Abstract
Plant pathogens deliver effector proteins into both the host apoplast and host cells. These effectors function to colonize the host typically by altering host physiology or by subverting plant immune responses. The host plants have evolved intracellular nucleotide-binding site leucine-rich repeat (NBS-LRR) immunoreceptors that directly or indirectly recognize specific effector(s) to trigger plant immunity that prevents colonization. To circumvent effector-triggered immunity, adapted pathogens rely on constantly effectors evolution to further enhance susceptible host colonization. During the past few years, evidence has arisen that many effectors containing tandem repeat modules are particularly prone to rapid evolution through module insertion/deletion/shuffling, point mutations or adoption of other function domains. In this review, we highlight the diverse function of two modular effectors: TAL effectors in prokaryotic bacteria, (L) WY effectors in eukaryotic oomycetes, focus on new insights and the potential role of modularity in effector evolution, and discuss avenues for future research.
Keywords: Xanthomonas, Phytophthora, Effectors, Tandem-repeat modules, TAL effectors, (L)WY effectors
Introduction
A number of specialized proteins, known as effectors, are released into the tissues of host plants by a wide range of plant-associated pathogens, including bacteria, fungi, oomycetes, nematodes and insects (Deslandes and Rivas 2012; Bozkurt et al. 2012). These effectors play key roles in facilitating colonization by altering physiological processes in the host plants or by manipulating their immune defenses (Hogenhout et al. 2009). In response, certain host plants have evolved complex immune systems with receptor proteins that are able to detect, either directly or indirectly, one or more of these effectors or the changes they cause in host targets (Cui et al. 2015; Böhm et al. 2014; Jones and Dangl 2006). This recognition triggers robust immune responses aimed at thwarting the pathogen's colonization efforts. To circumvent these immune barriers or to gain additional advantages in colonizing susceptible hosts, plant-associated pathogens often undergo effector modification through the process of adaptive evolution. This evolution is driven by the intense selective pressure of the host immune system, resulting in the appearance of novel, altered or enhanced effector functions that enhance the pathogen's colonizing capabilities (Dong et al. 2014; Win et al. 2007; Stergiopoulos et al. 2007; Deslandes and Rivas 2012; Bozkurt et al. 2012).
The effectors produced by plant-associated pathogens exhibit a diverse array of characteristics. Many of these proteins possess a signal peptide that directs their secretion or delivery into the host, ensuring their targeted deployment (Deslandes and Rivas 2012; Bozkurt et al. 2012). The tandem repeat module is a common feature of effector proteins produced by many pathogens (Franceschetti et al. 2017; Dong and Ma 2021). For example, effectors from the bacterial wilt pathogen, Ralstonia solanacearum, such as RipAP, RipBB, RipBC, and RipY, feature ankyrin repeats (Peeters et al. 2013). A number of effectors from R. solanacearum (RipG1-RipG7), and Xanthomonas species (XopAC, XopAE, and XopL) contain leucine-rich repeat (LRR) domains (Peeters et al. 2013; Xu et al. 2008; White et al. 2009). Effectors from both R. solanacearum and Xanthomonas species, including RipS1-RipS8, XopAD, and XopN, are characterized by the presence of HEAT/armadillo repeats (Peeters et al. 2013; White et al. 2009). In addition, the poplar leaf rust fungus Melampsora larici-populina produces three effectors known as Chloroplast-targeted protein 1–3 (CTP1-3). These proteins are characterized by carrying two or three amphipathic imperfect near-tandem repeats (Petre et al. 2016). A prominent example of modular effectors is the transcription activator-like (TAL) effector family produced by the bacterium Xanthomonas species. These effectors possess repeating modules, each containing variant residues that enable them to bind specifically to host DNA sequences of different lengths and nucleotide compositions (Timilsina et al. 2020; Perez-Quintero 2019). By rearranging these repeat units in different combinations, TAL effectors can target and manipulate specific genes in the host plant, underlining their remarkable versatility and adaptability (Timilsina et al. 2020; Perez-Quintero 2019). Another notable class of effectors that feature tandem repeats is the (L)WY effector family, which is highly abundant in oomycete species belonging to the Phytophthora and downy mildew lineages (Ma et al. 2018; Mesarich et al. 2015; Dong and Ma 2021). These modular proteins are distinguished by their RXLR motif, a characteristic sequence that facilitates their translocation into host cells after being released from the signal peptide (Dong and Ma 2021). The effector domains of (L)WY effectors with various tandem repeat (L)WY modules are designed to target diverse biological processes and subcellular compartments within the host plant, ultimately contributing to the infection strategy and success of the pathogen (Dong and Ma 2021; Li et al. 2023). Here, we focus on the TAL effectors in bacteria and (L)WY effectors in oomycete to discuss recent findings on the function and evolution of these two families of tandem repeat-containing effectors and to compare their commonalities amid differences.
TAL effectors in bacteria
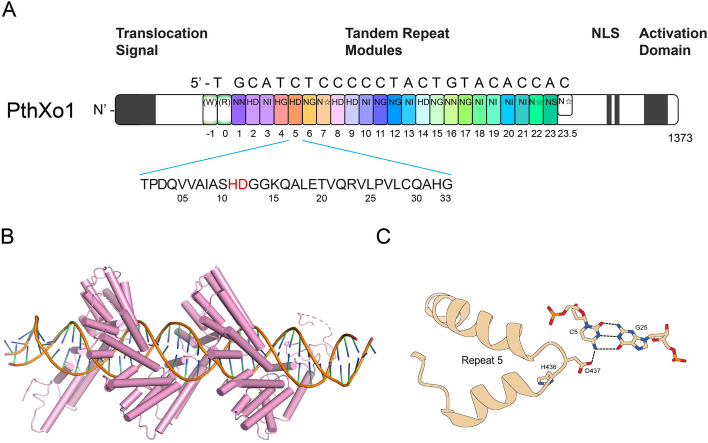
TAL effectors containing remarkable tandem repeat modules from the genus Xanthomonas species are proteins that are injected into the nucleus, bind to the specific sequence of the host genes and modulate their expression, which can either benefit the bacterial colonization or trigger host defense (Kay et al. 2007; Römer et al. 2007). These effectors usually contain an N-terminal translocation signal region that is required for their secretion via the type III secretion system (T3SS), a central DNA-binding domain responsible for sequence specificity, a C-terminal putative motif resembling monopartite nuclear localization signal (NLS), and a C-terminal acidic activation domain (AD) responsible for gene modulation (Boch and Bonas 2010) (Fig. 1A).
Fig. 1.
The domain organization and structure of PthXo1. A The domain organization of a TAL effector, PthXo1, contains N-terminal signals for bacterial type III secretion, tandem repeats specifying the target nucleotide sequence, nuclear localization signals (NLS) and a C-terminal region required for transcriptional activation. PthXo1 contains 23.5 canonical repeats. B The crystal structure of the PthXo1-dsDNA complex. The pink represents the PthXo1 protein. C Topology and contacts between repeat 5 of PthXo1 with a cytosine in the structure. HD (repeat 5) forms a steric and electrostatic contact with cytosine
In each TAL effector, the DNA-binding domain typically consists of a variable number of highly conserved tandem repeat modules of 33 to35 residues in length, which mediate DNA recognition. The nucleotide specificity of the repetitive module in TAL effector is encoded by two adjacent residues, positioned at the 12th and 13th amino acid positions, collectively known as the repeat variable diresidues (RVDs) (Boch et al. 2009; Boch and Bonas 2010). The recognition codes between RVDs and DNA bases have been established experimentally and computationally (Boch et al. 2009; Moscou and Bogdanove 2009). More than 20 types of RVDs have been identified in TAL effectors so far, but only seven types of repeats—His/Asp (HD), Asn/Gly (NG), Asn/Ile (NI), Asn/Asn (NN), Asn/Ser (NS), "N*" (representing a 33 residue repeat in which the RVD appears to lack its second residue), and Asn/Gly (HG)—together account for nearly 90% of all repeats and specifically encode for C, T, A, G/A, A/C/T/G, C/T, and T respectively (Boch et al. 2009; Boch and Bonas 2010). The crystal structure of the Xanthomonas oryzae TAL effector PthXo1 adopts a right-handed super helical structure with 11 repeat units per turn. Upon binding to DNA, it encircles the major groove, positioning the RVD loop on the inside of the helix in direct interaction with the DNA strands (Fig. 1B) (Mak et al. 2012). In the complexes of TAL proteins with double-stranded DNA, each TAL repeat is linked by two loops containing RVDs. The first RVD-containing loop stabilizes the protein backbone through hydrogen bonding, while the second loop establishes base-specific contacts with the dsDNA (Fig. 1C). These interactions enable TAL effectors to adopt a super helical conformation that tracks along the dsDNA (Mak et al. 2012). For now, several other crystal structures of TAL effectors have been solved, including PthA (Murakami et al. 2010), AvrBs3 (Stella et al. 2013), a TALE-like protein from Paraburkholderia rhizoxinica (Stella et al. 2014), which not only shed light on the mechanisms behind DNA-binding specificity, but also made TAL effectors one of artificially engineering tools (Deng et al. 2012b, 2012a; Yin et al. 2012; Gao et al. 2012).
TAL effectors bind the promoters of various host susceptibility (S) genes in a sequence-specific manner, making them an ideal probe for identifying physiological processes that control plant susceptibility to bacteria. Several TAL effectors-associated S genes have been identified (Yang et al. 2006; Antony et al. 2010). One of the best studied examples of TAL effectors and their corresponding S genes are the TAL effectors of X. oryzae pv. oryzae (Xoo) and the SWEET genes of rice. SWEET proteins act as sugar uniporters, which mediate the import and efflux of sugars into and out of animal and plant cells (Chen 2014). Several TAL effectors of Xoo are known to target one of the three SWEET genes in rice (Yu et al. 2011; Yang et al. 2006; Tran et al. 2018; Antony et al. 2010). More specifically, the expression of SWEET11 is induced by the strains encoding the TAL effector PthXo1, SWEET13 is induced by PthXo2 and SWEET14 is induced by any one of the following TAL effectors at distinct site of the SWEET14 promoter: AvrXa7, PthXo3, TalC and TalF (Doucouré et al. 2018; Zhou et al. 2015; Bing and White 2004; Yang et al. 2006; Yu et al. 2011; Tran et al. 2018; Antony et al. 2010; Streubel et al. 2013). The sequence-specific manner of TAL effectors offers the potential to edit the SWEET promoter using CRISPR-Cas9 and engineer broad-spectrum resistance in rice (Oliva et al. 2019). TAL effectors also can target crucial host transcription factors, including the ethylene response factor OsERF123 and the leucine zipper domain (bZIP) transcription factor OsTFX1 in rice (Wang et al. 2017; Sugio et al. 2007; Tran et al. 2018), the basic helix-loop-helix (bHLH) UPA20 in pepper (Kay et al. 2007), and the LOB (lateral organ boundary) family of the transcription factors in Citrus (Zhang et al. 2017; Hu et al. 2014; Doucouré et al. 2018), two bHLH transcription factors in tomato (Schwartz et al. 2017).
As expected in an arms race, plants develop different strategies to effectively reduce bacterial virulence. A simpler strategy, referred to as "loss-of-susceptibility," involves mutating the TAL effector binding element (EBE) of the S gene to prevent it binding of TAL effector. This resistance mechanism underlies some of the recessive resistant (R) genes already described, including xa13 (Yang et al. 2006; Chu et al. 2006), xa25 (Zhou et al. 2015) and xa41 (Hutin et al. 2015b). These mutations either occur within the EBE of the SWEET genes and block the binding of the TAL effector (Hutin et al. 2015a). In addition, TALE-mediated ETI has been used as another counter-attack strategy for plants with NBS-LRR resistance genes identified in tomato and rice. XA1 (Ji et al. 2016; Yoshimura et al. 1998) and Bs4 (Schornack et al. 2004), which are NBS-LRR resistance proteins from rice and tomato, recognize multiple TALEs.
(L)WY effectors in oomycete
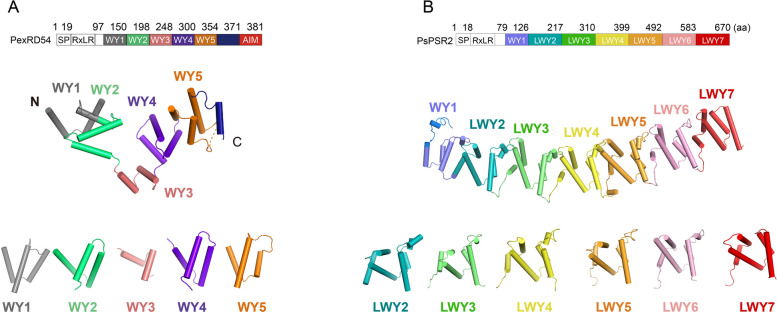
The well-known class of oomycete effectors containing tandem repeat modules are the RXLR effectors, which are strongly enriched in Phytophthora and downy mildew lineages. The family of modular effectors contains an N-terminal RXLR motif consisting of arginine (R), any amino acid (X), leucine (L) and arginine (R), which is critical for the translocation of these effectors (Whisson et al. 2007; Dou et al. 2008). The RXLR motif is followed by the C-terminal effector domain, which shares a set of structurally conserved but highly degenerate folds, termed the WY modules (Jiang et al. 2008). Protein structural analysis subsequently revealed that two hydrophobic residues (Trp (W) and Tyr (Y)) are buried within the core of the helical bundle in each WY module. The WY modules appear to be prevalent in the Peronosporales and were predicted by bioinformatic methods to be present in approximately 44% of Phytophthora infestans RXLR effectors and a quarter of Hyaloperonospora arabidopsidis RXLR effectors. (Goss et al. 2013; Franceschetti et al. 2017; Boutemy et al. 2011; Win et al. 2012). Effectors containing the WY module exist in a variety of forms including single, duplicated, dimeric or tandem repeats (Goss et al. 2013; Franceschetti et al. 2017; Boutemy et al. 2011; Win et al. 2012). Structural analysis of the P. infestans effector PexRD54 shows that it contains five tandem repeat WY modules (Fig. 2A), whereas the P. sojae effector PsAvh240 consists of two WY modules (Dagdas et al. 2016).
Fig. 2.
The crystal structure of two (L)WY effectors, PexRD54 (left) and PsPSR2 (right). A The crystal structure of WY effector, PexRD54, which contains five WY tandem repeat modules. Top: Schematic representation of the domain organization of PexRD54. Middle: The crystal structure of PexRD54. The individual WY module is displayed in a specific color. Bottom: The structure of individual WY module. B The crystal structure of WY effector, PsPSR2, which contains seven (L)WY tandem repeat modules. Top: Schematic representation of the domain organization of PsPSR2. Middle: The crystal structure of PsPSR2. The individual (L)WY module is displayed in a specific color. Bottom: The structure of individual LWY module. SP, secretion signal peptide; RXLR, a translocation motif of Phytophthora effectors
Recently, based on the structure of another WY effector Phytophthora suppressor of RNA silencing 2 (PsPSR2), a new LWY module was defined (Fig. 2B) (He et al. 2019). PsPSR2 is composed of seven tandem repeat modules including an N-terminal WY (WY1) and six LWY modules (LWY2-LWY7). Different from the WY module, each LWY module consists of a conserved five-helix fold with two hydrophobic pockets and the newly defined L motif, a region of about 45 amino acids, contains several conserved residues (often leucine) which are responsible for forming the additional hydrophobic core and interacts with an internal loop from the preceding module (He et al. 2019). This 'joint-like' connection provides directional links that stabilize the concatenation of individual modules, resulting in the highly organized, rod-shaped structure of PsPSR2 (Fig. 2B) (He et al. 2019). The effectors with only WY modules lack conservation in the internal loop due to the absence of the L motif, resulting in different linkage mechanisms between adjacent WY module pairs, such as in the case of PexRD54 (Fig. 2A). As a result, the effectors with only WY modules may exhibit variable protein shapes. Meanwhile, the effectors with LWY modules are generally longer in length than effectors without the (L)WY tandem repeat modules or with only WY modules, resulting in larger surface areas that promote the ability to interact with proteins or other molecules in the host. Approximately 15% of the total number of RXLR effectors consists of the LWY modules in five Phytophthora species, suggesting that this module is also prevalent in Phytophthora effectors (He et al. 2019). (L)WY modules share a low degree of amino acid sequence conservation, limited to a small number of buried residues that are essential for maintaining the fold of individual modules and the overall structure. Functional differentiation of (L)WY effectors may be facilitated by the high degree of sequence plasticity of surface residues and different chimeras of (L)WY modules. Indeed, effectors with different WY/LWY modules or combinations of modules have different molecular functions in the host plant.
A P. sojae effector PsAvh240 contains two WY modules in the C-terminal region (Guo et al. 2019). The first WY module is responsible for its plasma membrane localization and interaction with the soybean aspartic protease 1 (GmAP1), whereas the second WY module is responsible for homodimerization via a molecular handshake arrangement and the effector's repression of GmAP1 secretion to promote infection (Guo et al. 2019). The first two WY/LWY repeat modules in PsPSR2 are sufficient to mediate interaction with a host target protein, Double-Stranded RNA Binding Protein 4 (DRB4), which suppresses trans-kingdom RNAi to promote disease susceptibility (Hou et al. 2019). Recently, the function of other LWY modules of PsPSR2 has been demonstrated, indicating that the LWY2-LWY3 combination of PsPSR2 forms a functional module to recruit the serine/threonine protein phosphatase 2A (PP2A) core enzyme in plant hosts, and that the C-terminal LWY modules may be responsible for the recruitment of PSR2-PP2A complex substrates (Li et al. 2023). Interestingly, while a cluster of LWY effectors sharing PP2A-interacting LWY modules at the amino terminus all have the ability to hijack the host PP2A core enzyme and form functional holoenzymes, they possess C-terminus with a divergent combination of LWY modules and may recruit distinct sets of phosphoproteins to the effector-PP2A complexes in the host (Li et al. 2023). Hyaloperonospora parasitica effector ATR1, which contains two LWY modules, not only confers enhanced virulence to Pst DC3000 on susceptible Arabidopsis accessions, but could also be recognized by an Arabidopsis NLR Recognition of Peronospora parasitica 1 (RPP1) that carries Toll-like interleukin-1 (TIR) receptor domains and elicits a plant immune response (Sohn et al. 2007; Ma et al. 2020; Chou et al. 2011). The linear arrangement of the two tandem repeat LWY modules in ATR1, which folds it into an extended, non-globular overall structure, and the lack of significant sequence identity between the two repeats allow it to evade recognition by rapid evolution and adopt diverse virulence functions (Krasileva et al. 2010; Ma et al. 2020). The broadly conserved WY module-containing effectors in several Phytophthora species, AVRamr3 and its homologues, share similar structures despite extensive sequence variation, but all could be recognized by a potato late blight resistance protein Rpi-amr3, demonstrating that the common fold of the WY module together with sequence polymorphisms on the effector surface may determine the interaction with host proteins (Lin et al. 2022).
In addition, the WY module may employ functional domains to associate with host proteins to promote disease. The crystal structure of the P. infestans effector protein PexRD54 revealed that it contains five WY domains and a short linear motif known as the ATG8-interacting motif (AIM), which binds to the host autophagy protein ATG8CL to stimulate autophagosome formation and subverts host vesicle trafficking (Dagdas et al. 2016; Pandey et al. 2021). The structural arrangement of PexRD54 demonstrates that a tandem (L)WY module or multiple (L)WY combinations could serve as a scaffold to provide a functional domain for interaction with host proteins.
Tandem repeat modules may contribute to effectors diversity
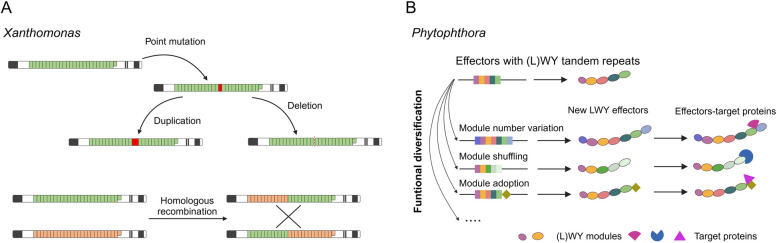
Tandem repeat modules can evolve in a number of ways, including by changes in the number or order of repeat modules (duplication, and insertions/deletions), and by amino acid substitutions, particularly of residues on the surface of the module (Fig. 3A) (He et al. 2019; Perez-Quintero 2019; Dong and Ma 2021). In addition, recombination appears to be particularly common, with multiple repeats set often swapped.
Fig. 3.
The evolutionary mechanisms of TAL effectors and LWY effectors. A Mechanisms described to generate repeated sequence variation in TAL effectors. B Tandem repeat modules -driven effector evolution through the arrangement of the WY/LWY modules. This modular architecture facilitates extensive polymorphism, arising from the insertion, deletion, and shuffling of modules, as well as the integration of additional functional domains
Indeed, there has since been evidence for intra- and inter- genic recombination events among TAL effectors (Yang et al. 2005; Yang and Gabriel 1995). The first example of how repeat module variability could confer an adaptive advantage on effectors was identified through the experimental manipulation of AvrBs3, a TAL effector from Xanthomonas euvesicatoria (Herbers et al. 1992). AvrBs3 binds to the promoter of UPA20, a host gene encoding a basic helix-loop-helix transcription factor, to induce hypertrophy of the plant cell in a compatible interaction with pepper plants (Moscou and Bogdanove 2009; Kay et al. 2007). Whereas AvrBs3 binds to the promoter of Bs3, a pepper gene that encodes an executor resistance protein to induce host immunity in an incompatible interaction with pepper plants (Römer et al. 2009, 2007). To explore whether the repeat module in AvrBs3 provide a source of functional diversity, many AvrBs3 deletion derivatives that differed in the number of their repeat modules were generated. And most of the deletion derivatives lost their ability to induce Bs3-dependent immunity, however, others could gain a new host specificity and induce immunity in the pepper plants with Bs3-E, an allele of Bs3 (Herbers et al. 1992). In addition, the repeat module swaps between AvrBs3 and AvrXa7 provide that the potential for a virulence effector to lose avirulence activity but retain effector virulence function, also suggesting that repeat module variability could confer TAL effectors with an adaptive advantage and rapid evolution (Yang et al. 2005). The recombination of repeat modules in/between TAL effectors could generate novel effectors that target different host genes by altering the DNA binding specificity (Yang et al. 2005; Yang and Gabriel 1995).
For (L)WY effectors, sequence diversification has been shown to play a particularly important role in driving the evolution of tandem repat modules, with sequence conservation of (L)WY modules restricted to a small number of buried residues (He et al. 2019; Jiang et al. 2008; Li et al. 2023). This feature provides a conservative structural framework for these effectors, enabling their divergent evolution and facilitating functional differentiation (Fig. 3B) (He et al. 2019; Jiang et al. 2008; Li et al. 2023). In addition, many (L)WY effectors are a combination of different (L)WY modules, adding another layer of potential to develop novel activities (He et al. 2019; Jiang et al. 2008; Li et al. 2023). The cluster of PP2A-interacting effectors elegantly demonstrated how (L)WY effectors combine the common module with different LWY modules, leading to the diversity in an effector repertoire. Moreover, (L)WY modules also combined with other functional motif(s), further expanding the diversity of effector function (Pandey et al. 2021; Dagdas et al. 2016).
Future perspectives — Precision engineering to create resistant crops
Despite the discovery of two families of tandem repeat-containing effectors: TAL effectors in prokaryotic bacteria, (L) WY effectors in eukaryotic oomycetes, and that many of these effector proteins enhance virulence, we are still in the early stages of understanding how these repetitive modules contribute to virulence. From the limited number of examples analyzed, it is becoming apparent that such repeats provide a mechanism for adaptation through changes in repeat order or number by intra- and inter-genic recombination, and slippage during replication, leading to insertions or deletions (Perez-Quintero 2019; Dong and Ma 2021). However, the current challenge is to develop assays that are sensitive sufficient to detect subtle differences in the function of tandem repeat-containing effectors that are caused by variations in individual repeats, repeats combination, or specific repeat arrangements. Understanding how these variations confer a selective advantage to the pathogen in its co-evolutionary arms race with the host is crucial for developing effective strategies to combat plant diseases caused by pathogens. This requires a deep understanding of the molecular mechanisms underlying effector function and how they evolve over time.
Currently, our understanding of the structural features and molecular mechanisms of TAL effectors and (L)WY effectors empower scientists the potential to precisely manipulate these effector proteins through techniques such as gene editing, protein direct evolution, de novo design, and artificial intelligence (AI). Protein direct evolution and de novo protein design methodologies enable the creation of proteins featuring novel folds not previously observed in nature. Recent advancements have led to the successful development of various tandem repeat proteins using protein direct evolution or de novo design techniques (Harris et al. 2013; Doyle et al. 2023; Jiang et al. 2024). Notably, TAL and (L)WY effectors, which are characterized by their specific repeat numbers, DNA binding sites, interacting proteins, and functional attributes, exhibit significant potential for being crafted through protein direct evolution, de novo design integrated with artificial intelligence. With the advancement of AI tools, such as AlphaFold, large-scale structure prediction and functional characterization of tandem repeat-containing effectors have become feasible. These AI tools have enabled a deeper understanding of the precise roles that repeat modules play in the function and adaptive evolution of these effectors. Recently, a significant number of short TALE-like repeat (STAR) DNA-binding proteins have been identified and functionally characterized, with AlphaFold2/3 playing a crucial role in this process (Hu et al. 2024). It is anticipated that as AI tools continue to evolve, they will play an increasingly pivotal role in future studies of effectors containing tandem repeat modules, which form conserved folds with varying levels of sequence similarity, such as the higher similarity observed in TAL effectors and the lower similarity in WY effectors. These multidisciplinary approaches not only enhance our ability to create disease-resistant crops but also accelerate the pace of agricultural innovation, ensuring global food security in the face of ever-evolving pathogen threats.
Acknowledgements
We apologize to researchers whose relevant studies were not cited in this review due to page limitations. Authors would like to thanks their colleagues, whose suggestions helped in improving the manuscript.
Abbreviations
- AD
Activation domain
- AI
Artificial intelligence
- AIM
ATG8-interacting motif
- bHLH
Basic helix-loop-helix
- DRB4
Double-Stranded RNA Binding Protein 4
- EBE
Effector binding element
- GmAP1
Soybean aspartic protease 1
- L
Leucine
- NBS-LRR
Nucleotide-binding site leucine-rich repeat
- NLS
Nuclear localization signal
- PP2A
Protein phosphatase 2A
- PsPSR2
Phytophthora Suppressor of RNA silencing 2
- R
Arginine
- R genes
Resistant genes
- RPP1
Recognition of Peronospora parasitica 1
- RVDs
Repeat variable diresidues
- S genes
Susceptibility genes
- STAR
Short TALE-like repeat
- TAL
Transcription Activator-like
- TIR
Toll-like interleukin-1
- T3SS
Type III secretion system
- W
Trp
- X
Amino acid
- Xoo
Xanthomonas. oryzae pv. oryzae
- Y
Tyr
Authors’ contributions
H.L. conceived and wrote the paper.
Funding
The work is supported by the National Natural Science Foundation of China (32470318 to H.L.).
Data availability
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
H.L. agrees for publication.
Competing interests
Authors declare that they have no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Antony G, Zhou JH, Huang S, Li T, Liu B, White F, Yang B (2010) Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22(11):3864–3876. 10.1105/tpc.110.078964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing Y, White FF (2004) Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol Plant Microbe interact 17(11):1192–1200. 10.1094/Mpmi.2004.17.11.1192 [DOI] [PubMed] [Google Scholar]
- Boch J, Bonas U (2010) Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 48:419–436. 10.1146/annurev-phyto-080508-081936 [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326(5959):1509–1512. 10.1126/science.1178811 [DOI] [PubMed] [Google Scholar]
- Böhm H, Albert I, Fan L, Reinhard A, Nürnberger T (2014) Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol 20:47–54. 10.1016/j.pbi.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Boutemy LS, King SRF, Win J, Hughes RK, Clarke TA, Blumenschein TMA, Kamoun S, Banfield MJ (2011) Structures of RXLR effector proteins. J Biol Chem 286(41):35834–35842. 10.1074/jbc.M111.262303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt TO, Schornack S, Banfield MJ, Kamoun S (2012) Oomycetes, effectors, and all that jazz. Curr Opin Plant Biol 15(4):483–492. 10.1016/j.pbi.2012.03.008 [DOI] [PubMed] [Google Scholar]
- Chen LQ (2014) SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol 201(4):1150–1155. 10.1111/nph.12445 [DOI] [PubMed] [Google Scholar]
- Chou S, Krasileva KV, Holton JM, Steinbrenner AD, Alber T, Staskawicz BJ (2011) ATR1 effector is a repeat protein with distributed recognition surfaces. Proc Natl Acad Sci U S A 108(32):13323–13328. 10.1073/pnas.1109791108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu ZH, Fu BY, Yang H, Xu CG, Li ZK, Sanchez A, Park YJ, Bennetzen JL, Zhang QF, Wang SP (2006) Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor Appl Genet 112(3):455–461. 10.1007/s00122-005-0145-6 [DOI] [PubMed] [Google Scholar]
- Cui HT, Tsuda K, Parker JE (2015) Effector-triggered immunity: From pathogen perception to robust defense. Annu Rev Plant Biol 66:487–511. 10.1146/annurev-arplant-050213-040012 [DOI] [PubMed] [Google Scholar]
- Dagdas YF, Belhaj K, Maqbool A, Chaparro-Garcia A, Pandey P, Petre B, Tabassum N, Cruz-Mireles N, Hughes RK, Sklenar J, Win J, Menke F, Findlay K, Banfield MJ, Kamoun S, Bozkurt TO (2016) An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. Elife 5:e10856. 10.7554/eLife.10856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D, Yan CY, Pan XJ, Mahfouz M, Wang JW, Zhu JK, Shi YG, Yan NE (2012a) Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335(6069):720–723. 10.1126/science.1215670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D, Yin P, Yan CY, Pan XJ, Gong XQ, Qi S, Xie T, Mahfouz M, Zhu JK, Yan N, Shi YG (2012b) Recognition of methylated DNA by TAL effectors. Cell Res 22(10):1502–1504. 10.1038/cr.2012.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Rivas S (2012) Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci 17(11):644–655. 10.1016/j.tplants.2012.06.011 [DOI] [PubMed] [Google Scholar]
- Dong SM, Ma WB (2021) How to win a tug-of-war: the adaptive evolution of effectors. Curr Opin Plant Biol 62:102027. 10.1016/j.pbi.2021.102027 [DOI] [PubMed] [Google Scholar]
- Dong SM, Stam R, Cano LM, Song J, Sklenar J, Yoshida K, Bozkurt TO, Oliva R, Liu ZY, Tian MY, Win J, Banfield MJ, Jones AME, van der Hoorn RAL, Kamoun S (2014) Effector specialization in a lineage of the Irish potato famine pathogen. Science 343(6170):552–555. 10.1126/science.1246300 [DOI] [PubMed] [Google Scholar]
- Dou DL, Kale SD, Wang X, Jiang RHY, Bruce NA, Arredondo FD, Zhang XM, Tyler BM (2008) RXLR-mediated entry of effector into soybean cells does not require pathogen-encoded machinery. Plant Cell 20(7):1930–1947. 10.1105/tpc.107.056093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucouré H, Pérez-Ouintero AL, Reshetnyak G, Tekete C, Auguy F, Thomas E, Koebnik R, Szurek B, Koita O, Verdier V, Cunnac S (2018) Functional and genome sequence-driven characterization of effector gene repertoires reveals novel variants with altered specificities in closely related Malian pv. strains. Front Microbiol 9:1657. 10.3389/fmicb.2018.01657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle LA, Takushi B, Kibler RD, Milles LF, Orozco CT, Jones JD, Jackson SE, Stoddard BL, Bradley P (2023) De novo design of knotted tandem repeat proteins. Nat Commun 14(1):6746. 10.1038/s41467-023-42388-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschetti M, Maqbool A, Jiménez-Dalmaroni MJ, Pennington HG, Kamoun S, Banfield MJ (2017) Effectors of filamentous plant pathogens: Commonalities amid diversity. Microbiol Mol Biol Rev 81(2):e00066. 10.1128/MMBR.00066-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao HS, Wu XJ, Chai JJ, Han ZF (2012) Crystal structure of a TALE protein reveals an extended N-terminal DNA binding region. Cell Res 22(12):1716–1720. 10.1038/cr.2012.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss EM, Press CM, Grünwald NJ (2013) Evolution of RXLR-class effectors in the oomycete plant pathogen. Plos One 8(11):e79347. 10.1371/journal.pone.0079347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo BD, Wang HN, Yang B, Jiang WJ, Jing MF, Li HY, Xia YA, Xu YP, Hu QL, Wang FF, Yu F, Wang Y, Ye WW, Dong SM, Xing WM, Wang YC (2019) Effector PsAvh240 inhibits host aspartic protease secretion to promote infection. Mol Plant 12(4):552–564. 10.1016/j.molp.2019.01.017 [DOI] [PubMed] [Google Scholar]
- Harris CJ, Slootweg EJ, Goverse A, Baulcombe DC (2013) Stepwise artificial evolution of a plant disease resistance gene. Proc Natl Acad Sci U S A 110(52):21189–21194. 10.1073/pnas.1311134110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JQ, Ye WW, Choi DS, Wu BX, Zhai Y, Guo BD, Duan SY, Wang YC, Gan JH, Ma WB, Ma JB (2019) Structural analysis of suppressor of RNA silencing 2 (PSR2) reveals a conserved modular fold contributing to virulence. Proc Natl Acad Sci U S A 116(16):8054–8059. 10.1073/pnas.1819481116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbers K, Conradsstrauch J, Bonas U (1992) Race-specificity of plant-resistance to bacterial spot disease determined by repetitive motifs in a bacterial avirulence protein. Nature 356(6365):172–174. 10.1038/356172a0 [Google Scholar]
- Hogenhout SA, Van der Hoorn RAL, Terauchi R, Kamoun S (2009) Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe in 22(2):115–122. 10.1094/Mpmi-22-2-0115 [DOI] [PubMed] [Google Scholar]
- Hou YN, Zhai Y, Feng L, Karimi HZ, Rutter BD, Zeng LP, Choi DS, Zhang BL, Gu WF, Chen XM, Ye WW, Innes RW, Zhai JX, Ma WB (2019) A effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe 25(1):153. 10.1016/j.chom.2018.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhang JL, Jia HG, Sosso D, Li T, Frommer WB, Yang B, White FF, Wang NA, Jones JB (2014) Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc Natl Acad Sci U S A 111(4):E521–E529. 10.1073/pnas.1313271111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Zhang X, Sun W, Liu C, Deng P, Cao Y, Zhang C, Xu N, Zhang T, Zhang YE, Liu JG, Wang H (2024) Systematic discovery of DNA-binding tandem repeat proteins. Nucleic Acids Res 52(17):10464–10489. 10.1093/nar/gkae710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin M, Pérez-Quintero AL, Lopez C, Szurek B (2015) MorTAL Kombat: the story of defense against TAL effectors through loss-of-susceptibility. Front Plant Sci 6:647. 10.3389/fpls.2015.00647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin M, Sabot F, Ghesquière A, Koebnik R, Szurek B (2015b) A knowledge-based molecular screen uncovers a broad-spectrum resistance allele to bacterial blight from wild rice. Plant J 84(4):694–703. 10.1111/tpj.13042 [DOI] [PubMed] [Google Scholar]
- Ji ZY, Ji CH, Liu B, Zou LF, Chen GY, Yang B (2016) Interfering TAL effectors of neutralize R-gene-mediated plant disease resistance. Nat Commun 7:13435. 10.1038/ncomms13435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang RHY, Tripathy S, Govers F, Tyler BM (2008) RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci U S A 105(12):4874–4879. 10.1073/pnas.0709303105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Jude KM, Wu K, Fallas J, Ueda G, Brunette TJ, Hicks DR, Pyles H, Yang A, Carter L, Lamb M, Li X, Levine PM, Stewart L, Garcia KC, Baker D (2024) De novo design of buttressed loops for sculpting protein functions. Nat Chem Biol 20(8):974–980. 10.1038/s41589-024-01632-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kay S, Hahn S, Marois E, Hause G, Bonas U (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318(5850):648–651. 10.1126/science.1144956 [DOI] [PubMed] [Google Scholar]
- Krasileva KV, Dahlbeck D, Staskawicz BJ (2010) Activation of an resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22(7):2444–2458. 10.1105/tpc.110.075358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang J, Kuan TA, Tang B, Feng L, Wang J, Cheng Z, Sklenar J, Derbyshire P, Hulin M, Li Y, Zhai Y, Hou Y, Menke FLH, Wang Y, Ma W (2023) Pathogen protein modularity enables elaborate mimicry of a host phosphatase. Cell 186(15):3196-3207 e3117. 10.1016/j.cell.2023.05.049 [DOI] [PubMed] [Google Scholar]
- Lin X, Olave-Achury A, Heal R, Pais M, Witek K, Ahn HK, Zhao H, Bhanvadia S, Karki HS, Song TQ, Wu CH, Adachi H, Kamoun S, Vleeshouwers VGAA, Jones JDG (2022) A potato late blight resistance gene protects against multiple species by recognizing a broadly conserved RXLR-WY effector. Mol Plant 15(9):1457–1469. 10.1016/j.molp.2022.07.012 [DOI] [PubMed] [Google Scholar]
- Ma LS, Pellegrin C, Kahmann R (2018) Repeat-containing effectors of filamentous pathogens and symbionts. Curr Opin Microbiol 46:123–130. 10.1016/j.mib.2018.01.007 [DOI] [PubMed] [Google Scholar]
- Ma SC, Lapin D, Liu L, Sun Y, Song W, Zhang XX, Logemann E, Yu DL, Wang J, Jirschitzka J, Han ZF, Schulze-Lefert P, Parker JE, Chai JJ (2020) Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370(6521):1184 eabe3069. 10.1126/science.abe3069 [DOI] [PubMed] [Google Scholar]
- Mak ANS, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL (2012) The Crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335(6069):716–719. 10.1126/science.1216211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesarich CH, Bowen JK, Hamiaux C, Templeton MD (2015) Repeat-containing protein effectors of plant-associated organisms. Front Plant Sci 6:872. 10.3389/fpls.2015.00872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ (2009) A simple cipher governs DNA recognition by TAL effectors. Science 326(5959):1501–1501. 10.1126/science.1178817 [DOI] [PubMed] [Google Scholar]
- Murakami MT, Sforça ML, Neves JL, Paiva JH, Domingues MN, Pereira ALA, Zeri ACD, Benedetti CE (2010) The repeat domain of the type III effector protein PthA shows a TPR-like structure and undergoes conformational changes upon DNA interaction. Proteins 78(16):3386–3395. 10.1002/prot.22846 [DOI] [PubMed] [Google Scholar]
- Oliva R, Ji CH, Atienza-Grande G, Huguet-Tapia JC, Perez-Quintero A, Li T, Eom JS, Li CH, Nguyen H, Liu B, Auguy F, Sciallano C, Luu V, Dossa GS, Cunnac S, Schmidt SM, Slamet-Loedin IH, Cruz CV, Szurek B, Frommer WB, White FF, Yang B (2019) Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat Biotechnol 37(11):1344. 10.1038/s41587-019-0267-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P, Leary AY, Tumtas Y, Savage Z, Dagvadorj B, Duggan C, Yuen ELH, Sanguankiattichai N, Tan E, Khandare V, Connerton AJ, Yunusov T, Madalinski M, Mirkin FG, Schornack S, Dagdas Y, Kamoun S, Bozkurt TO (2021) An oomycete effector subverts host vesicle trafficking to channel starvation-induced autophagy to the pathogen interface. Elife 10:e65285. 10.7554/eLife.65285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N, Carrere S, Anisimova M, Plener L, Cazale AC, Genin S (2013) Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics 14:859. 10.1186/1471-2164-14-859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Quintero AL (2019) Szurek B (2019) A decade decoded: Spies and hackers in the history of TAL effectors research. Annu Rev Phytopathol 57(57):459–481. 10.1146/annurev-phyto-082718-100026 [DOI] [PubMed] [Google Scholar]
- Petre B, Lorrain C, Saunders DGO, Win J, Sklenar J, Duplessis S, Kamoun S (2016) Rust fungal effectors mimic host transit peptides to translocate into chloroplasts. Cell Microbiol 18(4):453–465. 10.1111/cmi.12530 [DOI] [PubMed] [Google Scholar]
- Römer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T (2007) Plant pathogen recognition mediated by promoter activation of the pepper resistance gene. Science 318(5850):645–648. 10.1126/science.1144958 [DOI] [PubMed] [Google Scholar]
- Römer P, Strauss T, Hahn S, Scholze H, Morbitzer R, Grau J, Bonas U, Lahaye T (2009) Recognition of AvrBs3-Like proteins is mediated by specific binding to promoters of matching pepper alleles. Plant Physiol 150(4):1697–1712. 10.1104/pp.109.139931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack S, Ballvora A, Gürlebeck D, Peart J, Baulcombe D, Ganal M, Baker B, Bonas U, Lahaye T (2004) The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severly truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J 37(5):46–60. 10.1111/j.1365-313X.2004.02045.x [DOI] [PubMed] [Google Scholar]
- Schwartz AR, Morbitzer R, Lahaye T, Staskawicz BJ (2017) TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proc Natl Acad Sci U S A 114(5):E897–E903. 10.1073/pnas.1620407114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn KH, Lei R, Nemri A, Jones JDG (2007) The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell 19(12):4077–4090. 10.1105/tpc.107.054262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella S, Molina R, Yefimenko I, Prieto J, Silva G, Bertonati C, Juillerat A, Duchateau P, Montoya G (2013) Structure of the AvrBs3-DNA complex provides new insights into the initial thymine-recognition mechanism. Acta Crystallogr D 69:1707–1716. 10.1107/S0907444913016429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella S, Molina R, López-Méndez B, Juillerat A, Bertonati C, Daboussi F, Campos-Olivas R, Duchateau P, Montoya G (2014) BuD, a helix-loop-helix DNA-binding domain for genome modification. Acta Crystallogr D 70:2042–2052. 10.1107/S1399004714011183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos I, De Kock MJD, Lindhout P, De Wit PJGM (2007) Allelic variation in the effector genes of the tomato pathogen reveals different modes of adaptive evolution. Mol Plant Microbe Interact 20(10):1271–1283. 10.1094/Mpmi-20-10-1271 [DOI] [PubMed] [Google Scholar]
- Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B (2013) Five phylogenetically close rice genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol 200(3):808–819. 10.1111/nph.12411 [DOI] [PubMed] [Google Scholar]
- Sugio A, Yang B, Zhu T, White FF (2007) Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAγ1 and OsTFX1 during bacterial blight of rice. Proc Natl Acad Sci U S A 104(25):10720–10725. 10.1073/pnas.0701742104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timilsina S, Potnis N, Newberry EA, Liyanapathiranage P, Iruegas-Bocardo F, White FF, Goss EM, Jones JB (2020) Xanthomonas diversity, virulence and plant-pathogen interactions. Nat Rev Microbiol 18(8):415–427. 10.1038/s41579-020-0361-8 [DOI] [PubMed] [Google Scholar]
- Tran TT, Pérez-Quintero AL, Wonni I, Carpenter SCD, Yu YH, Wang L, Leach JE, Verdier V, Cunnac S, Bogdanove AJ, Koebnik R, Hutin M, Szurek B (2018) Functional analysis of African Xanthomonas oryzae pv. oryzae TALomes reveals a new susceptibility gene in bacterial leaf blight of rice. Plos Pathog 14(6):e1007092. 10.1371/journal.ppat.1007092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Rinaldi FC, Singh P, Doyle EL, Dubrow ZE, Tran TT, Pérez-Quintero AL, Szurek B, Bogdanove AJ (2017) TAL effectors drive transcription bidirectionally in plants. Mol Plant 10(2):285–296. 10.1016/j.molp.2016.12.002 [DOI] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, Hein I, Toth IK, Pritchard L, Birch PRJ (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450(7166):115. 10.1038/nature06203 [DOI] [PubMed] [Google Scholar]
- White FF, Potnis N, Jones JB, Koebnik R (2009) The type III effectors of Xanthomonas. Mol Plant Pathol 10(6):749–766. 10.1111/j.1364-3703.2009.00590.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win J, Morgan W, Bos J, Krasileva KV, Cano LM, Chaparro-Garcia A, Ammar R, Staskawicz BJ, Kamoun S (2007) Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell 19(8):2349–2369. 10.1105/tpc.107.051037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win J, Krasileva KV, Kamoun S, Shirasu K, Staskawicz BJ, Banfield MJ (2012) Sequence divergent RXLR effectors share a structural fold conserved across plant pathogenic Oomycete species. Plos Pathog 8(1):e1002400. 10.1371/journal.ppat.1002400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RQ, Blanvillain S, Feng JX, Jiang BL, Li XZ, Wei HY, Kroj T, Lauber E, Roby D, Chen B, He YQ, Lu GT, Tang DJ, Vasse J, Arlat M, Tang JL (2008) AvrAC(Xcc8004), a type III effector with a leucine-rich repeat domain from Xanthomonas campestris pathovar campestris confers avirulence in vascular tissues of Arabidopsis thaliana ecotype Col-0. J Bacteriol 190(1):343–355. 10.1128/JB.00978-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gabriel DW (1995) Intragenic recombination of a single plant pathogen gene provides a mechanism for the evolution of new host specificities. J Bacteriol 177(17):4963–4968. 10.1128/jb.177.17.4963-4968.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF (2005) Avoidance of host recognition by alterations in the repetitive and C-terminal regions of AvrXa7, a type III effector of Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact 18(2):142–149. 10.1094/Mpmi-18-0142 [DOI] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF (2006) Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci U S A 103(27):10503–10508. 10.1073/pnas.0604088103 [DOI] [PMC free article] [PubMed]
- Yin P, Deng D, Yan CY, Pan XJ, Xi JJ, Yan N, Shi YG (2012) Specific DNA-RNA hybrid recognition by TAL effectors. Cell Rep 2(4):707–713. 10.1016/j.celrep.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang ZX, Kono I, Kurata N, Yano M, Iwata N, Sasaki T (1998) Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc Natl Acad Sci U S A 95(4):1663–1668. 10.1073/pnas.95.4.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YH, Streubel J, Balzergue S, Champion A, Boch J, Koebnik R, Feng JX, Verdier V, Szurek B (2011) Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice Nodulin-3 Os11N3 gene. Mol Plant Microbe Interact 24(9):1102–1113. 10.1094/Mpmi-11-10-0254 [DOI] [PubMed] [Google Scholar]
- Zhang JL, Huguet-Tapia JC, Hu Y, Jones J, Wang N, Liu SZ, White FF (2017) Homologues of in citrus function as disease susceptibility genes in citrus canker. Mol Plant Pathol 18(6):798–810. 10.1111/mpp.12441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JH, Peng Z, Long JY, Sosso D, Liu B, Eom JS, Huang S, Liu SZ, Cruz CV, Frommer WB, White FF, Yang B (2015) Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J 82(4):632–643. 10.1111/tpj.12838 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.