboj/19.21.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sherman F, Hicks J. Micromanipulation and dissection of asci. Methods Enzymol. 1991;194:21–37. doi: 10.1016/0076-6879(91)94005-w. [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Smith CD, Blackburn EH. Uncapping and deregulation of telomeres lead to detrimental cellular consequences in yeast. J Cell Biol. 1999;145:203–14. doi: 10.1083/jcb.145.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedecor GW, Cocharan WG. Statistical Methods. Ed. 7. Ames: The Iowa State University Press; 1980. [Google Scholar]
- Teng SC, Chang J, McCowan B, Zakian VA. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Taggart AK, Zakian VA. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol. 2001;11:1328–35. doi: 10.1016/s0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- Vaziri H. Critical telomere shortening regulated by the ataxia-telangiectasia gene acts as a DNA damage signal leading to activation of p53 protein and limited life-span of human diploid fibroblasts. A review. Biochemistry (Mosc) 1997;62:1306–1310. [PubMed] [Google Scholar]
- Wakayama T, Kondo T, Ando S, Matsumoto K, Sugimoto K. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:755–764. doi: 10.1128/MCB.21.3.755-764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA. Dual cell cycle checkpoints sensitive to chromosome replication and DNA damage in the budding yeast Saccharomyces cerevisiae. Radiat Res. 1992;132:141–143. [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA. Origin activation and formation of single-strand TG1–3 tails occur sequentially in late S phase on a yeast linear plasmid. Mol Cell Biol. 1993a;13:4057–4065. doi: 10.1128/mcb.13.7.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell. 1993b;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- Zhang DK, Ngan HY, Cheng RY, Cheung AN, Liu SS, Tsao SW. Clinical significance of telomerase activation and telomeric restriction fragment (TRF) in cervical cancer. Eur J Cancer. 1999;35:154–160. doi: 10.1016/s0959-8049(98)00303-7. [DOI] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]