Abstract
The soil fungus Trichoderma atroviride, a mycoparasite, responds to a number of external stimuli. In the presence of a fungal host, T. atroviride produces hydrolytic enzymes and coils around the host hyphae. In response to light or nutrient depletion, asexual sporulation is induced. In a biomimetic assay, different lectins induce coiling around nylon fibers; coiling in the absence of lectins can be induced by applying cyclic AMP (cAMP) or the heterotrimeric G-protein activator mastoparan. We isolated a T. atroviride G-protein α-subunit (Gα) gene (tga1) belonging to the fungal subfamily with the highest similarity to the Gαi class. Generated transgenic lines that overexpress Gα show very delayed sporulation and coil at a higher frequency. Furthermore, transgenic lines that express an activated mutant protein with no GTPase activity do not sporulate and coil at a higher frequency. Lines that express an antisense version of the gene are hypersporulating and coil at a much lower frequency in the biomimetic assay. The loss of Tga1 in these mutants correlates with the loss of GTPase activity stimulated by the peptide toxin Mas-7. The application of Mas-7 to growing mycelial colonies raises intracellular cAMP levels, suggesting that Tga1 can activate adenylyl cyclase. In contrast, cAMP levels and cAMP-dependent protein kinase activity drop when diffusible host signals are encountered and the mycoparasitism-related genes ech42 and prb1 are highly expressed. Mycoparasitic signaling is unlikely to be a linear pathway from host signals to increased cAMP levels. Our results demonstrate that the product of the tga1 gene is involved in both coiling and conidiation.
Members of the genus Trichoderma belong to an aggregate of Deuteromycetes species whose teleomorphs, where identified, belong to the Ascomycetes (Hypocrea) (35). A number of soil isolates are being studied because of their ability to antagonize soilborne plant pathogens, and these Trichoderma species are often referred to as biocontrol fungi (9). During the initial stages of this interfungal interaction, Trichoderma atroviride responds to the presence of the host by coiling around the host hyphae. At later stages, the mycoparasite produces hydrolytic enzymes, such as β-1,3-glucanases, β-1,6-glucanases, chitinases, and proteases, to penetrate the host and use its cellular content as a source of nutrients. Coiling occurs in response to signals that presumably originate from the host, its surface structure, or the by-products of its initial degradation by enzymes secreted by Trichoderma (36).
Recognition between a parasite and its host appears to be an essential step for successful continuation of the parasitic process (46). Surface molecules such as lectins are good candidates for the recognition process in Trichoderma. Evidence for this supposition comes from experiments on the proposed specific recognition of Rhizoctonia mediated by lectins present on its cell wall (3, 4). Further support for this hypothesis was generated by use of a biomimetic system based on the use of nylon fibers coated with lectins developed to detect attachment and coiling similar to those observed during the Rhizoctonia-Trichoderma interaction (23, 24, 25). In addition, Inbar and Chet (25) showed an increase in the activity of an intracellular 102-kDa glucosaminidase by using the biomimetic system, suggesting that the recognition event mediated by lectins serves as a signal that triggers a general antifungal response in Trichoderma. In this sense, the expression of genes for at least some cell wall-degrading enzymes is induced by a soluble signal from the host with no coiling (11). Thus, the coiling response appears to follow signaling pathways different from those involved in the induction of the expression of these genes; alternatively, the coiling response involves multiple signals. However, the intracellular signaling pathways downstream of surface recognition and the detection of the soluble molecule are still unclear.
Another interesting developmental process in this fungus is asexual sporulation, which is induced by light or nutrient depletion. Asexually produced spores (conidia) are useful as inocula in the field or the greenhouse; however, sporulation is not conducive to efficient biocontrol because the resources of the culture are diverted to spore production rather than attack of the host. An understanding of the switch determining the relative priorities of conidiation and mycoparasitic coiling has intrinsic interest, as a rather unique example of fungal development. Furthermore, such an understanding may help in the selection of improved biocontrol strains.
Loss-of-function and gain-of-function experiments with G-protein α-subunit (Gα) and G-protein β-subunit (Gβ) genes have indicated that heterotrimeric G proteins are involved in sporulation, mating, pathogenicity, secondary metabolite production, and vegetative incompatibility in a variety of ascomycete (5, 22, 28, 29, 42, 51,) and basidiomycete (50) fungi.
In plant pathogens, loss-of-function mutations introduced into genes encoding heterotrimeric G proteins (14, 28, 38, 50) prevent the mutants from efficiently attacking their hosts. Mycoparasitism may well depend on similar, conserved eukaryotic signaling pathways. In Trichoderma, there is biochemical evidence for the participation of Gα in coiling. Omero et al. have reported that the application of exogenous activators of G-protein-mediated signaling, the peptide toxin mastoparan and fluoroaluminate (AlF4−), results in an increase in coiling around nylon fibers of more than twofold compared to the results obtained for controls (47). Furthermore, the application of exogenous dibutyryl-cyclic AMP (cAMP) also increases coiling. In addition, cAMP affects another developmental process in Trichoderma; it bypasses the requirement for light for sporulation, while atropine, a compound known to block cAMP production in fungi, prevents sporulation even after photoinduction.
We have isolated from T. atroviride a Gα gene belonging to the well-conserved fungal subfamily with the highest similarity to Gαi. Sense and antisense expression in transgenic lines shows that the product of this gene is involved in at least two developmental pathways.
MATERIALS AND METHODS
Strains and culture conditions.
T. atroviride (formerly T. harzianum) IMI 206040 was used in this work. Escherichia coli strains JM103, MC1061, and TOP10 (53) (Invitrogen) were used for DNA manipulations. The plasmids used were pBluescript (Stratagene), pCR2.1 (Invitrogen), pSPORT1 (Gibco-BRL, Madison, Wis.), pTZ18 (Pharmacia), pHatα (20), and pCB1004 (Fungal Genetics Stock Center). Plasmids pHatα and pCB1004 carry the E. coli hygromycin phosphotransferase gene under the control of the Aspergillus nidulans gpdA and trpC promoters, respectively.
Trichoderma was grown on potato dextrose agar (PDA; Difco) unless otherwise indicated. For transformation, soft agarose selection medium (1% agarose, 200 μg of hygromycin/ml) was used as an overlay, and PDA with 200 μg of hygromycin/ml was used as a base selection medium. For photoinduction experiments, T. atroviride was grown on complete medium (24 g of potato dextrose broth/liter, 2 g of yeast extract/liter, 1.2 g of casein hydrolysate/liter; PDYC; Difco).
DNA and RNA manipulations.
Fungal chromosomal DNA was isolated as previously described (49). All other DNA manipulations were performed by standard techniques (53). Fungal RNA was isolated according to the protocol described by Jones et al. (27). Southern blotting and Northern blotting were performed by standard procedures (53).
Cloning of tga1.
A DNA fragment of about 165 bp was amplified by PCR with T. atroviride genomic DNA as a template and degenerate primer oligonucleotides designed on the basis of conserved regions of known Gα genes (56). The PCR cycles were as previously described (63). The products were subcloned into pTZ18 and sequenced by the dideoxynucleotide chain termination method of Sanger et al. (54) with a Sequenase kit (version 2.0; U.S. Biochemicals). The short fragment sequence showed high similarity to the sequences of many Gα genes. The fragment was labeled with a random priming DNA labeling system and used to screen a cDNA library constructed from a Trichoderma culture grown in the presence of Rhizoctonia solani cell walls as a single carbon source in plasmid pSPORT1. Hybridization was carried out under high-stringency conditions as previously described (53). Several positive clones were identified, and their sequences were analyzed. The sequence data indicated that all clones corresponded to the same gene and that none was full length.
A full cDNA clone was obtained later by reverse transcription (RT)-PCR with a forward primer (5′-GGATGAGACTGCTAACCGAATGC-3′) and a reverse primer (5′-GTCGTCTAAATGAGAGACCGC-3′) designed based on the genomic sequence. The amplification product was cloned into pCR2.1 and later isolated as a 280-bp AatII-BamHI fragment, which was inserted into the cDNA clone at the same sites. The resulting plasmid (pTGAC) contained a cDNA clone with the complete open reading frame. The cDNA clone was used to screen a λEMBL3 genomic library. Four positive phage clones were identified, and DNAs were purified.
Southern analysis allowed the identification of 2.4-kb BamHI and 2.5-kb XhoI DNA-hybridizing fragments, which were subcloned, respectively, into BamHI- and SalI-digested pBluescript KS(+). Sequencing of the BamHI fragment indicated that it contained the sequence coding for the carboxy terminus of the protein (Tga1). The sequence of the SalI fragment contained the region of the gene coding for the amino terminus. A plasmid (pTGAG) containing the full-length tga1 genomic sequence was constructed by ligating the 2.4-kb BamHI fragment to the BamHI-digested 2.5-kb clone. Correct orientation was determined by restriction mapping and sequencing across the BamHI site.
Constructs.
Plasmids for antisense and sense expression were constructed as follows. pTGAC was digested with EcoRI; this fragment contained the complete cDNA and was ligated to the EcoRI site of plasmid pPR2, which carries the promoter of the T. atroviride prb1 gene (15). The two resulting plasmids were pAsTGA, which is an antisense construct, and pSTGA, which is a sense construct; both are controlled by the prb1 promoter.
A Q203L mutant gene was generated by using a PCR-based strategy. Three oligonucleotides were designed based on the genomic sequence to generate the desired mutation in a two-step procedure. A primer (5′-CGGTGCCCAGCGATC-3′) containing a single base modification (marked in bold) which results in a single amino acid modification of Q to L and a primer (5′-GGACGTCTTGATGAACC-3′) containing an AatII site were used to amplify a 187-bp fragment. The resulting mutant product was used as a reverse primer for a second round of PCR amplification with a forward primer having the sequence 5′-CGCGTCATTCTCGA-3′. The PCR product (344 bp), which includes an NcoI site next to the reverse primer, was ligated to plasmid pCRII and used for the transformation of E. coli. Plasmids containing the expected insert were confirmed by digestion with BalI (MscI) (this site is lost due to the introduced mutation) and sequenced. Finally, the AatII-NcoI fragment (325 bp) cloned into pCRII was used to replace the same region in plasmid pTGAG, which carries the complete genomic clone, resulting in a modified tga1 carrying the desired point mutation.
Transformation.
Biolistic transformation of Trichoderma was carried out essentially as described by Lorito et al. (41), with minor modifications. Briefly, particle preparation was done according to the protocol described by Tomes et al. (59). Conidia (108) were plated on PDA supplemented with 0.4 M sorbitol, and plates were allowed to dry for 4 h. Projectiles carrying the DNA were delivered at a helium pressure of 800 lb/in2. The fungus was allowed to recover for 7 h and was overlaid with soft agarose selection medium. Approximately 48 h later, single colonies reaching the surface were cut out and transferred to selection medium. For purification of transformants, selected colonies were allowed to sporulate, conidia were collected, and serial dilutions were plated on PDA containing 100 μg of hygromycin/ml and 0.5% Triton X-100.
Photoinduction.
Colonies were grown on filter paper soaked with 3 ml of PDYC: an 8-cm disk of Whatman 50 overlaying a 7-cm disk of Whatman 1 in a 9-cm plastic petri dish. Experiments were begun after 36 h of growth in total darkness, when colony diameter had reached 40 to 50 mm. The cultures were photoinduced by exposure to a standard blue source consisting of light from a cool-white fluorescent tube filtered through a blue acrylic filter (maximum transmission, 442 nm; fluence rate, 2 μmol m−2 s−1) or two cool-white fluorescent tubes filtered with LEE filter no. 183 (fluence rate, 3 μmol m−2,s−1). At various times, the mycelia or conidiophore rings were scraped from the surface of the filter paper under weak yellow or red safelight (0.1 μmol m−2 s−1). Samples for RNA extraction were immediately frozen in liquid nitrogen.
Protein analysis.
Cultures grown on PDYC in the dark at 28°C for 48 h were harvested, frozen, and ground in liquid nitrogen. The powder was suspended in homogenization buffer A (3 ml/g), which contained 50 mM HEPES (pH 7.5), 330 mM sucrose, 5 mM EDTA, 0.2% bovine serum albumin (BSA), 1 mM phenylmethylsulfonyl fluoride (PMSF), and 5 mM dithiothreitol (DTT). The suspension was then sonicated for 1 min at 4°C with 50% pulses by using a Sonicator model W-375 microtip (Heat Systems Ultrasonics Inc., Farmingdale, N.Y.). Samples were centrifuged for 10 min at 4°C and 10,000 × g. The supernatant was collected and centrifuged for 1 h at 4°C and 100,000 × g. Finally, the pellet obtained was suspended in homogenization buffer. The protein concentration was determined by the Bradford method with assay dye reagent from Bio-Rad Laboratories, Richmond, Calif., and BSA as a protein standard. Western blotting was performed according to standard procedures (53). Equal loading of the lanes was confirmed before probing by staining with Ponceau S. The anti-Gna1 and anti-Gna2 antibodies were kindly provided by K. Borkovich and were used at a 1:7,000 dilution. Goat peroxidase-coupled secondary antibody (Amersham) was used at a 1:12,000 dilution, followed by chemiluminescent signal detection in 1.25 mM luminol (Sigma)-400 μM p-coumaric acid (Sigma)- 2.7 mM H2O2 (Merck)- 100 mM Tris base (pH 8.5).
Biomimetics.
The culture conditions and the biomimetic system used were as described previously (23). For the biomimetic system, T. atroviride was grown on a synthetic medium (SM) with 0.2% (wt/vol) glucose (23) and covered with a cellophane (permeable cellulose) membrane. Nylon 66 fibers (approximate diameter, 14 μm; kindly supplied by Nilit, Migdal-Haemek, Israel) were extended across grids (square polyester nets measuring 7 by 7 mm and having a pore size of 420 μm), and the edges were fastened with cyanoacrylate glue (Loctite). The grids were washed several times with chloroform for 30 min and then were washed with 70% ethanol for 30 min. Finally, the grids were washed with sterile distilled water for a further 30 min.
Binding of lectins and BSA to the nylon fibers was brought about as described by Inbar and Chet (24). The various lectins bound to the nylon fibers were peanut (Arachis hypogaea), wheat germ (Triticum vulgaris), gorse (Ulex europaeus), soybean (Glycine max), and concanavalin A (Sigma catalog no. L-0881, L-9640, L-5505, L-1395, and C-2631, respectively). Cultures were started by inoculation of an agar block with mycelium on top of the cellophane membrane. Sixteen hours later, the grids were placed 5 mm in front of the advancing colony. Experiments were performed 9 h after placement of the grids, when the front had reached one-third to one-half way across the grids. Incubation was done in total darkness at 28 to 30°C, and all manipulations of growing colonies were performed under a red safelight to avoid photoinduction of sporulation. The grids were removed aseptically and vapor fixed with 5% (wt/vol) OsO4 and 25% glutaraldehyde (electron microscopy grade) for 48 h.
Specimens were dried for 24 h, coated with gold palladium in a model E-500 sputter coater (Polaron Equipment Ltd., Watford, United Kingdom), and observed under a scanning electron microscope (JEOL JSM 35C). For quantitative analysis of the results, the number of coils per milliliter of nylon fiber was counted in replicate scanning electron microscope images (23). Quantitative data represent the averages of data pooled from 9 to 11 replicate fields pooled from at least two independent sets of experiments. Statistical significance was assessed by Student's t test, and error bars in the figures indicate standard errors of the means.
GTPase activity assays.
High-affinity membrane GTPase was measured by the assay of Cassel and Selinger (8). The reaction mixture contained 50 mM morpholinepropanesulfonic acid (MOPS) (pH 7.0), 1 mM MgCl2, 0.2 mM EGTA, 0.5 μM [γ-32P]GTP (BLU/NEG/004; New England Nuclear) diluted with unlabeled GTP to a final activity of 5 to 20 Ci/mmol, 0.5 mM ATP, 2 mM DTT, 2 mM PMSF, 2 mM creatine phosphate, and creatine phosphokinase freshly added at 0.06 U/μl. The reaction was started by the addition of 50 μl of the reaction mixture into 25 μg of membrane protein in 50 μl of homogenization buffer B, which contained 50 mM HEPES (pH 7.5), 330 mM sucrose, 5 mM EDTA, 5 mM EGTA, 0.2% (wt/vol) BSA, 0.2% (wt/vol) casein hydrolysate (acid), 5 mM DTT, and protease inhibitor cocktail (complete EDTA free; Roche). Incubation was done at room temperature and was terminated after 20 to 30 min by the addition of 500 μl of ice-cold and well-stirred 6% activated charcoal (Sigma) in 50 mM phosphate buffer (pH 7.0). Samples were centrifuged for 5 min at 12,000 × g, 440 μl of the supernatant was added to 3 ml of a POP scintillation liquid containing 50% (vol/vol) toluene and 50% (vol/vol) Triton X-100, and counts were determined (1600 TR analyzer; Packard). Means and standard errors of the means are the combined results of at least two independent assays.
cAMP measurements.
Mycelia were ground in liquid nitrogen to a fine powder and resuspended in ice-cold 6% trichloroacetic acid. Trichloroacetic acid extracts were incubated on ice for 20 min with frequent vortexing. A 1-ml aliquot of each sample was centrifuged for 25 min at 16,000 × g in a microcentrifuge at 4°C. The pellet was solubilized in 0.1 N NaOH-5% sodium dodecyl sulfate at 60°C, and protein was quantitated with bicinchoninic acid reagent (Sigma) and BSA as a standard. This solution was extracted four times with 5 ml of ice-cold water-saturated ether and stored at −20°C. Radioimmunoassays were performed as described by Harper and Brooker (18). Results are representative of at least two independent experiments. Means and standard errors of the means are reported.
For induction with diffusible host signals, T. atroviride was pregrown for 3 days on SM plates covered with a cellophane membrane and then placed on an R. solani colony that had been pregrown for 2 days on the same medium. The pregrown Trichoderma colony was transferred to SM medium on which Rhizoctonia had been cultivated for 2 days and removed. In both instances, Trichoderma was incubated for a further 24 h after being transferred.
cAMP-dependent protein kinase activity.
Approximately 50 mg of mycelium was frozen in liquid nitrogen, ground to a fine powder, and resuspended in 200 μl of a buffer containing 20 mM Tris-HCl (pH 7.4), 1 mM EDTA (pH 8.0), and 0.5 mM PMSF. Samples were then vortexed three times for 10 s each time and kept at 4°C. The homogenate was centrifuged for 10 min at 12,000 rpm and 4°C in an Eppendorf Microfuge. Supernatants were transferred to fresh Eppendorf tubes, and the protein content was determined by the Bradford method with a commercial product (Bio-Rad). Glycerol was added to each of the extracts to a final concentration of 20%. The protein concentration was then brought to 1 μg/μl. The extracts were kept at −70oC until used.
Protein kinase A (PKA) activity was determined by the incorporation of 32P into the specific substrate kemptide from [γ-32P]ATP. The reaction was carried out in a 20-μl mixture containing 50 mM Tris-HCl (pH 7.5), 1.5 mM EGTA, 5.8 mM MgSO4, 50 μM kemptide, 1 mM ATP, and 0.1 mM [γ-32P]ATP (0.2 μCi) and with and without 10 μM cAMP. The reaction was carried out with 10 μg of protein, and the reaction mixture was incubated for 5 min at 30°C. After incubation, the reaction mixture was spotted on phosphocellulose filters (Whatman P-81), and the filters were washed three times with 75 mM phosphoric acid for 10 min with agitation and once with ethanol for 5 min. The phosphocellulose filters were allowed to dry, and radioactivity was measured with scintillation liquid in a 1600 TR liquid scintillation analyzer. The experiments were repeated three times.
RESULTS
Role of lectins in host recognition.
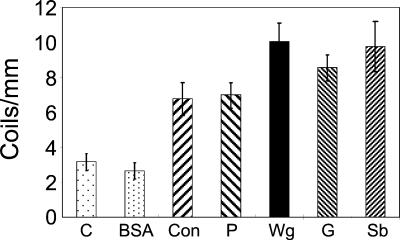
To investigate whether lectins determine a specific recognition event between Trichoderma and its host, as previously proposed (25), a series of commercially available lectins with different sugar moieties covalently linked to nylon fibers were tested in biomimetic assays. Previous work (25) was done with a different Trichoderma strain and only two lectin types. As shown in Fig. 1, we confirmed and extended the results (25) with the strain used here. All tested lectins induced coiling of Trichoderma around the nylon fibers to different degrees. Wheat germ, gorse, and soybean lectins induced slightly higher coiling levels, reaching a maximum of 10 coils/mm, compared to concanavalin A and peanut lectins, which induced coiling levels of about 7 coils/mm. In contrast, the use of nylon fibers coated with BSA resulted in coiling levels (two or three coils per milliliter) similar to those obtained in control experiments with noncoated nylon fibers (three coils per milliliter). These results suggest that lectins play a role in host recognition.
FIG. 1.
Induction of coiling of Trichoderma IMI 206040 with different lectins. The commercial plant lectins used were peanut (P), wheat germ (Wg), gorse (G), soybean (Sb), and concanavalin A (Con). The lectins were coupled to nylon fibers, and the rate of coiling of Trichoderma around them was measured. Unmodified nylon fibers (C) or BSA-coupled nylon fibers (BSA) served as controls.
Attachment and coiling are stimulated by activation of a G-protein signaling pathway.
It has been suggested that a cAMP signaling pathway participates in a lectin-mediated host recognition process (47). Thus, we decided to examine the effect of the exogenous application of cAMP and the G-protein activator Mas-7 in a biomimetic system with noncoated nylon fibers by scanning electron microscopy. As shown in Fig. 2, the application of 5 mM cAMP (Fig. 2B) and 2 μM Mas-7 (Fig. 2C) resulted in Trichoderma attachment to and coiling around the nylon fibers. In contrast, Trichoderma did not attach to or coil around noncoated nylon fibers at a high frequency without the application of the mentioned compounds (Fig. 2A). These results suggest that a G-protein-mediated signaling pathway and cAMP are involved in both attachment and coiling.
FIG. 2.
Effect of the mastoparan analog Mas-7 and cAMP on coiling. The indicated concentration of Mas-7 or cAMP was applied under red safelight to the perimeter of growing colonies in the dark as described previously (47). After 6 h, the grids were removed and fixed for scanning electron microscopy.
Structure and expression of tga1, a T. atroviride Gα gene.
In order to determine whether heterotrimeric G proteins participate in coiling and conidiation, a Gα gene was isolated. For this purpose, degenerate oligonucleotides were designed on the basis of conserved regions of other Gα sequences in publically available data banks (see Materials and Methods). Using T. atroviride genomic DNA, we obtained a 165-bp PCR fragment. Sequence analysis indicated that this fragment corresponded to a portion of a Gα gene. The cloned PCR fragment was used as a probe to screen a cDNA library. Several positive clones were obtained, and sequence analysis indicated that all clones corresponded to the same gene and that none of them was full length. A cDNA clone (1,332 bp) containing the complete open reading frame of 1,049 bp was later assembled (see Materials and Methods), resulting in plasmid pTGAC. The partial cDNA clone was used to screen a λEMBL3 genomic library (see Materials and Methods). A 4.9-kb clone containing the full-length tga1 genomic sequence was then also assembled (see Materials and Methods), resulting in plasmid pTGAG.
Comparison of the complete cDNA and genomic sequences of tga1 GenBank accession number AY036905) indicated the presence of four introns. The first intron is located 128 bp upstream of the ATG, and the other three introns are located 114, 873, and 1,065 bp downstream of the ATG, matching the intron positions of genes encoding proteins that belong to the Gαi superfamily (39). The deduced protein sequence of Tga1 corresponds to a peptide of 353 amino acids with a calculated molecular mass of 40.8 kDa, similar to that deduced from the Neurospora crassa gna1 gene. gna1 was the first gene cloned from a lower eukaryote encoding Gα and was classified as part of the mammalian Gαi family (61). Both tga1 and gna1 belong to the same ascomycete group proposed by Horwitz et al. (22), based on the three consensus sequences. It is noteworthy that all members of this group contain the consensus sequence for potential ADP ribosylation by pertussis toxin (62) and cholera toxin and have an N-myristoylation site (MGXXXS) in the amino-terminal region (6). The promoter region of tga1 contains potential binding sites for several regulatory proteins, such as Stre, PacC, and StuAp (30, 58). Perhaps the most relevant elements are the 10 StuAp response elements detected in the tga1 promoter. StuAp is an important regulator of multicellular development in A. nidulans (12). Response elements for this transcription factor have been found upstream of important developmental and cell cycle genes in A. nidulans (12). Southern analysis of T. atroviride genomic DNA under high-stringency hybridization conditions suggested the presence of a single copy of the gene. However, low-stringency hybridization showed one or two additional bands, suggesting the presence of at least one additional homologous gene in the T. atroviride genome (data not shown).
Generation of Trichoderma transgenic lines overexpressing tga1 and expressing an antisense version.
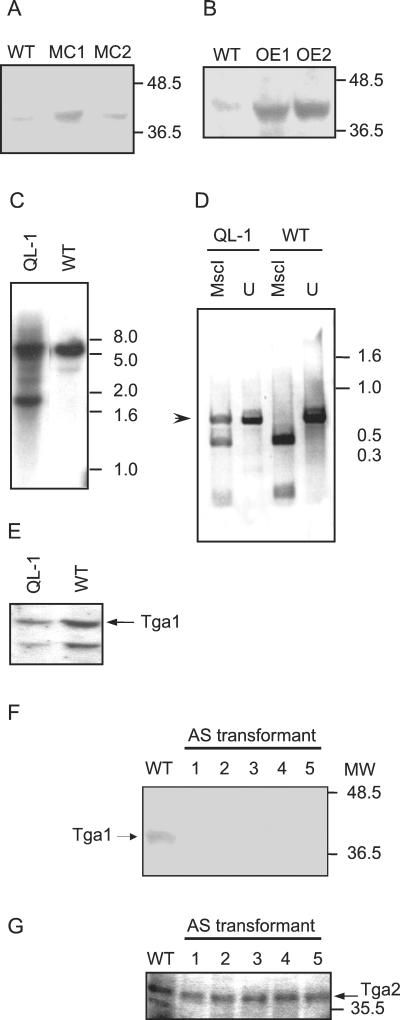
To elucidate the function of Tga1 in Trichoderma, we overexpressed the protein and expressed a mutant protein in transgenic lines. Two strategies were adopted with the objective of overexpressing Tga1. First, we generated transformants (MC1 and MC2) carrying multiple copies of vector pTGAG, which contains the complete genomic clone. With the same objective, we introduced multiple copies of plasmid pSTGA into transformants OE1 and OE2, in which the expression of the tga1 coding sequence is driven by the derepressible promoter of the prb1 gene from T. atroviride. This promoter is switched off when the fungus is grown in the presence of ammonium and is switched on when the fungus is grown on nitrate or deprived of nitrogen (unpublished data). All transformants obtained showed higher levels of Tga1 protein than did the wild type (Fig. 3A and B), had similar fluffy phenotypes, in contrast to the wild type, and showed no obvious alteration of their growth rates.
FIG. 3.
Expression of Tga1 in wild-type and transgenic Trichoderma strains. (A and B) Western blots of membrane fractions from the indicated strains probed with antibody to Gna1. WT, wild type. (A) Lines MC1 and MC2, carrying multiple copies of tga1. (B) Lines OE1 and OE2, expressing tga1 from the prb1 promoter. (C) Southern analysis of wild-type and transgenic strains carrying a mutant allele of tga1. Genomic DNAs from the two strains were digested with EcoRI, and the blot was probed with a 1.2-kb EcoRV genomic fragment. Thepositions of molecular size markers are indicated at the right in kilobases. (D) Detection of expression of the mutant tga1 allele. RNA was extracted and used as a template for RT and PCR amplification as described in Materials and Methods. The products were digested to completion with MscI or left undigested (U). The arrowhead indicates the expected cDNA product of the mutant gene, which cannot be digested with MscI. DNA molecular size markers are indicated at the right in kilobases. (E) Immunoblot analysis of transformants carrying the mutant tga1 allele. Membrane protein fractions from wild-type and transformant strains were analyzed as described in panel A. The arrow indicates the signal corresponding to Tga1. (F and G) Immunoblot analyses of transformants expressing tga1 in the antisense (AS) form. Shown are Western blots of membrane fractions from the indicated transformants probed with antibody to Gna1 (F) or Gna2 (G) of N. crassa. MW, molecular weight, in thousands.
In order to gain further insight into the role of Tga1, a point mutation in the corresponding gene (tga1) was introduced (see Materials and Methods), resulting in the production of an activated mutant protein with a single amino acid change at position 203 (Q to L). This mutation was previously reported to eliminate the intrinsic GTPase activity of Gα, switching on constitutive signaling in the absence of receptor activation in animal cells (19). This construct was used to transform Trichoderma. Southern blot (Fig. 3C) and RT-PCR (Fig. 3D) analyses of two lines from a total of about 10 different hygromycin-resistant colonies confirmed integration and expression of the introduced construct, respectively. The RT-PCR strategy used was designed to amplify a region which includes an intron, enabling us to distinguish between amplification products derived from DNA and RNA. Additionally, the mutation generated results in the loss of an MscI site, allowing us to distinguish between the products of the wild-type and mutant genes and at the same time to detect its expression. MscI digestion of the RT-PCR products obtained from the mutant showed three bands (Fig. 3D). The upper band corresponds to the expected cDNA product of the mutant gene, which cannot be digested with MscI; the two additional bands correspond to the product of the wild-type gene. Colonies of these types of transformants showed a fluffy phenotype similar to that of the overexpressing strains. The Tga1 protein, however, was detected at lower levels than in the wild type, so that the phenotype of these strains must be attributable to the mutant gene rather than to overexpression (Fig. 3E).
In a first attempt to elucidate the function of Tga1, we used a gene disruption strategy. For this purpose, we constructed a plasmid derived from pTGAG in which the central coding region of tga1 was replaced by a hygromycin resistance cassette from plasmid pCB1004. However, even after multiple transformation experiments and the use of different purification strategies, all transformants obtained either carried the construct integrated ectopically or contained more than one nucleus with at least one carrying a wild-type copy of the gene. Thus, to gain further insight into the role of Tga1 in T. atroviride, a vector carrying an antisense tga1 cDNA under the control of the derepressible promoter of the prb1 gene was constructed. This plasmid was used in cotransformation experiments together with plasmid pHatα, which confers hygromycin resistance. Ten hygromycin-resistant transformants were purified through monoconidial cultures and used for further characterization. Southern analysis confirmed the integration of the construct in the Trichoderma genome, characterized by the presence of the expected 1.3-kb EcoRI band in addition to the 9-kb band corresponding to the wild-type copy of the gene (data not shown).
An antibody raised against N. crassa Gα Gna1 (26) was used for immunoblot analysis of membrane fractions prepared from the different transformants (Fig. 3F). Antisense transformants (AS1, AS2, AS3, AS4, and AS5) showed no detectable Tga1 levels. There are only 17 amino acid substitutions between Gna1 and Tga1, so we expect the cross-reacting signal, which comigrates with the signal from Neurospora membranes at 40 kDa (data not shown), to correspond to Tga1. Probing of replicate blots with antibodies against Gna2, another N. crassa Gα (1), gave similar signals in all the transformants (Fig. 3G). The signal obtained with anti-Gna2 indicates that Trichoderma has at least one more Gα gene in addition to that for Tga1. Furthermore, this signal serves as an internal control for the amount of membrane protein loaded in each lane.
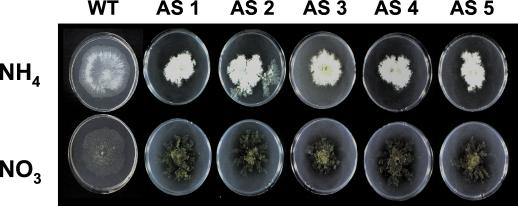
All 10 transformants showed a clearly aberrant phenotype, characterized by the formation of rather dense and small colonies, continuous sporulation, and the production of a yellowish pigment (data not shown). The phenotype of Gα antisense transformants was most extreme on medium containing nitrate as a sole nitrogen source and was alleviated to some extent by growth on ammonium, as expected (Fig. 4).
FIG. 4.
Nitrogen source effects on phenotypes of Trichoderma transformants expressing tga1 in the antisense (AS) form. T. atroviride and Gα antisense transformants were grown on SM medium with 12.5 mM NH4Cl or 12.5 mM KNO3 as the sole nitrogen source and 0.5% glucose as the carbon source. Cultures were kept in light for 3 days at 28°C and photographed.
Mas-7 activates Tga1 and raises cAMP levels.
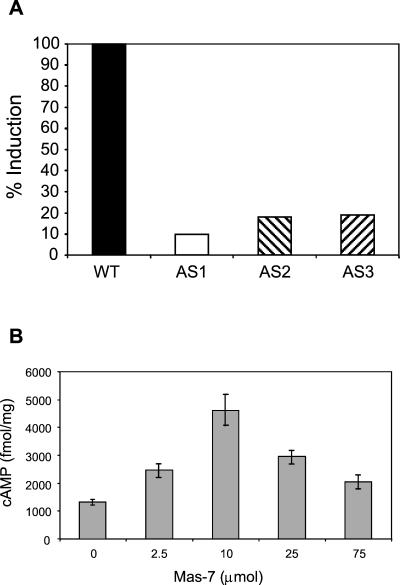
The activation of heterotrimeric G proteins can be monitored by measuring the intrinsic GTPase activity of Gα. By following the method used for animal cells (8), we developed an assay to detect high-affinity GTPase in fungal membrane fractions (C. Omero et al., unpublished data). In order to further characterize Tga1 and because mycoparasitic coiling is induced by the specific peptide toxin Mas-7, the membrane-associated GTPase activity in Trichoderma and its response to Mas-7 were analyzed. Figure 5A shows that GTPase activity increased in response to the application of Mas-7 in the wild-type strain, while no significant GTPase activity increase was detected for any of the antisense transgenic lines tested. Furthermore, basal GTPase activity was considerably lower (30 to 40%) in all antisense transformants than in the wild type. These experiments suggest that Tga1 is responsible for most of the Mas-7-inducible GTPase activity, since it was strongly decreased in antisense transformants (Fig. 5A). Mastoparan and Mas-7 are specific for the Gi and Go classes of mammalian G-protein subunits, and the loss of Mas-7-stimulated GTPase activity in the antisense transformants is consistent with this fact, since Tga1 is most homologous to Gαi.
FIG. 5.
Mas-7 activates Tga1 and raises cAMP levels. (A) Mas-7 activates GTPase activity. Membrane fractions from wild-type (WT) or Tga1 antisense (AS) lines (25 μg of protein) were assayed for high-affinity GTPase activity with or without the G-protein activator Mas-7. Radioactive phosphate released from [γ-32P]GTP was assayed as described in Materials and Methods. Bars represent the difference between induced (with Mas-7) and noninduced (without Mas-7) GTPase activities relative to the wild type and indicate means of two or three independent experiments. (B) G-protein activators raise cAMP levels. The G-protein activator Mas-7 was applied to growing colonies under red safelight. After 2 h, the material was frozen and cAMP was assayed as described in Materials and Methods.
Having obtained genetic evidence that Mas-7 is primarily a specific activator of Tga1, we tested the effect of Mas-7 on cAMP levels. The application of 7 μM to growing mycelial colonies resulted in more than a twofold increase in cAMP levels (Fig. 5B). By analogy with what is known for animal cells, this result suggests that the activation of Tga1 raises the activity of adenylyl cyclase, although other interpretations are possible (for example, activation of adenylyl cyclase by the Gβγ heterodimer released by the activation of Tga1 or inhibition of phosphodiesterase).
Transgenic Trichoderma lines with modified tga1 expression levels or function show altered conidiation patterns.
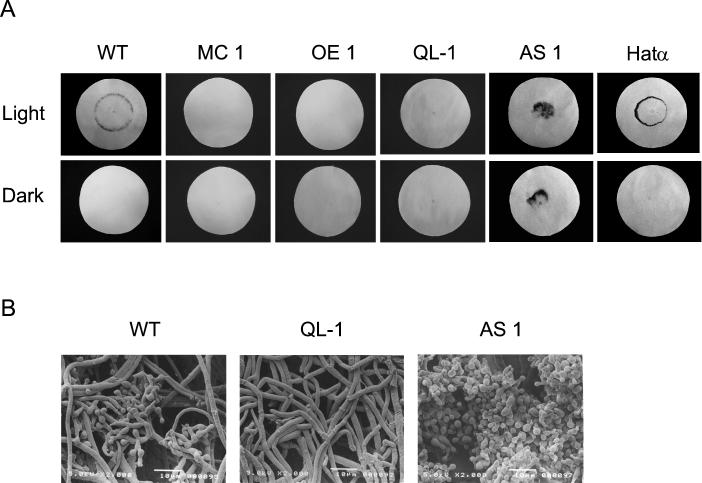
All transformants carrying multiple copies of tga1 either under the control of its own promoter (MC1 and MC2) or under the control of the prb1 promoter (OE1 and OE2) were used in photoinduced conidiation assays. In a typical assay, the wild-type strain produces a conidiation ring where the colony perimeter was located at the time of the light pulse. Conidia are mature by 36 h after the light pulse, while cultures kept in the dark do not produce conidia. None of the overexpressing lines produced a conidiation ring (Fig. 6A, MC1 and OE1). In contrast, the wild type and transformants carrying only the selectable vector (pHatα) formed conidia normally in response to light (Fig. 6A). Transgenic lines carrying the wild-type tga1 gene and at least one copy of the mutant gene (QL-1) showed the same behavior as the overexpressing transformants in light-induced conidiation experiments (Fig. 6A). These data suggest that Tga1 plays an important role in light-induced conidiation. A closer inspection of conidiation by scanning electron microscopy allowed us to determine that multicopy transformants produce a few conidiophores in response to the light stimulus, with a normal number of phyalides per stalk and with a single conidium per phyalide. Multicopy transformants eventually produced fully developed conidiophores and conidia. In contrast, transformants carrying the mutant tga1 allele (QL-1) did not produce conidiophores at all (Fig. 6B).
FIG. 6.
Growth and developmental alterations in transgenic Trichoderma lines. (A) Photoinduction of conidiation in transgenic lines. Each sample was inoculated in the center of filter paper saturated with liquid medium (see Materials and Methods). After 36 h, the cultures were given a saturating pulse of blue light (900 μmol m−2) (Light) or kept in total darkness (Dark); the cultures were then grown for a further 36 h in the dark, fixed in ethanol, and photographed. Shown are the wild type (WT); transgenic lines carrying multiple copies of tga1 (MC 1), overexpressing tga1 from the prb1 promoter (OE 1), expressing a mutant Tga1 protein (QL-1), and expressing tga1 in the antisense form (AS 1); and a control transgenic line transformed only with plasmid pHatα (Hatα). (B) Scanning electron microscopy analysis of transgenic lines expressing a mutant Tga1 protein (QL-1) and expressing tga1 in the antisense form (AS 1) and the wild type (WT) as a control.
Analyses of antisense transformants resulted in two distinct phenotypes. The most common phenotype was a constitutive sporulation pattern, with the presence of conidia over the whole colony, regardless of light stimulation (Fig. 6A, AS1) (data not shown for AS2, AS3, AS4, and AS5). The second phenotype was similar to that of the wild type, with a conidiation ring at the periphery of the colony (data not shown for AS6 and AS7). The transformants that produced a light-induced conidiation ring had Tga1 levels that appeared only slightly reduced compared to the wild-type level (data not shown). The phenotypes thus correlated well with the immunoblot analysis results and suggested variable levels of antisense expression and a consequent reduction of sense expression in different transformants. In addition, antisense transformants sporulated in both shaken and stationary cultures, conditions under which the wild-type strain does not produce spores. In a closer analysis of conidiation by scanning electron microscopy, we were able to corroborate the hypersporulating phenotype of antisense transformants (Fig. 6B, AS1) and to determine that they produced many more phyalides per stalk (data not shown).
Altered cAMP levels in Tga1 transgenic lines.
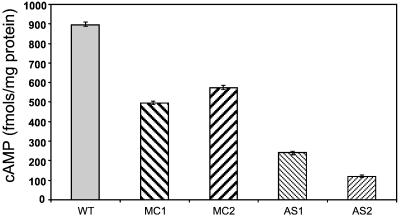
A likely effector of Tga1 is cAMP, since the application of cAMP stimulated coiling (47) (Fig. 2) and the activation of Tga1 with Mas-7 raised cAMP levels (Fig. 5). We therefore tested whether genetic manipulation of Tga1 affected cAMP levels. Both overexpressing and antisense lines had lower steady-state levels of cAMP than the wild type, but the overexpressing lines had moderately decreased cAMP levels while the antisense lines had much lower cAMP levels (Fig. 7). The reason for the moderately decreased cAMP levels in the overexpressing lines is not clear, but it is important to note that the overexpressed alpha subunit may titrate the beta-gamma heterodimer, which could also be an activator of adenylyl cyclase. The very low cAMP levels in the antisense lines may be due to a loss of adenylyl cyclase activation by Tga1 but may also be a secondary effect as well—a result of, rather than a cause of, the intense sporulation of these lines.
FIG. 7.
Altered steady-state cAMP levels in transgenic lines. Bars indicate the cAMP levels detected in cultures of wild-type Trichoderma (WT) and Trichoderma transgenic lines carrying multiple copies of tga1 (MC1 and MC2) or expressing tga1 in the antisense form (AS1 and AS2).
Role of Tga1 in mycoparasitism.
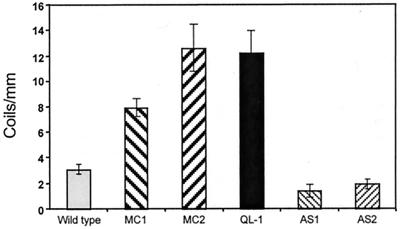
We have shown that Mas-7 (or mastoparan) (47) induces the coiling of Trichoderma around nylon fibers (Fig. 2) and that Mas-7-induced GTPase activity is strongly decreased in antisense transformants (Fig. 4). Thus, we used the biomimetic system to test the possible participation of Tga1 in mycoparasitic coiling. Overexpressing transformants (MC1 and MC2) showed an increase of more than twofold in coiling, compared to the wild type (Fig. 8). Furthermore, coiling was also increased in transformants carrying the constitutively active Q203L allele (Fig. 6). In contrast, two antisense transformants that were typical of the extreme hypersporulation phenotype and whose Tga1 levels were below detection by immunoblotting (Fig. 3F) showed markedly decreased coiling around the nylon fibers (Fig. 8). Although their intense hypersporulation, even in the presence of ammonium in the medium, might have masked any response, these data strongly suggest that tga1 plays an important role in mycoparasitic coiling.
FIG. 8.
Hyphal coiling around nylon fibers. The wild type and transformants overexpressing tga1 (MC1 and MC2) or carrying the constitutively active Q203L allele (QL-1) or an antisense copy of tga1 (AS1 and AS2) were grown on cellophane overlaying standard complete medium and allowed to overgrow grids with nylon fibers. The number of coils per millimiter of fiber was counted (see Materials and Methods) on scanning electron microscopy images of the cultures.
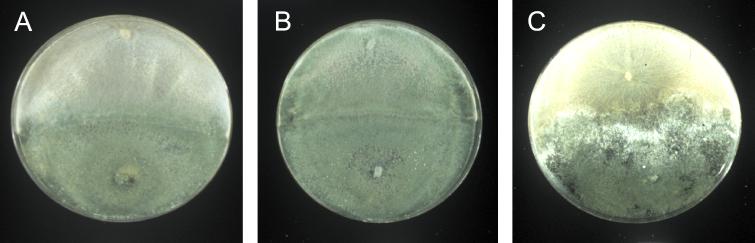
A common test for the selection of Trichoderma strains with high biocontrol capacity is a direct confrontation with the host in dual cultures. To test the impact of the alteration of the expression of Tga1 in vivo, we established direct confrontation assays with Trichoderma and R. solani. Figure 9 shows that there was an impressive increase in the capacity of the parasite to overgrow its host when Tga1 was overexpressed (Fig. 9B), compared to the results for the nontransformed strain (Fig. 9A). In contrast, antisense transformants with no detectable levels of Tga1 protein (Fig. 3F) were unable to overgrow R. solani (Fig. 9C). These data suggest that Tga1 plays a major role in mycoparasitism.
FIG. 9.
Direct confrontation assays of wild-type and transgenic Trichoderma against R. solani. Overgrowth of Rhizoctonia by the different Trichoderma strains is evident from the production of green conidia. (A) Wild type. (B and C) Transgenic lines carrying multiple copies of tga1 (MC1) or expressing tga1 in the antisense form (AS1), respectively.
Higher Tga1 levels are thus correlated with an increased capacity to overgrow the host. If Tga1 stimulates adenylyl cyclase, then one might predict that in the presence of the host, cAMP levels, PKA activity, and mycoparasitism-related gene expression all will be increased.
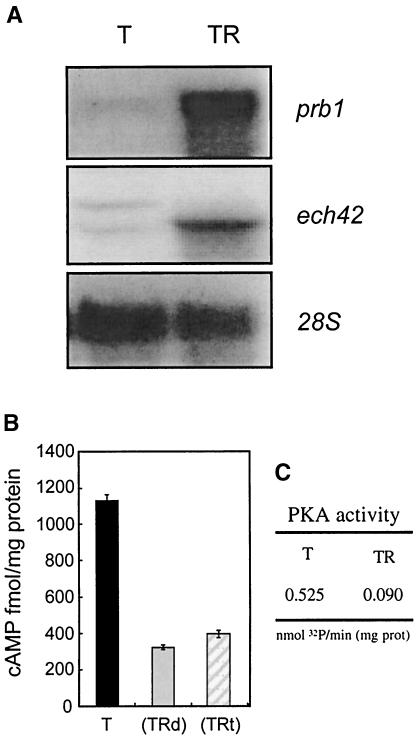
It has been shown that the genes prb1 (encoding a basic protease) and ech42 (encoding an endochitinase) play relevant roles in the biocontrol capacity of Trichoderma and that their expression is triggered during the interaction with the host (7, 13) Thus, we decided to test whether the activation of the expression of these genes correlated with the activation of a cAMP signaling pathway. Figure 10A shows the analysis of the expression of prb1 and ech42 during a direct confrontation assay with R. solani, where contact was avoided by the use of a cellophane membrane placed between the two fungi. As indicated in the autoradiographs, high levels of expression of both genes were obtained at the time of sampling, while their expression was nearly undetectable when Trichoderma was grown alone. Measurements of cAMP levels in the samples showed high cAMP levels when Trichoderma was grown in the absence of the host (Fig. 10B, T). Interestingly, cAMP levels dropped to nearly one-fourth when the parasite was grown in the presence of the host or just diffusible factors produced by it (Fig. 10B, TRt and TRd). Furthermore, cAMP-dependent protein kinase activity levels decreased in proportion to cAMP levels when Trichoderma was grown in the presence of the host, compared to the activity levels detected when the mycoparasite was grown alone (Fig. 10C). If Tga1 is responsible for the induction of ech42 and prb1 gene expression, it does not do so by increasing intracellular cAMP levels. Our data therefore suggest that these two responses to the host use distinct signaling pathways.
FIG. 10.
Analysis of cAMP levels and cAMP-dependent protein kinase activity in confrontation experiments. (A) Northern analysis showing the levels of expression of two mycoparasitism-related genes (prb1 and ech42) at the time when samples were collected for measurements of cAMP levels and cAMP-dependent protein kinase activity. T, Trichderma growing alone; TR, Trichoderma growing on top of Rhizoctonia. (B) cAMP levels detected in cultures of Trichoderma growing alone (T), Trichoderma exposed to diffusible factors produced by Rhizoctonia (TRd), or Trichoderma growing on top of Rhizoctonia (TRt). (C) cAMP-dependent protein kinase activity measured in cultures of Trichoderma growing alone (T) or Trichoderma growing on top of Rhizoctonia (TR). PKA, protein kinase A; prot, protein. The results shown are those of a typical experiment out of three carried out.
DISCUSSION
During the initial stages of mycoparasitism, Trichoderma attaches to and coils around the host. The signals for this developmental route are presumably cues immobilized on the host surface (23, 24), diffusible factors released from the host upon attack by Trichoderma (11), or both. While the receptors for these signals are likely to be very specific, eukaryotic signal transducers such as heterotrimeric G proteins are evolutionarily conserved from fungi to humans (5, 16, 44). In phytopathogenic fungi, G-protein and cAMP signaling pathways have been shown to play a major role in pathogenicity and, in particular, in the formation of invasive structures. Trichoderma hyphae coil around lectin-coated nylon fibers in the absence of a host (23). The G-protein activators fluoroaluminate, mastoparan, and MAS-7 trigger Trichoderma attachment to and coiling around noncoated nylon fibers (47) (Fig. 2). Since mastoparan is specific for the Gi and Go classes of Gα in animal cells, we postulated that a G protein of this type could transduce the signals for coiling. Furthermore, because it replaced the lectin signal, cAMP might be the second messenger.
We have cloned, characterized, and obtained loss- and gain-of-function mutations for the tga1 gene encoding Gα of T. atroviride. A sequence comparison shows that the T. atroviride Tga1 protein belongs to the Gαi superfamily. This group includes the corresponding Gα of Coprinus congregatus (Gpa1) (32), N. crassa (Gna1) (61), A. nidulans (FadA) (64), Cryphonectria parasitica (Cpg-1) (10), Magnaporthe grisea (MagB) (38), Ustilago maydis (Gpa1) (50), Cochliobolus heterostrophus (Cga1) (22), and Colletotrichum trifolii (60). All of these proteins show a molecular mass (determined by immunoblotting and/or calculated from the sequence) of approximately 40 kDa, and all of the corresponding genes have at least two introns in identical positions which are conserved in mammalian Gαi genes (22). In addition, all of them contain putative N-myristoylation and ADP-ribosylation sites for pertussis and cholera toxins. Southern analysis under high-stringency conditions indicated the presence of a single Gα gene, but low-stringency hybridization suggested the presence of at least one more homologous gene (data not shown). Further evidence for another Gα gene comes from immunoblot analysis, which revealed one protein similar to Gna1 of N. crassa and a second reacting with antiserum raised against Gna2 of N. crassa.
Since we have no viable deletion mutants, sense expression and antisense expression were used to manipulate the level of Tga1. Loss of Tga1 promoted intense sporulation, while overexpression or an activated allele inhibited sporulation and promoted coiling. Activation of the A. nidulans homolog of Tga1 or loss of its regulator, FlbA, likewise inhibits sporulation and promotes hyphal proliferation (55). In A. nidulans, activated Gα (FadA) promotes active growth and inhibits sporulation (21, 55, 64). Our findings are consistent with the model proposed for Aspergillus. Loss of Tga1 would release the inhibition of sporulation normally mediated by the active GTP-bound form of Gαi. The activated Q203L allele or overexpression of free Tga1 would have the opposite effect, promoting vegetative proliferation. Decreased sporulation and increased proliferation of hyphae may themselves be sufficient to account for the increase in coiling in both kinds of gain-of-function experiments. In contrast, loss of Tga1 due to antisense expression results in profusely sporulating and slowly growing colonies.
To reach a better understanding of how Tga1 acts to modulate sporulation and coiling, we adapted a high-affinity GTPase assay from the animal cell literature. This assay, combined with Western blot data, confirmed that Tga1 is reduced to very low levels in the antisense transformants, that other types of Gα are probably unaffected by the antisense transgene, and that Mas-7 is specific to the Gαi class, as for mammalian G proteins. If Tga1 can activate adenylyl cyclase, then one would expect that the application of Mas-7 can raise intracellular cAMP levels, and this is indeed the case.
Steady-state cAMP levels have been used to infer whether fungal Gα activates or inhibits adenylyl cyclase. The cAMP levels were moderately reduced in Trichoderma overexpressing lines and greatly reduced in antisense lines. Studies with other fungi, such as M. grisea (38), N. crassa (61), and C. trifolii (60), have suggested positive regulation of adenylyl cyclase by Gαi homologs as well as by some members of the other fungal Gα classes (33, 42). Gna3 of N. crassa appears to regulate posttranscriptionally the level of adenylyl cyclase (29). C. parasitica is an exception, in that CPGA1 null mutants display an increase in cAMP levels, suggesting that the G protein inhibits adenylyl cyclase or that this enzyme is stimulated by Gβγ (14). Mammalian adenylyl cyclase isoforms differ in their regulatory sites and properties (57), so more information about the fungal adenylyl cyclases is needed before a general model is produced.
Although steady-state cAMP levels are likely to depend on cellular factors in addition to Gα signaling, we can infer that the decreased sporulation and the increased coiling in overexpressing lines cannot be the result of increased steady-state levels of cAMP, because cAMP levels are somewhat lower in these strains than in wild-type controls. Thus, Tga1 has a cAMP-independent role in promoting coiling, consistent with the current model for Aspergillus (55). Precedent for this interpretation can be found in the animal cell literature; for example, the overexpression of stimulatory Gα in transgenic mice enhanced L-type calcium channels through an adenylyl cyclase-independent route (37). Increased sporulation in our antisense lines could, on the other hand, result from the drastically decreased cAMP levels, consistent with hypersporulation in Aspergillus mutants lacking the catalytic PKA subunit gene pkaA (55). The hallmark of all the cases where the loss of a fungal Gα gene decreases cAMP levels is that exogenous cAMP may rescue some or all of the Gα mutant phenotypes. We found that exogenous cAMP did not rescue the phenotypes of the Trichoderma Tga1 antisense lines (data not shown). Again, this finding could have been the result of cAMP-independent signaling by Tga1. The corresponding experiments with Aspergillus, i.e., the application of cAMP or PKA overexpression in a Gαi (FadA) loss-of-function background, could shed light on this question in a genetically well-defined model system. A further element in the scheme is the release of Gβγ upon activation of the heterotrimer. Overexpression of Gα would be expected to titrate Gβγ, diminishing any Gβγ signal. Expression of the activated Q203L allele, in contrast, should not affect the availability of Gβγ. The similarity in the phenotypes conferred by overexpression and by the activated allele suggests that Gβγ does not play a major role in sporulation and coiling.
In Trichoderma, light, which induces sporulation, triggers a transient biphasic oscillation in intracellular cAMP levels followed by the start of an upward trend (17). Exogenous cAMP promotes sporulation in the dark (45), and light results in the activation of adenylyl cyclase (31). This scenario appears difficult to reconcile with the decreased cAMP levels in the hypersporulating antisense lines. It is important to note, however, that transient activation and a steady-state low level are quite different effects. The induction and development of sporulation in Trichoderma are distinct events, separable by low temperature or nutrient limitation, and each may be under the control of different cAMP signals. It is development, rather than induction, that is studied in Aspergillus, as well as in our antisense lines.
All the data relevant to mycoparasitism might be arranged in a linear pathway: lectins or other signals, activation of Tga1, increased cAMP levels, activation of PKA, coiling, and gene expression. Some experiments were done as an initial test of this simple model. At the input of the postulated pathway, it was suggested that host lectins play a major role in attachment and that lectins may determine host specificity (24). Further research directed to investigating the role of host lectins in the mycoparasitic process showed that the expression of at least one chitinase is triggered by the detection of lectins in a biomimetic assay (25). These data led to the suggestion that a lectin-carbohydrate recognition event could be the signal that triggers a general parasitic response. Using a variety of lectins to coat nylon fibers for use in biomimetic experiments, we have been able to show that lectins indeed promote attachment to and coiling around the fibers. The fact that all lectins tested induced coiling, regardless of their sugar moiety, suggests that they are not the specificity-determining factor in host recognition by Trichoderma. However, different lectins induced to different degrees (Fig. 1). Thus, it is still possible that saturating amounts of any lectin can produce similar effects but that, under nonsaturating conditions, the lectins may show higher degrees of specificity. Attachment requires the use of a solid support, and under these conditions, it is, unfortunately, not possible to control the amount of lectin linked to the support. Furthermore, Trichoderma may carry receptors capable of recognizing a wide spectrum of lectins; it has a wide host range and attacks fungi with disparate cell surface compositions.
Because one of our goals was to determine the role of tga1 in mycoparasitism, we used our transgenic lines in direct confrontation assays with a host, a common procedure in the selection of strains with high biocontrol capacity. Transformed lines overexpressing Tga1 showed an impressive increase in the capacity of the fungus to overgrow the host, compared to the capacity of the wild type. In contrast, lines blocked in the production of Tga1 were unable to overgrow the host (Fig. 9). Taken together, our data suggest a major role for Tga1 in mycoparasitism.
Furthermore, we determined the levels of cAMP and cAMP-dependent protein kinase activity under conditions where mycoparasitism-related genes are expressed (Fig. 10A). Our results showed in both cases behavior exactly opposite that predicted from the simple linear model outlined above, with decreasing levels of cAMP (and, correspondingly, decreasing PKA activity) during the interaction with the host. Thus, either coiling uses a signaling pathway different from that used to trigger the expression of mycoparasitism-related genes or there is a sequential activation allowing gene expression only when cAMP levels and PKA activity are low again. We can conclude that, although several more signaling elements would need to be studied in Trichoderma, the signaling pathway to mycoparasitic development is not a single linear sequence. This conclusion is expected by analogy with genetic model systems such as Aspergillus and yeasts.
In budding yeast, a combination of cAMP-dependent (heterotrimeric G protein) and cAMP-independent (Ras-mitogen-activated protein kinase) pathways controls filamentous growth (34, 40, 43, 48, 52). Although a budding yeast cannot follow as many morphogenetic pathways as can a filamentous fungus, its signaling pathways can provide a basic biochemical model with which to understand the development of filamentous fungi, as proposed by Banuett (2). From an evolutionary point of view, invasive growth of yeasts and mycoparasitic coiling by filamentous fungi may be related. Both may be triggered by nutrient status and host signals.
Acknowledgments
We are grateful to Edna Cukierman for some of the initial PCR experiments, to Katherine Borkovich for antisera to Neurospora G proteins, and to June Simpson and Plinio Guzmán for critical reading of the manuscript.
This work was supported in part by grants from Conacyt (31790N) and ICGEB (CRP/MEX99-02) to A.H.-E., from the Israel Academy of Sciences (ISF) to B.A.H., and from BARD to I.C.
Víctor Rocha-Ramírez and Carmi Omero contributed equally to this work.
REFERENCES
- 1.Baasiri, R. A., X. Lu, P. S. Rowley, G. E. Turner, and K. A. Borkovich. 1997. Overlapping functions for two G protein α subunits in Neurospora crassa. Genetics 1 47:137-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banuett, F. 1998. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol. Mol. Biol. Rev. 62:249-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barak, R., and I. Chet. 1990. Lectin of Sclerotium rolfsii: its purification and possible function in fungal interaction. J. Appl. Bacteriol. 69:101-112. [Google Scholar]
- 4.Barak, R., Y. Elad, D. Mirelman, and I. Chet. 1985. Lectins: a possible basis for specific recognition in the interaction of Trichoderma and Sclerotium rolfsii. Phytopathology 75:458-462. [Google Scholar]
- 5.Boelker, M. 1998. Sex and crime: heterotrimeric G protein in fungal mating and pathogenesis. Fungal Genet. Biol. 25:143-156. [DOI] [PubMed] [Google Scholar]
- 6.Buss, J. E., S. E. Mumby, P. J. Casey, A. G. Gilman, and B. M. Sefton. 1987. Myristoylated alpha subunits of guanine nucleotide-binding regulatory proteins. Proc. Natl. Acad. Sci. USA 84:7493-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carsolio, C., N. Benhamou, S. Haran, C. Cortés, A. Gutiérrez, I. Chet, and A. Herrera-Estrella. 1999. Role of the Trichoderma harzianum endochitinase gene, ech42, in mycoparasitism. Appl. Environ. Microbiol. 65:929-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassel, D., and Z. Selinger. 1976. Catecholamine-stimulated GTPase activity in turkey erythrocyte membranes. Biochim. Biophys. Acta 452:538-551. [DOI] [PubMed] [Google Scholar]
- 9.Chet, I. 1987. Trichoderma—application, mode of action, and potential as a biocontrol agent of soilborn plant pathogenic fungi, p. 137-160. In I. Chet (ed.), Innovative approaches to plant disease control. John Wiley & Sons, Inc., New York, N.Y.
- 10.Chen, B., S. Gao, G. H. Choi, and D. L. Nuss. 1996. Extensive alteration of fungal gene transcript accumulation and elevation of G-protein-regulated cAMP levels by a virulence-attenuating hypovirus. Proc. Natl. Acad. Sci. USA 93:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortés, C., A. Gutiérrez, V. Olmedo, J. Inbar, I. Chet, and A. Herrera-Estrella. 1998. The expression of genes involved in parasitism by Trichoderma harzianum is triggered by a diffusible factor. Mol. Gen. Genet. 260:218-225. [DOI] [PubMed] [Google Scholar]
- 12.Dutton, J. R., S. Johns, and B. L. Miller. 1997. StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 16:5710-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores, A., I. Chet, and A. Herrera-Estrella. 1997. Improved biocontrol activity of Trichoderma harzianum strains by overexpression of the proteinase encoding gene prb1. Curr. Genet. 31:30-37. [DOI] [PubMed] [Google Scholar]
- 14.Gao, S., and D. L. Nuss. 1996. Distinct roles for two G protein α subunits in fungal virulence, morphology, and reproduction reveled by targeted gene disruption. Proc. Natl. Acad. Sci. USA 93:14122-14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geremia, R. A., G. Goldman, D. Jacobs, W. Ardiles, M. Vila, M. Van Montagu, and A. Herrera-Estrella. 1993. Molecular caracterization of a proteinase-encoding gene, prb1, related to mycoparasitism by Trichoderma harzianum. Mol. Microbiol. 6:1231-1242. [DOI] [PubMed] [Google Scholar]
- 16.Gilman, A. G. 1987. G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 56:615-649. [DOI] [PubMed] [Google Scholar]
- 17.Gresik, M., N. Kolarova, and V. Farkas. 1988. Membrane potential, ATP, and cyclic AMP changes induced by light in Trichoderma viride. Exp. Mycol. 12:295-301. [Google Scholar]
- 18.Harper, J. F., and G. Brooker. 1975. Femtomole sensitive radioimmunoassay for cyclic AMP and cyclic GMP after 2′-O-acetylation by acetic anhydride in aqueous solotions. J. Cyclic Nucleotide Res. 1:207-218. [PubMed] [Google Scholar]
- 19.Hermouet, S., J. J. Merendino, J. S. Gutkind, and A. M. Spiegel. 1991. Activating and inactivating mutations of the α subunit of Gi2 protein have opposite effects on proliferation of NIH 3T3 cells. Proc. Natl. Acad. Sci. USA 88:10455-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrera-Estrella, A., G. H. Goldman, and M. Van Montagu. 1990. High-efficiency transformation system for the biocontrol agents, Trichoderma spp. Mol. Microbiol. 4:839-843. [DOI] [PubMed] [Google Scholar]
- 21.Hicks, J. K., J.-H. Yu, N. P. Keller, and T. H. Adams. 1997. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G protein-dependent signaling pathway. EMBO J. 16:4916-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horwitz, B. A., A. Sharon, S. Lu, V. Ritter, T. M. Sandrock, O. C. Yoder, and B. G. Turgeon. 1999. A G protein alpha subunit from Cochliobulus heterostrophus involved in mating and appressorium formation. Fungal Genet. Biol. 26:19-32. [DOI] [PubMed] [Google Scholar]
- 23.Inbar, J., and I. Chet. 1992. Biomimics of fungal cell-cell recognition by use of lectin-coated nylon fibers. J. Bacteriol. 174:1055-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inbar, J., and I. Chet. 1994. A newly isolated lectin from the plant pathogenic fungus Sclerotium rolfsii: purification, characterization and role in mycoparasitism. Microbiology 140:651-657. [DOI] [PubMed] [Google Scholar]
- 25.Inbar, J., and I. Chet. 1995. The role of recognition in the induction of specific chitinases during mycoparasitism by Trichoderma harzianum. Microbiology 141:2823-2829. [DOI] [PubMed] [Google Scholar]
- 26.Ivey, F. D., P. N. Hodge, G. E. Turner, and K. A. Borkovich. 1996. The Gαi homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell 7:1283-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, J. D. G. P. Dunsmuir, and J. Bedbrook. 1985. High level expression of induced chimeric genes in regenerated transformed plants. EMBO J. 4:2411-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasahara, S., and D. L. Nuss. 1997. Targed disruption of a fungal G-protein α subunit gene results in incresed vegetative growth but reduced virulence. Mol. Plant-Microbe Interact. 10:984-993. [DOI] [PubMed] [Google Scholar]
- 29.Kays, A. M., P. S. Ruwley, R. A. Baasiri, and K. A. Borkovich. 2000. Regulation of conidiation and adenylyl cyclase levels by the G alpha protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 20:7693-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi, N., and K. McEntee. 1993. Identification of cis and trans components of a novel heat shock stress regulatory pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:248-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolarova, N., J. Haplova, and M. Gresik. 1992. Light-activated adenyl cyclase from Trichoderma viride. FEMS Microbiol. Lett. 72:275-278. [DOI] [PubMed] [Google Scholar]
- 32.Kozak, K. R., M. F. Lyon, I. K. Ross, and K. R. Ian. 1995. Cloning and characterization of a G protein α-encoding gene from the basidiomycete, Coprinus congregatus. Gene 163:133-137. [DOI] [PubMed] [Google Scholar]
- 33.Kruger, J., G. Loubradou, E. Regenfelder, A. Hartmann, and R. Kahmann. 1998. Crosstalk between cAMP and pheromone signalling pathwys in Ustilago maydis. Mol. Gen. Genet. 260:193-198. [DOI] [PubMed] [Google Scholar]
- 34.Kubler, E., H. U. Mosch, S. Rupp, and M. P. Lisanti. 1997. Gpa2p, a G-protein α-subunit, regulateds growth and pseudohyphal development in Sacharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272:20321-20323. [DOI] [PubMed] [Google Scholar]
- 35.Kuhls, K., E. Lieckfeldt, G. J. Samuels, W. Kovacs, W. Meyer, O. Petrini, W. Gams, T. Borner, and C. P. Kubicek. 1996. Molecular evidence that the asexual industrial fungus Trichoderma reesei is a clonal derivative of the ascomycete Hypocrea jecorina. Proc. Natl. Acad. Sci. USA 93:7755-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kullnig, C., R. L. Mach, M. Lorito, and C. P. Kubicek. 2000. Enzyme diffusion from Trichoderma atroviride (= T. harzianum P1) to Rhizoctonia solani is a prerequisite for triggering of Trichoderma ech42 gene expression before mycoparasitic contact. Appl. Environ. Microbiol. 66:2232-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lader, A. S., Y. F. Xiao, Y. Ishikawa, Y. Cui, D. E. Vatner, S. F. Vatner, C. J. Homcy, and H. F. Cantiello. 1998. Cardiac Gsalpha overexpression enhances L-type calcium channels through an adenylyl cyclase independent pathway. Proc. Natl. Acad. Sci. USA 95:9669-9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, S., and R. A. Dean. 1997. G protein α subunit genes control growth:development, and pathogenicity of Magnaporthe grisea. Mol. Plant-Microbe Interact. 10:1075-1086. [DOI] [PubMed] [Google Scholar]
- 39.Lochrie, M. A., J. E. Mendel, P. W. Sterberg, and M. I. Simon. 1991. Homologous and unique G protein alpha subunits in the nematode Caenorhabditis elegans. Cell Regul. 2:135-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein homolog. EMBO J. 16:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorito, M., C. K. Hayes, A. Di Pietro, and G. E. Harman. 1993. Biolistic transformation of Trichoderma harzianum and Gliocladium virens using plasmid and genomic DNA. Curr. Genet. 24:349-356. [DOI] [PubMed] [Google Scholar]
- 42.Loubradou, G., J. Beguret, and B. Turcq. 1999. MOD-D, a G alpha subunit of fungus Podospora anserina, is involved in both regulation of development and vegetative incompatibility. Genetics 152:519-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mösch, H. U., R. L. Roberts, and G. R. Fink. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5352-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neer, E. J., and D. E. Clapham. 1988. Roles of G protein subunits in transmembrane signalling. Nature 333:129-134. [DOI] [PubMed] [Google Scholar]
- 45.Nemcovic, M., and V. Farkas. 1998. Stimulation of conidiation by derivatives of cAMP in Trichoderma viride. Folia Microbiol. 43:399-402. [Google Scholar]
- 46.Ofek, I., I. Kahane, and N. Sharon. 1996. Toward anti-adhesion therapy for microbial diseases. Trends Microbiol. 4:297-299. [DOI] [PubMed] [Google Scholar]
- 47.Omero, C., J. Inbar, V. Rocha-Ramírez, A. Herrera-Estrella, I. Chet, and B. A. Horwitz. 1999. G protein activators and cAMP promote mycoparasitic behavior in Trichoderma harzianum. Mycol. Res. 103:1637-1642. [Google Scholar]
- 48.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raeder, U., and P. Broda. 1989. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1:17-20. [Google Scholar]
- 50.Regenfelder, E., T. Spellig, A. Hartmann, S. Lauenstein, M. Boelker, and R. Kahmann. 1997. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 16:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosen, S., J. H. Yu, and T. H. Adams. 1999. The Aspergillus nidulans sfaD gene encodes a G protein beta subunit that is required for normal growth and repression of sporulation. EMBO J. 18:5592-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rupp, S., E. Summers, H. J. Lo, H. Madhani, and G. Fink. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strathmann, M., T. M. Wilkie, and M. I. Simon. 1989. Diversity of the G-protein family: sequences from five additional α subunits in the mouse. Proc. Natl. Acad. Sci. USA 86:7407-7409. [DOI] [PMC free article] [PubMed]
- 57.Taussig, R., W. J. Tang, J. R. Hepler, and A. G. Gilman. 1994. Distinct patterns of bidirectional regulation of mammalian adenylyl cyclases. J. Biol. Chem. 269:6093-6100. [PubMed]
- 58.Tilburn, J., S. Sarkar, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Peñalva, and H. N. Arst. 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomes, D. T., M. C. Ross, and D. D. Songstad. 1995. Direct DNA transfer into intact plant cells via microprojectile bombardment, p. 197-213. In O. L. Gamborg and G. C. Phillips (ed.), Plant cell, tissue and organ culture: fundamental methods. Springer-Verlag, Berlin, Germany.
- 60.Truesdell, G. M., Z. Yang, and M. B. Dickman. 2000. A Gα subunit gene from the phytopathogenic fungus Colletotrichum trifolii is required for conidial germination. Physiol. Mol. Plant Pathol. 56:131-140. [Google Scholar]
- 61.Turner, G. E., and K. A. Borkovich. 1993. Identification of a G protein α subunit from Neurospora crassa that is a member of the Gi family. J. Biol. Chem. 268:14805-14811. [PubMed] [Google Scholar]
- 62.West, J. R., E. J. Moss, M. Vaughan, and T. Liu. 1985. Pertussis toxin-catalyzed ADP-ribosylation of transducin. J. Biol. Chem. 260:14428-14430. [PubMed] [Google Scholar]
- 63.Wilkie, T. M., P. A. Scherle, M. P. Strathmann, V. Z. Slepak, and M. I. Simon. 1991. Characterization of G-protein alpha subunits in the Gq class: expression in murine tissues and in stromal and hematopoietic cell lines. Proc. Natl. Acad. Sci. USA 88:10049-10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, J. H. J. Wieser, and T. H. Adams. 1996. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 15:5184-5190. [PMC free article] [PubMed] [Google Scholar]