Abstract
The Schizosaccharomyces pombe fbp1 gene, encoding fructose-1,6-bisphosphatase, is transcriptionally repressed by glucose. Mutations that confer constitutive fbp1 transcription identify git (glucose-insensitive transcription) genes that encode components of a cyclic AMP (cAMP) signaling pathway required for adenylate cyclase activation. Four of these genes encode the three subunits of a heterotrimeric G protein (gpa2, git5, and git11) and a G protein-coupled receptor (git3). Three additional genes, git1, git7, and git10, act in parallel to or downstream from the G protein genes. Here, we describe the cloning and characterization of the git7 gene. The Git7p protein is a member of the Saccharomyces cerevisiae Sgt1p protein family. In budding yeast, Sgt1p associates with Skp1p and plays an essential role in kinetochore assembly, while in Arabidopsis, a pair of SGT1 proteins have been found to be involved in plant disease resistance through an interaction with RAR1. Like S. cerevisiae Sgt1p, Git7p is essential, but this requirement appears to be due to roles in septation and cell wall integrity, which are unrelated to cAMP signaling, as S. pombe cells lacking either adenylate cyclase or protein kinase A are viable. In addition, git7 mutants are sensitive to the microtubule-destabilizing drug benomyl, although they do not display a chromosome stability defect. Two alleles of git7 that are functional for cell growth and septation but defective for glucose-triggered cAMP signaling encode proteins that are altered in the highly conserved carboxy terminus. The S. cerevisiae and human SGT1 genes both suppress git7-93 but not git7-235 for glucose repression of fbp1 transcription and benomyl sensitivity. This allele-specific suppression indicates that the Git7p/Sgt1p proteins may act as multimers, such that Git7-93p but not Git7-235p can deliver the orthologous proteins to species-specific targets. Our studies suggest that members of the Git7p/Sgt1p protein family may play a conserved role in the regulation of adenylate cyclase activation in S. pombe, S. cerevisiae, and humans.
Transcription of the Schizosaccharomyces pombe fbp1 gene is repressed by glucose (23, 48). Previous studies showed that glucose detection triggers a cyclic AMP (cAMP) signal responsible for the activation of cAMP-dependent protein kinase A, which acts to repress fbp1 transcription (11, 20, 26). Glucose starvation leads to the activation of the Spc1p stress-activated mitogen-activated protein kinase pathway, which in turn activates the heterodimeric bZIP transcriptional activator Atf1p-Pcr1p (27, 44, 45, 51). These two signaling pathways regulate fbp1 transcription via at least two distinct mechanisms, with Atf1p-Pcr1p acting both directly at one cis-acting element (UAS1) and indirectly at a second element (UAS2) (25). Transcriptional activation from UAS2 appears to be carried out by the Rst2p zinc finger protein (19). Additional regulators of fbp1 transcription include Tup11p and Tup12p, homologs of the Saccharomyces cerevisiae Tup1p global corepressor, which are required for fbp1 repression, and the CCAAT box binding factor, which is required in concert with Atf1p-Pcr1p for fbp1 derepression (25).
The S. pombe adenylate cyclase (Git2p/Cyr1p) activation mechanism resembles that of the mammalian enzyme (46) in that it involves the activity of a heterotrimeric G protein coupled to a seven-transmembrane domain receptor protein. In S. pombe, the gpa2, git5, and git11 genes encode the Gα, Gβ, and Gγ subunits, respectively, while the git3 gene encodes the putative G protein-coupled receptor (GPCR) (13, 19, 20, 23, 36). The role of these four genes is to activate Gpa2p Gα, as mutational activation of Gpa2p or overexpression of the wild-type gpa2+ gene bypasses the need for Git5p Gβ, Git11p Gγ, and Git3p GPCR (31, 32, 37, 52). The Git3p, Gpa2p, and Git2p/Cyr1p proteins also display sequence homology to S. cerevisiae Gpr1p GPCR, Gpa2p Gα, and Cyr1p adenylate cyclase, which act in the glucose-triggered cAMP signaling pathway (12, 30, 53), and are involved in the regulation of pseudohyphal growth (33, 40). Unlike S. pombe, S. cerevisiae does not appear to express a Gβγ dimer that functions in this signaling pathway. Related cAMP signaling pathways are also found in a number of fungal pathogens. For example, the human fungal pathogen Cryptococcus neoformans uses Gα (Gpa1) (2) and adenylate cyclase (Cac1) (3), which resemble S. pombe Gpa2p and Git2p, to detect nutrient conditions and to regulate aspects of differentiation and virulence.
Three additional S. pombe genes, git1, git7, and git10, required for glucose-stimulated adenylate cyclase activation (11, 21), are still required for glucose repression of fbp1 transcription in a strain carrying an activated allele of gpa2 or overexpressing gpa2+ (37, 52). Thus, Git1p, Git7p, and Git10p presumably regulate adenylate cyclase activation either downstream from or in parallel to Gpa2p Gα. In addition, git7-235 strains display a temperature-sensitive lethal growth phenotype (21), suggesting an essential role for this gene independent of cAMP signaling, as both adenylate cyclase and protein kinase A are dispensable in S. pombe (20, 34, 35).
In this article, we describe the cloning and characterization of the S. pombe git7 gene. Sequence analysis shows that Git7p is a member of the Sgt1p protein family found in many eukaryotes, including the budding yeast S. cerevisiae, plants, and mammals. S. cerevisiae Sgt1p associates with Skp1p and is required for kinetochore assembly (29), while Arabidopsis SGT1a and SGT1b associate with RAR1, playing a role in disease resistance (4, 5). Skp1p is a component of the SCF (Skp1p- Cullin-F-box) E3 ubiquitin ligase responsible for the polyubiquitination of proteins that are subsequently degraded by the proteasome (41); however, there is no evidence that the role of Skp1p and Sgt1p in kinetochore assembly involves ubiquitin ligase activity. Conservation of Sgt1p function in eukaryotes is suggested by the ability of the human ortholog HuSgt1p to restore viability to a strain lacking S. cerevisiae Sgt1p activity (29) and by the ability of the Arabidopsis orthologs to restore temperature-resistant growth to S. cerevisiae sgt1-3 and sgt1-5 mutant strains (5). Our work shows that, like S. cerevisiae Sgt1p, S. pombe Git7p is an essential protein, although cell death in S. pombe is associated with cell lysis along with cell division and septation defects. Two git7 alleles that alter the carboxy terminus of Git7p confer a defect in fbp1 regulation without affecting cell wall integrity and septation, suggesting that Git7p possesses discrete functional domains. Finally, we show that the expression of the human and S. cerevisiae Sgt1p proteins can restore fbp1 transcriptional regulation in a git7-93 mutant but not in a git7-235 mutant. Such allele-specific suppression suggests that the Git7p/Sgt1p proteins form multimeric complexes that play conserved roles in a variety of cellular processes, including the regulation of adenylate cyclase activity, cell division, septation, and kinetochore assembly.
MATERIALS AND METHODS
S. pombe strains and growth media.
The S. pombe strains used in this study are listed in Table 1. The fbp1::ura4+ and ura4::fbp1-lacZ reporters (21) are translational fusions integrated in single copies at the fbp1 and ura4 loci, respectively. Rich media YEA and YEL (16) were supplemented with 2% Casamino Acids. Defined medium PM (50) was supplemented with required nutrients at 75 mg/liter, except for l-leucine, which was used at 150 mg/liter. Solid medium SC containing 0.4 g of 5-fluoro-orotic acid (5-FOA)/liter and 8% glucose was used to determine 5-FOA resistance (21), which is a reflection of glucose repression of the fbp1-ura4 reporter. Strains were grown at 30°C unless otherwise indicated.
TABLE 1.
Strain list
| Strain | Genotype |
|---|---|
| CHP27 | h+fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M210 his7-366 git7-27 |
| CHP93 | h+fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M210 his7-366 git7-93 |
| CHP449 | h+fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M216 his7-366 git7-235 |
| CHP556 | h+fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M216 his3-D1 git7-93 |
| CHP558 | h90fbp1::ura4 leu1-32 ade6-M216 git2-1::LEU2+ |
| CHP578 | h+fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M210 his 3-D1 |
| CHP594 | h−fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M210 his3-D1 |
| CHP595 | h−fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M216 his3-D1 |
| CHP718 | h+fbp1::ura4 ura4-D18 leu1-32 ade6-M210 his3-D1 git7-235 |
| CHP758 | h+fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M210 git7-GFP::kan |
| CHP767 | h+fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M210 git7-GFP::kan skp1::ura4+ars1(MluI)::pREP41-skp1-3×HA LEU2+ |
| CHP792 | h90 fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M216 his7-366 git7-GFP::kan |
| CHP795 | h90 fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M216 his7-366 |
| CHP803 | h−ura4::fbp1-lacZ leu1-32 ade6-M210 skp1::ura4+ars1(MluI)::pREP41-skp1-3×HA LEU2+ |
| FWP72 | h−fbp1::ura4 ura4::fbp1-lacZ leu1-32 |
| FWP101 | h+fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M210 his7-366 |
| FWP145 | h−fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M216 git7-235 |
| KSP1 | h−fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M210 his3-D1 git7-27 |
| KSP2 | h−ura4::fbp1-lacZ leu1-32 ade6-M210 git7-GFP::kan (Ch16 ade6-M216 m23::ura4+) |
| KSP5 | h+fbp1::ura4 ura4::fbp1-lacZ leu1-32 ade6-M210 his7-366 git7-V5::intLEU2+ |
| PBP1 | h−fbp1::ura4 ura4::fbp1-lacZ leu1-32 git7-GFP::kan |
Recombinant DNA Methods.
All DNA manipulations were performed, unless otherwise stated, by using reagents and protocols from New England Biolabs. Escherichia coli transformations were done by using XL1-Blue electroporation-competent cells (Stratagene). The Failsafe PCR system (Epicentre Technologies) was used for PCR according to the manufacturer's instructions. Oligonucleotides were obtained from Integrated DNA Technologies. S. pombe plasmid transformations were performed by overnight incubation with polyethylene glycol-lithium acetate-Tris-EDTA buffer (13) or as described by Bähler et al. (6).
Cloning of the git7 gene.
A pBG2-based S. pombe genomic DNA library (38) was screened for clones that confer 5-FOA resistance (5-FOAr) to host strain CHP556 (git7-93), indicating the restoration of glucose repression of the fbp1-ura4 reporter. From 33,000 transformants, 37 5-FOAr candidates were identified. Plasmids from these strains were rescued in E. coli (22) and screened for their ability to restore growth at 37°C to S. pombe strain CHP718, which is temperature sensitive for growth due to the git7-235 allele. Two of the 37 plasmid candidates conferred temperature-resistant growth to strain CHP718. These two plasmids, one of which was designated pHF1, contained identical inserts. Plasmid pHF1 was linearized in the insert with NheI and used to transform strain CHP718 (his3-D1 git7-235) to His+. A stable integrant was identified and determined to be temperature resistant and 5-FOAr, indicating that integrated plasmid pHF1 suppresses the git7-235 mutation. This strain was then crossed with strain CHP595 (his3-D1 git7+). All 13 tetrads examined displayed a parental ditype pattern (4 5-FOAr:0 5-FOAs [5-FOA sensitive] progeny), indicating that plasmid pHF1 had integrated in or near the git7 locus. DNA sequence analysis of the two ends of the insert DNA from plasmid pHF1 revealed that the insert contains the sequence from positions 30,876 to 35,540 of cosmid c36 (GenBank accession number AL023589), which carries a portion of S. pombe chromosome 2, including two candidate open reading frames. Subcloning analyses demonstrated that the git7 gene is open reading frame SPBC36.12c.
Disruption and green fluorescent protein (GFP) tagging of git7.
The git7 gene was disrupted by using a PCR-based approach as described by Bähler et al. (6). Oligonucleotides git7delta (5′-TCTGGCAAATAGTAATGCATTCGCGTAATGACGTCTTTGTTTCAAAATTTGCGAAGCACGCTTCGCATCGGATCCCCGGGTTAATTAA-3′) and git7revkan (5′-TTGGCACCAATTCCAGGAAGGCGACCCTTATCCATATCTTCGGATTTGTATAATGACTCGGAATTCGAGCTCGTTTAAAC-3′]) were used to PCR amplify a kanMX6-containing fragment from pFA6a-GFP-kanMX6. The amplified fragment was used to transform a diploid S. pombe strain, constructed by mating CHP556 (git7-93) with CHP594 (git7+), to G418 resistance, replacing the wild-type git7+ allele with a kanMX6-marked disruption allele. Homologous recombination at the git7 locus was confirmed by Southern blot analysis. Azygotic asci, dissected on YEA medium, produced tetrads with only two viable progeny that were 5-FOAs G418 sensitive, indicating that the git7 deletion strain was nonviable. Progeny carrying the git7 disruption could be rescued by transforming the diploid strain with plasmid pHF1 (git7+) prior to tetrad dissection.
A git7-GFP fusion was created at the git7 locus in strain FWP72 by homologous recombination with a PCR product made by PCR amplification of plasmid pFA6a-GFP-kanMX6 with primers git7GFPtag (5′-CAACTAACTGGAAAGATGTGAAAAGCAAAACATTTGAAACAAAGCCTCCACAGGGAATGGAACCAAAAAAATTTCGGATCCCCGGGTTAATTAA-3′) and git7revkan (see above). The resulting strain, PBP1, was defective in glucose repression of fbp1 transcription, as indicated by its 5-FOAs phenotype and by elevated fbp1-lacZ expression in glucose-grown cells (see Results). Strain PBP1 was confirmed to have the homologous recombination event by Southern blot analysis and by a cross with strain CHP93 (git7-93), demonstrating that the kanMX6 marker was linked to the git7 locus (all 24 tetrads displayed the parental ditype pattern of 0 5-FOAr:4 5-FOAs progeny).
Construction of a transcriptionally regulated allele of skp1.
The S. pombe skp1+ gene (GenBank accession number AF071066) was precisely replaced with the ura4+ marker in a diploid strain (h+/h− ura4-D18 ade6-210/ade6-216 leu1-32) by transformation with a PCR-generated cassette bearing the 1.7-kb ura4+ gene flanked by 84 and 58 bases of homology to the skp1+ locus. This cassette was generated by PCR with oligonucleotides A (5′-CAGCATAACTAGAAATGCTAACAGCTTAACTTTCATTCATCCATTACTTACATACATCAACGCTTAGCTACAAATCCCACTGG-3′) and B (5′-GGCGCATGATGAGGTGGATGGGAAGATTCCCATTTTACACTGAAAACTACACTTAATGTCCAACACCAATGTTTATAACC-3′) to amplify ura4+ with 58 bases of homology to the skp1+ locus. Homology to the 5′ end of skp1+ was extended by subsequent amplification of the primary product with oligonucleotides B and C (5′-GAGTTCTGCCACTGTAGGAATATCAGCATAACTAGAAATGC-3′). Transformants were selected by growth on medium lacking uracil, and the proper disruption was verified by PCR.
A transcriptionally regulated, hemagglutinin (HA)-tagged skp1 allele (nmt41-HA-skp1) was generated by insertion of the skp1+ open reading frame (residues 2 to 161) into the SalI and BglII sites of pSLF173 (15) after PCR amplification by using oligonucleotides incorporating an XhoI site and a BglII site at the 5′ and 3′ends, respectively. PCR was performed by using a plasmid (c5) containing S. pombe skp1 cDNA as a template. This HA-tagged gene was then shuttled into a lower-level expression construct that uses the nmt41 promoter by release with XhoI and BglII from pSLF173 and cloning into the XhoI and BamHI sites of pREP41x (14). This vector was linearized by digestion with MluI and integrated into the ars1 locus of the skp1Δ::ura4+/skp1+diploid strain. The diploid strain was induced to sporulate by growth on minimal medium lacking thiamine, and skp1Δ::ura4+ ars1(MluI)::pREP41-skp1+-3×HA LEU2+ spores were selected on minimal medium lacking uracil and leucine. Integration at ars1 was confirmed by PCR. The sequence of the tagging vector was verified prior to integration.
β-Galactosidase assays.
Strains were cultured overnight under repression conditions (8% glucose) in YEL or PM medium. Cultures were grown overnight to a final cell density of approximately 107 cells/ml. Protein lysates were prepared on ice and assayed for β-galactosidase activity as previously described (37).
Gap repair and DNA sequencing.
Mutant alleles of git7 were cloned by gap repair (39) by transforming strains KSP1 (git7-27), CHP556 (git7-93), and CHP718 (git7-235) to His+ with EcoRV-linearized plasmid pHF5. Plasmids were rescued in E. coli (22), and the open reading frames of the rescued git7 mutant alleles were sequenced by using custom oligonucleotides on one strand. Mutations were confirmed by sequencing of the complementary strand.
Construction of vectors expressing S. pombe git7 and human and S. cerevisiae SGT1 in S. pombe.
The S. pombe git7, S. cerevisiae SGT1, and human SGT1 genes were cloned into vector pNMT81-TOPO (Invitrogen Corp.) according to the manufacturer's instructions, placing the expression of these genes under the control of the thiamine-repressible low-level-expression nmt81 promoter (14). The git7 gene was amplified from plasmid pHF1 by PCR with primers git7ATG1-For (5′-TGATAAAATATTCACATGAAACCCATAGCC-3′) and git7-V5-Rev (5′-AAATTTTTTTGGTTCCATTCCCTGTGGAGG-3′) to produce an insert encoding the originally predicted 444-residue protein. This sequence was translationally fused to a carboxy-terminal V5 epitope tag (43) upon insertion into pNMT81-TOPO to create plasmid pKS1. A git7 promoter-driven git7-V5 fusion was constructed by homologous integration of a PacI-linearized derivative of pKS1 (an SphI fragment containing the git7 5′ coding sequence was removed to avoid having git7+ expressed from the nmt81 promoter) into the git7+ genomic locus in strain FWP101 to create strain KSP5. A second, nmt81-driven git7-V5 plasmid-borne construct was made by PCR amplification from the SPLE-2 cDNA library (25) with primers git7ATG3-For (5′-CCATAAAATGGGTGTAGATCTTTCTGA-3′) and git7-V5-Rev, resulting in plasmid pKS2, which expresses the 379-residue Git7p protein (which would be expressed from the third potential start codon in git7) fused to a carboxy-terminal V5 epitope tag (see Results). Plasmids expressing untagged forms of the human SGT1 gene (pKS3) and the S. cerevisiae SGT1 gene (pKS4) were constructed to carry PCR products amplified from plasmid BKK43 with primers HuSGT1-For (5′-ATGGCGGCGGCTGCAGCAGGA-3′) and HuSGT1-Rev (5′-TTAGTACTTTTTCCATTCCATATCATCAGG-3′) and from plasmid BKK9 with primers ScSGT1-For (5′-ATGCCTGTTGAAAAAGATTTAAAAACTGCCTAC-3′) and ScSGT1-Rev (5′-TTACCAATGTTTAGGTTCCATGCCTTC-3′), respectively (29).
Protein isolation and immunoblotting.
Strains were cultured in PM liquid medium (lacking leucine for strains carrying autonomous plasmids) to an optical density at 600 nm of 0.6. Total protein extracts were prepared by trichloroacetic acid precipitation as described by Volland et al. (49). Protein extracts were electrophoresed on a sodium dodecyl sulfate-10% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. Immunodetection was carried out by using an anti-V5-horseradish peroxidase antibody (Invitrogen) according to the manufacturer's instructions, and visualization was carried out by using LumiGLO chemiluminescence (Kirkegaard & Perry Laboratories).
Immunofluorescence microscopy.
Strains were cultured overnight in PM liquid medium and grown to a final cell density of approximately 107 cells/ml. Cells were fixed by using paraformaldehyde for 30 min and prepared as described by Hagan and Hyams (17). Git7p-V5 was localized by using an anti-V5 antibody (Invitrogen) according to the manufacturer's instructions and visualized by using Alexa Fluor 488-labeled immunoglobulin G as a secondary antibody (Molecular Probes) at 10 μg/ml for 1 h. Cells were resuspended in mounting medium containing 1 μg of Hoechst 33342/ml.
RESULTS
Cloning of the S. pombe git7 gene.
The git genes, including git7, were originally identified in a genetic screen for mutations that confer a defect in the glucose repression of both fbp1-ura4 and fbp1-lacZ reporters in S. pombe (21). Thus, while wild-type (git+) strains carrying these reporters are 5-FOAr in a glucose-rich medium, git mutant strains are 5-FOAs. By screening plasmid libraries for clones that restore a 5-FOAr phenotype to git mutant hosts, we have been able to clone git genes as well as multicopy suppressors (13, 20, 26, 32). To clone git7+, strain CHP556 (git7-93) was transformed to His+ with a pBG2-based S. pombe genomic DNA library (38). From 33,000 transformants, we identified 37 5-FOAr candidates. These candidate plasmids were subsequently screened for their ability to suppress the temperature-sensitive growth of strain CHP718 conferred by the git7-235 allele. The original library screening was not carried out with this strain due to its poor transformation efficiency. Two of the 37 rescued plasmid candidates conferred temperature-resistant growth to strain CHP718. These two plasmids possess identical inserts, and one was designated pHF1 (Fig. 1). Sequence analysis across the two ends of the insert showed that pHF1 carries a portion of chromosome 2 from bp 30,876 to 35,540 of cosmid c36. Subclones from pHF1 demonstrated that the git7-complementing activity is encoded by ORF SPBC36.12c (GenPept accession number CAA19060; Fig. 1), which is divided into three exons and encodes a 379-residue protein. Integration of plasmid pHF1 into the S. pombe genome by homologous recombination followed by genetic linkage analysis indicated that pHF1 carries the git7 gene and not a multicopy suppressor (see Materials and Methods). In addition, plasmid pHF1 failed to act as a multicopy suppressor when introduced into strains carrying mutations in any of five git genes required for adenylate cyclase activation: git1, git3 (GPCR), git5 (Gβ), gpa2 (Gα), and git10 (data not shown).
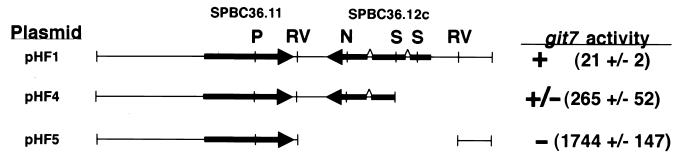
FIG. 1.
Restriction map of insert DNA and git7 suppression analysis for plasmid pHF1 and related plasmids. Restriction sites for PstI (P), EcoRV (RV), NheI (N), and SpeI (S) are shown. git7 activity is based on the ability of the plasmids to restore glucose repression of fbp1-lacZ expression when introduced into strain CHP556 (git7-93) and to restore growth at 37°C to strain CHP718 (git7-235). β-Galactosidase activity (in parentheses) in CHP556 transformants represents the average specific activity and standard error for three independent transformants grown under glucose repression conditions and assayed twice each. Under the same conditions, strain CHP578 (git7+) carrying plasmid pHF5 possessed 5 ± 1 U of activity.
Git7p is a member of the S. cerevisiae Sgt1p protein family.
A BLASTP analysis shows that Git7p is a member of a small family of proteins related to the S. cerevisiae Sgt1p protein (Fig. 2). Members of this family possess three distinct domains: a weakly conserved amino-terminal domain that appears to be a tetratricopeptide domain (5); a moderately conserved central domain referred to as the CS domain, due to its presence in some cysteine- and histidine-rich domain-containing proteins (42); and a highly conserved carboxy-terminal domain (Fig. 2). During our subcloning analysis of git7, we constructed plasmid pHF4, which lacks exon 1 and part of exon 2 and presumably expresses only the carboxyl-terminal 216 residues of Git7p. This plasmid partially restores both fbp1 regulation in a git7-93 strain (Fig. 1, plasmid pHF4) and 37°C growth in a git7-235 strain (data not shown).
FIG. 2.
Amino acid sequence alignment of S. pombe Git7p and related proteins. The Git7p protein (GenPept accession number CAA19060) was aligned with the S. cerevisiae Sgt1p protein (ScSgt1p) (GenPept accession number NP_014700), the Oryza sativa Sgt1p protein (OsSgt1p) (GenPept accession number BAB19060), the Arabidopsis thaliana Sgt1p protein (AtSgt1p) (GenPept accession number T05589), and the human Sgt1p protein (huSgt1p) (GenBank accession number AF132856) by using the Clustal W (version 1.8) sequence alignment program (47); the alignment was displayed by using BOXSHADE. Identical residues are shown as white letters shaded in black, while conserved residues are shown as white letters shaded in gray. The methionine encoded by the putative start codon used by the partially functional, truncated git7 open reading frame carried on plasmid pHF4 (Fig. 1) is indicated by an asterisk. Alterations to the Git7p protein conferred by the git7-27, git7-93, and git7-235 alleles are also indicated.
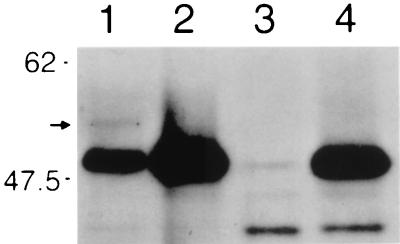
The Git7p protein was originally predicted to be 444 residues long due to the fact that exon 1 potentially includes 150 codons. However, a Clustal W alignment (47) of the putative 444-residue Git7p protein and other members of the family suggested that Git7p possesses a 50-residue amino-terminal domain not found in the other proteins (data not shown). The identity of the git7 translational start site was brought into question when we cloned this presumed open reading frame into an nmt81-driven expression vector (nmt stands for “no message on thiamine”; see Materials and Methods). Plasmid pKS1 expresses a smaller-than-expected Git7p-V5 product whose activity is not repressed by thiamine (Fig. 3; CHP93/pKS1 cells express 12 ± 7 U of β-galactosidase activity after 48 h of thiamine repression, indicating plasmid suppression of the host git7-93 mutation). Thus, it appears that the 5′ end of the cloned sequence includes the git7 promoter. The most likely candidate TATA box is immediately upstream of the second putative ATG start codon, too close to allow transcription of this ATG. As translation from the third available start codon would produce a 379-residue protein, a size similar to those of other members of the Sgt1p/Git7p family (Fig. 2), we cloned the git7 open reading frame starting with this third potential start codon into the nmt81-driven vector. Plasmid pKS2 expresses a Git7p-V5 protein with the same mobility as that of plasmid pKS1; however, this activity is now thiamine repressible (Fig. 3; CHP93/pKS2 cells express 1,884 ± 184 U of β-galactosidase activity after 48 h of thiamine repression, indicating a loss of suppression of the host git7-93 mutation). As cells expressing Git7p-V5 from the git7 genomic locus (see Materials and Methods) produce a protein with the same mobility as those of the plasmid-expressed constructs (Fig. 3), the authentic Git7p protein appears to be 379 residues long.
FIG. 3.
Immunoblot of Git7p-V5 proteins. Protein extracts were prepared from CHP93 (git7-93) cells carrying either plasmid pKS1 (including 444 potential codons of git7) or plasmid pKS2 (including 379 potential codons of git7) and grown in the absence of thiamine (to promote expression from the plasmid-borne nmt81 promoter). Additional extracts were made from strains FWP101 (untagged git7+) and KSP5 (git7-V5 integrated at the git7 locus). Extracts were subjected to a Western blot analysis to detect V5-tagged (43) proteins. Lane 1, 20 μg of extract from CHP93/pKS1; lane 2, 7 μg of extract from CHP93/pKS2; lane 3, 60 μg of extract from FWP101; lane 4, 60 μg of extract from KSP5. Note that the CHP93/pKS1 extract in lane 1 contains a 54-kDa protein (indicated by an arrow) which was not seen in the other lanes and which may represent the protein translated from the first available start codon in pKS1.
S. pombe Skp1p does not appear to regulate fbp1-lacZ expression.
It has been shown that the S. cerevisiae Sgt1p protein interacts with Skp1p of the SCF E3 ubiquitin ligase. We therefore set out to determine whether the S. pombe homolog of Skp1p plays a role in fbp1 regulation and cAMP signaling, as does Git7p. Surprisingly, thiamine repression of nmt41-HA-skp1 expression in a git7+ strain (CHP803) had almost no effect on fbp1-lacZ expression. Cells continued to display glucose-repressed β-galactosidase levels (25 ± 8 U) after 16 h in the presence of thiamine. After 48 h of thiamine repression, cells showed arrested growth, yet β-galactosidase levels were only 207 ± 64 U. As we have observed a higher level of derepression of fbp1-lacZ expression in cells grown to stationary phase under glucose-rich conditions, this small degree of derepression does not suggest a role for S. pombe Skp1p in fbp1 regulation. Therefore, it appears that the role of Git7p in cAMP signaling is Skp1p independent.
The git7 gene is essential for cell wall integrity and septation.
We constructed a git7-null allele (git7Δ) by homologous recombination (6) in an effort to study the role of git7 in cAMP signaling and other processes. The sporulation of a diploid strain carrying the git7Δ allele showed that git7 is essential for viability. git7Δ spores germinated to produce microcolonies of approximately 200 to 300 cells, at which point all the cells appeared to undergo lysis (data not shown). git7Δ spore viability was rescued by either plasmid pHF1 or plasmid pHF4 (Fig. 1). Microscopic examination of cells from such transformants revealed the presence of both lysed cells and multinucleate cells, which likely resulted from plasmid loss (data not shown). Thus, Git7p seems to be required for both cell wall integrity and septation.
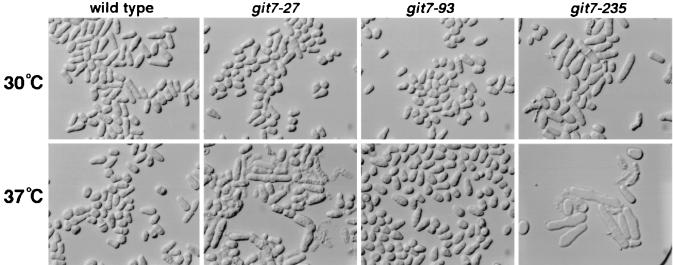
The original collection of git mutant strains included three independent git7 mutants (21). At the time, it was noted that git7-235 strains were temperature sensitive for growth. We therefore reexamined strains carrying the git7-27, git7-93, or git7-235 allele for defects in growth and septation at 30 and 37°C. As shown in Fig. 4, most git7-235 cells underwent lysis or failed to septate after 24 h at 37°C on solid medium. Even at 30°C, these defects were evident in git7-235 cells. Strains carrying the git7-27 allele displayed lysis and septation defects similar to those of git7-235 strains (Fig. 4), although to a lesser degree. Strains carrying the git7-93 allele appeared to have no growth defects, although they resembled other cAMP pathway mutants in that they divided at a slightly reduced cell length (26). Surprisingly, of the three original mutant alleles, the git7-93 allele conferred the most severe defect with regard to glucose repression of an fbp1-lacZ reporter (Table 2). Thus, Git7p has distinct roles in cAMP signaling and in cell wall integrity and septation.
FIG. 4.
Growth defects associated with git7 mutations. Cells from strains FWP101 (git7+; wild type), CHP27 (git7-27), CHP93 (git7-93), and CHP449 (git7-235) were grown for 24 h on YEA plates at either 30 or 37°C before being photographed.
TABLE 2.
Defect in glucose repression of fbp1-lacZ expression in git7 mutant strainsa
| Strain | Relevant genotype | β-Galactosidase activity |
|---|---|---|
| FWP101 | git7+ | 5 ± 0 |
| CHP27 | git7-27 | 1,124 ± 149 |
| CHP93 | git7-93 | 2,130 ± 301 |
| CHP449 | git7-235 | 1,600 ± 63 |
| PBP1 | git7-GFP | 1,288 ± 61 |
Cells were grown in YEL medium to exponential phase under repression conditions (8% glucose) and assayed as described in Materials and Methods. The values given represent mean specific activities and standard errors from two to four independent cultures of each strain.
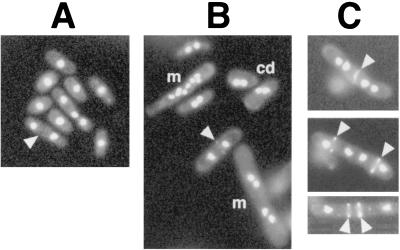
To better characterize the growth defects conferred by the git7-235 allele, a temperature shift experiment was performed with two git7-235 strains growing in liquid medium. Strains FWP145 and CHP449 differ by the presence of a his7-366 allele in the latter strain. We have noted in the past that the his7-366 mutant allele enhances growth defects that are conferred by mutations in other genes (unpublished data). Consistent with this observation, a greater percentage of cells in the CHP449 culture than in the FWP145 culture exhibited one of three growth defects at all time points (Table 3). These included multinucleate cells (cells with four or more nuclei; this phenotype was more common in cells growing on solid medium than in liquid medium), binucleate cells that appeared to be arrested during cell division, and lysed cells (Table 3 and Fig. 5). In addition, some cells that did form septa appeared to fail to complete cytokinesis (Fig. 5C). Further examination of the CHP449 cells failed to show any actin delocalization (data not shown). These results confirm a role for Git7p in cell wall integrity, cell division, and septation.
TABLE 3.
Effect of growth at 36°C on git7-235 cellsa
| h at 36°C | % of cells (CHP449/FWP145) with the following morphology:
|
|||
|---|---|---|---|---|
| Wild type | Multinucleate | Cell division | Lysed | |
| 0 | 90/100 | 10/0 | 0/0 | 0/0 |
| 4 | 70/89 | 13/5 | 14/6 | 3/0 |
| 6 | 68/81 | 12/4 | 16/14 | 4/1 |
| 8 | 36/65 | 14/2 | 33/24 | 17/9 |
| 27 | 12/27 | 15/6 | 37/46 | 36/22 |
Cells were grown in YEL medium, stained with Hoechst 33342 to detect nuclei and Calcofluor to detect septa, and examined by microscopy. Two hundred cells were examined for each time point.
FIG. 5.
Various morphologies associated with the git7-235 allele. CHP449 (git7-235) cells were grown for 24 h at 37°C, stained with Hoechst 33342 and Calcofluor, and photographed. Septa are indicated by arrowheads. (A) Cells displaying the wild-type phenotype, being either uninucleate or binucleate with a septum. (B) Multinucleate (m) and cell division-arrested (cd) binucleate cells. (C) Cells containing septa but failing to complete cytokinesis.
A Git7p-GFP fusion is defective in cAMP signaling.
In an effort to study the localization of Git7p in the cell, we constructed a git7-GFP fusion at the git7 locus by homologous recombination (see Materials and Methods) (6). Cells expressing this fusion as the sole source of Git7p activity were viable and showed no growth defects (data not shown); however, these cells were defective in the glucose repression of fbp1-lacZ, indicating a defect in cAMP signaling (Table 2). Thus, like the git7-93 allele, the git7-GFP fusion allele confers a defect in the glucose/cAMP signaling pathway but has no effect on cell wall integrity or on septation.
Git7p is required for nutrient regulation of mating.
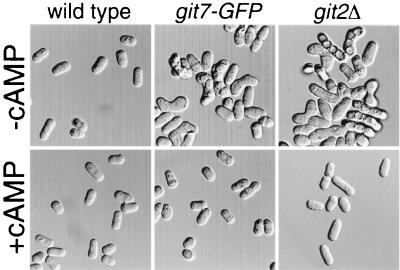
S. pombe cells normally require either a glucose or a nitrogen starvation signal to initiate mating and meiotic entry (44). Therefore, cells carrying mutations in genes required for the glucose-triggered cAMP signal will mate and sporulate in nutrient-rich media (24, 31, 32, 34, 52). Consistent with the defect in the glucose repression of fbp1-lacZ expression (Table 2), the git7-GFP allele allows homothallic (h90) cells to mate in a glucose-rich medium (Fig. 6). This starvation-independent mating is similar to that conferred by a deletion of the git2 adenylate cyclase gene (git2Δ), as the addition of 5 mM cAMP to the medium prevented conjugation in both git7-GFP and git2Δ cells (Fig. 6). Thus, git7 is required for the cAMP-dependent regulation of conjugation as well as fbp1 transcriptional regulation.
FIG. 6.
Starvation-independent sexual development in git7-GFP and git2Δ (adenlyate cyclase) homothallic strains. Cells of homothallic strains CHP795 (wild type), CHP792 (git7-GFP), and CHP558 (git2Δ) were pregrown at 37°C (to inhibit conjugation) in PM medium (8% glucose) to a concentration of 107 cells/ml, diluted to 106 cells/ml in the presence or absence of 5 mM cAMP, and incubated overnight at 30°C without shaking before being photographed.
The carboxy-terminal domain of Git7p is specifically involved in cAMP signaling.
Both the git7-GFP and the git7-93 alleles confer a defect in fbp1 regulation but not in cell growth and septation (Table 2, Fig. 4, and data not shown). We therefore cloned all three spontaneous git7 alleles by gap repair of EcoRV-linearized plasmid pHF5 (Fig. 1). The git7-93 allele contains a 54-bp duplication of codons 345 to 363, a sequence that is flanked in the wild-type allele by an 8-bp direct repeat. This leads to a duplication of 18 amino acids (YTESNGTALSTNWKDVKS; Fig. 2) in the carboxy-terminal domain of Git7p; this domain is highly conserved in other members of this protein family. Thus, both the git7-93 and the git7-GFP alleles encode proteins that are altered at their carboxy termini. In contrast, both the git7-27 and the git7-235 alleles contain single missense mutations in the region encoding the amino terminus of Git7p. The mutation in git7-27 changes a histidine to an aspartic acid at residue 42, while the mutation in git7-235 changes an alanine to a glutamic acid at residue 69 (Fig. 2). As these two mutant alleles confer temperature-sensitive lethality, it is possible that these mutations result in a general instability of the Git7p protein. Alternatively, they may identify a domain required for all Git7p functions, including cell wall integrity and septation.
git7 mutants do not display a kinetochore assembly defect.
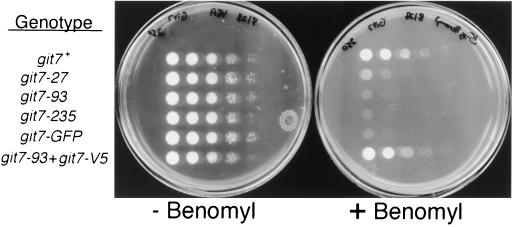
As S. cerevisiae Sgt1p is required for kinetochore assembly (29), we tested whether any phenotypes associated with git7 mutations would suggest a similar role for Git7p in S. pombe. We therefore examined the effect of the microtubule-destabilizing drug benomyl on strains carrying various git7 alleles. Strains carrying any of the three spontaneous mutant alleles or the git7-GFP allele were benomyl sensitive (Fig. 7). The introduction of plasmid pKS1 (git7-V5) into a git7-93 strain restored benomyl-resistant growth, providing further evidence that Git7p-V5 is fully functional for all Git7p activities of which we are aware, unlike Git7p-GFP. However, when assayed for mitotic stability of the Ch16 minichromosome (1), strain KSP2 (git7-GFP) did not display a chromosome stability defect (data not shown). Thus, the benomyl sensitivity is not likely due to a defect in kinetochore assembly, as is seen in S. cerevisiae sgt1 mutants (29), but may reflect other interactions with spindle or cytoplasmic microtubules. To test whether Git7p plays a role in spindle assembly, we constructed a double mutant carrying git7-235 and a deletion of the mad2 spindle checkpoint gene (mad2Δ) (18). The git7-235 mad2Δ double mutant failed to display any synthetic growth defects that would be expected if Git7p were required for spindle assembly (data not shown).
FIG. 7.
Benomyl sensitivity in git7 mutant strains. Serial dilutions (1:5) of strains FWP101 (git7+), CHP27 (git7-27), CHP93 (git7-93), CHP449 (git7-235), CHP758 (git7-GFP), and CHP93 carrying plasmid pKS1 (git7-93 + git7-V5) were spotted on YEA plates containing 75 μg of adenine/ml and either 0 or 5 μg of benomyl/ml. Growth was recorded after 3 days at 25°C.
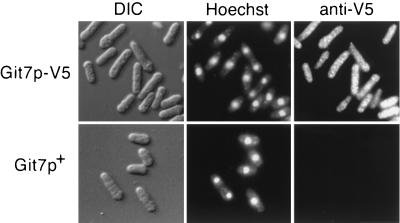
Localization of a Git7p-V5 fusion.
Because the Git7p-GFP fusion protein is not functional in the glucose-cAMP pathway, we examined the localization of the fully functional Git7p-V5 protein (Fig. 3 and Table 4) expressed from the git7 promoter in a fusion integrated in a single copy (see Materials and Methods). The Git7p-V5 protein is seen as punctate staining in both the nucleus and the cytoplasm of cells (Fig. 8), while little or no staining is seen in untagged cells. A similar localization pattern has been observed for S. cerevisiae Sgt1p (C. Dubacq and C. Mann, personal communication).
TABLE 4.
Suppression of constitutive fbp1-lacZ expression by human Sgt1p and S. cerevisiae Sgt1p in git7-93 but not git7-235 cellsa
| Plasmid-expressed protein | β-Galactosidase activity in the following host:
|
|
|---|---|---|
| git7-93 | git7-235 | |
| None | 2,428 ± 488 | 1,581 ± 198 |
| Git7p-V5 | 21 ± 3 | 17 ± 4 |
| Human Sgt1p | 21 ± 9 | 1,919 ± 214 |
| S. cerevisiae Sgt1p | 11 ± 3 | 1,176 ± 37 |
β-Galactosidase activity was assayed in CHP93 (git7-93) and CHP449 (git7-235) transformants grown in PM medium lacking thiamine (to derepress expression from the nmt81 promoter in the vector) under glucose-rich conditions. The plasmids used were pNMT81-TOPO (none; empty vector control), pKS2 (Git7p-V5), pKS3 (human Sgt1p), and pKS4 (S. cerevisiae Sgt1p). Values represent mean specific activities and standard errors from two to four independent cultures.
FIG. 8.
Localization of Git7p-V5. Indirect immunofluorescence was used to detect Git7p-V5 localization in KSP5 cells (carrying an integrated git7-V5 fusion) and in FWP101 cells (untagged git7+). Cells were stained with Hoechst 33342 to detect DNA prior to microscopy. KSP5 cells display a punctate signal throughout the nucleus and the cytoplasm. DIC, differential interference contrast microscopy.
Complementation of git7-93 by plasmid-expressed S. cerevisiae and human Sgt1p proteins.
We tested whether the human Sgt1p (Sgt1p) or S. cerevisiae Sgt1p proteins could restore transcriptional regulation of the fbp1-lacZ reporter when expressed in git7 mutant strains. Indeed, the expression of either the human or the S. cerevisiae SGT1 gene from the nmt81 promoter completely suppressed the constitutive fbp1-lacZ expression observed in a git7-93 mutant strain (Table 4). However, the expression of these genes in a git7-235 strain had little or no effect on the constitutive expression of the fbp1-lacZ reporter. As expected, the expression of the Git7p-V5 fusion from the same vector suppressed both git7-93 and git7-235 mutations (Table 4). Consistent with this allele-specific suppression, we observed that the git7-93 transformants were benomyl resistant, while the git7-235 transformants remained benomyl sensitive (data not shown). Therefore, the ability of the human and budding yeast proteins to restore Git7p functions to S. pombe depends upon the expression of the Git7-93p mutant protein.
DISCUSSION
In this study, we have cloned and carried out an initial characterization of the S. pombe git7 gene, which is required for the glucose repression of fbp1 transcription as part of the glucose-triggered cAMP signaling pathway. In previous studies, it was shown that git7 is required for both the maintenance of basal cAMP levels and the generation of the glucose-stimulated cAMP signal (11). It was also shown that while mutational activation of Gpa2p Gα bypasses the requirement for the Git3p GPCR or Git5p-Git11p Gβγ, it fails to suppress mutations in git1, git7, or git10 (52). Therefore, while the role of the Git3p GPCR and Git5p-Git11p Gβγ is to activate Gpa2p Gα, Git1p, Git7p, and Git10p appear to function either downstream from or in parallel to Gpa2p Gα.
It was surprising to discover that git7 is homologous to the S. cerevisiae SGT1 gene, whose best-characterized role had been in kinetochore assembly (29). However, Sgt1p must play multiple roles in S. cerevisiae, as different temperature-sensitive sgt1 alleles result in either G1 or G2 arrest. In addition, while Sgt1p physically interacts with Skp1p of the SCF E3 ubiquitin ligase, Sgt1p may carry out at least one Skp1p-independent function in G1. The S. cerevisiae skp1-11 allele causes G1 arrest due to the accumulation of the Sic1p inhibitor of the Cdc28p-Clb kinase required for G1 exit (7). As such, the arrest point for skp1-11 cells is after the pheromone arrest point in G1. Meanwhile, the arrest point for sgt1-5 cells in G1 precedes the pheromone arrest point (29), indicating a role for Sgt1p distinct from that of Skp1p in G1. Our studies show little or no connection between Git7p and Skp1p in S. pombe. While both Git7p and Skp1p are essential in S. pombe, cells depleted of Skp1p do not show the same phenotypes as git7 mutants. Thiamine repression of nmt41-HA-skp1 cells causes cell growth arrest but not a defect in septation or an increase in cell lysis (data not shown). In addition, fbp1-lacZ expression remains glucose repressed upon Skp1p depletion, with only partial derepression as cell growth is arrested, indicating that S. pombe Skp1p is not involved in the glucose-triggered cAMP signaling pathway.
Our characterization of a git7 deletion allele and other mutant alleles revealed a large number of mutant phenotypes, including constitutive fbp1 transcription, starvation-independent conjugation and sporulation, cell lysis, a septation defect, a cell division defect, and sensitivity to the microtubule-destabilizing drug benomyl. These phenotypes suggest at least three distinct Git7p functions. The inability to repress fbp1 transcription and conjugation is due to a defect in the cAMP signaling pathway, as has been shown for other git genes (26, 31, 32, 34, 52). The septation and lysis defects are most likely related, since lysis appears to occur at the septum during cytokinesis (data not shown). Along with a failure to form septa, git7-27 and git7-235 cells also form defective septa that lead to either cell lysis during cytokinesis or a failure to undergo cytokinesis, as shown in Fig. 5C. Finally, the cell division defect and benomyl sensitivity may be related, since microtubules are involved in the migration and positioning of the nuclei during mitosis in S. pombe (17). The lack of a mitotic chromosome stability defect in git7 mutants or synthetic growth defects in a git7-235 mad2Δ strain indicates that, unlike S. cerevisiae Sgt1p (29), Git7p plays no role in kinetochore assembly. However, as only a subset of S. cerevisiae sgt1 mutants display a kinetochore assembly defect, our failure to observe a similar defect in git7 mutants does not rule out a similar role for Git7p. The characterizations described here represent an initial investigation of a protein that is involved in at least three distinct processes, and further detailed studies are required in each area.
Protein modeling work carried out on members of the Sgt1p protein family by Azevedo et al. (5) suggests that there are three distict domains, with the poorly conserved amino-terminal domain acting as a tetratricopeptide domain (9). Our studies show that alteration of the highly conserved carboxy terminus by a short duplication in the git7-93 allele (Fig. 2) or by the addition of a large GFP tag to the carboxy terminus leads to defects in the glucose-triggered cAMP signaling pathway (Table 2) and benomyl sensitivity (Fig. 7) but has no effect on either septation or cell wall integrity (Fig. 4). Meanwhile, the Git7-27p and Git7-235p proteins both have alterations in the amino-terminal domain and share similar phenotypes that include cell lysis and septation defects when grown at 37°C (Fig. 4 and 5), in addition to the cAMP signaling and benomyl sensitivity phenotypes. These results indicate that Git7p is composed of distinct functional domains that act in a modular fashion such that the git7-93 mutation that interferes with cAMP signaling does not affect other essential Git7p functions. In support of this notion, we observed allele-specific suppression of the git7-93 mutation by the expression of human or budding yeast SGT1 genes (Table 4). Because such expression fails to suppress the same constitutive fbp1-lacZ expression or benomyl-sensitive growth conferred by a git7-235 allele, it appears that Git7-93p provides a function that is defective in Git7-235p and that is needed to allow human or S. cerevisiae Sgt1p to act in S. pombe. Thus, Git7-235p may be defective in forming complexes with either Sgt1p or other components of a protein complex, while Git7-93p can interact with Sgt1p, which then provides a functional carboxy-terminal domain to the complex. However, since we have not observed intragenic complementation between git7-93 and git7-235, it is possible that Git7-235p cannot interact with human or budding yeast Sgt1p because it is unstable.
The ability of human and S. cerevisiae SGT1 genes to suppress a git7-93 mutation suggests that this gene family may have a conserved role in cAMP signaling, although it need not be as a function of glucose detection. Indeed, S. cerevisiae Sgt1p has also been shown to interact both physically and genetically with adenylate cyclase in budding yeast (Dubacq and Mann, personal communication), supporting the idea that Sgt1p is involved in cAMP signaling. However, it remains to be seen whether or not members of the Git7p/Sgt1p family act to regulate adenylate cyclase in mammalian cells, even though human Sgt1p can provide that function in S. pombe. For example, although mammalian Ras proteins are able to replace S. cerevisiae Ras proteins in the budding yeast cAMP signaling pathway (10, 28), they do not appear to regulate cAMP signaling in mammalian cells (8). Thus, additional studies are needed to determine whether members of the Git7p/Sgt1p family are universally involved in cAMP signaling in yeasts, plants, and animals.
Acknowledgments
We thank Carl Mann for sharing unpublished results and for stimulating discussions. We also thank David Burgess and Brad Shuster for assistance with some of the microscopy.
This work was supported by National Institutes of Health grant GM46226 to C.S.H.
REFERENCES
- 1.Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. [DOI] [PubMed] [Google Scholar]
- 2.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alspaugh, J. A., R. Pukkila-Worley, T. Harashima, L. M. Cavallo, D. Funnell, G. M. Cox, J. R. Perfect, J. W. Kronstad, and J. Heitman. 2002. Adenylyl cyclase functions downstream of the G protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1:75-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austin, M. J., P. Muskett, K. Kahn, B. J. Feys, J. D. Jones, and J. E. Parker. 2002. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295:2077-2080. [DOI] [PubMed] [Google Scholar]
- 5.Azevedo, C., A. Sadanandom, K. Kitagawa, A. Freialdenhoven, K. Shirasu, and P. Schulze-Lefert. 2002. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295:2073-2076. [DOI] [PubMed] [Google Scholar]
- 6.Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, 3rd, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 7.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. [DOI] [PubMed] [Google Scholar]
- 8.Beckner, S. K., S. Hattori, and T. Y. Shih. 1985. The ras oncogene product p21 is not a regulatory component of adenylate cyclase. Nature 317:71-72. [DOI] [PubMed] [Google Scholar]
- 9.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 10.Broek, D., N. Samiy, O. Fasano, A. Fujiyama, F. Tamanoi, J. Northup, and M. Wigler. 1985. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell 41:763-769. [DOI] [PubMed] [Google Scholar]
- 11.Byrne, S. M., and C. S. Hoffman. 1993. Six git genes encode a glucose-induced adenylate cyclase activation pathway in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 105:1095-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colombo, S., P. Ma, L. Cauwenberg, J. Winderickx, M. Crauwels, A. Teunissen, D. Nauwelaers, J. H. de Winde, M. F. Gorwa, D. Colavizza, and J. M. Thevelein. 1998. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 17:3326-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dal Santo, P., B. Blanchard, and C. S. Hoffman. 1996. The Schizosaccharomyces pombe pyp1 protein tyrosine phosphatase negatively regulates nutrient monitoring pathways. J. Cell Sci. 109:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsburg, S. L. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21:2955-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsburg, S. L., and D. A. Sherman. 1997. General purpose tagging vectors for fission yeast. Gene 191:191-195. [DOI] [PubMed] [Google Scholar]
- 16.Gutz, H., H. Heslot, U. Leupold, and N. Loprieno. 1974. Schizosaccharomyces pombe, p. 395-446. In R. C. King (ed.), Handbook of genetics. Plenum Press, New York, N.Y.
- 17.Hagan, I. M., and J. S. Hyams. 1988. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 89:343-357. [DOI] [PubMed] [Google Scholar]
- 18.He, X., T. E. Patterson, and S. Sazer. 1997. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 94:7965-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higuchi, T., Y. Watanabe, and M. Yamamoto. 2002. Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol. Cell. Biol. 22:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman, C. S., and F. Winston. 1991. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev. 5:561-571. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman, C. S., and F. Winston. 1990. Isolation and characterization of mutants constitutive for expression of the fbp1 gene of Schizosaccharomyces pombe. Genetics 124:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 23.Hoffman, C. S., and F. Winston. 1989. A transcriptionally regulated expression vector for the fission yeast Schizosaccharomyces pombe. Gene 84:473-479. [DOI] [PubMed] [Google Scholar]
- 24.Isshiki, T., N. Mochizuki, T. Maeda, and M. Yamamoto. 1992. Characterization of a fission yeast gene, gpa2, that encodes a G alpha subunit involved in the monitoring of nutrition. Genes Dev. 6:2455-2462. [DOI] [PubMed] [Google Scholar]
- 25.Janoo, R. T., L. A. Neely, B. R. Braun, S. K. Whitehall, and C. S. Hoffman. 2001. Transcriptional regulators of the Schizosaccharomyces pombe fbp1 gene include two redundant Tup1p-like corepressors and the CCAAT binding factor activation complex. Genetics 157:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, M., M. Fujita, B. M. Culley, E. Apolinario, M. Yamamoto, K. Maundrell, and C. S. Hoffman. 1995. sck1, a high copy number suppressor of defects in the cAMP-dependent protein kinase pathway in fission yeast, encodes a protein homologous to the Saccharomyces cerevisiae SCH9 kinase. Genetics 140:457-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanoh, J., Y. Watanabe, M. Ohsugi, Y. Iino, and M. Yamamoto. 1996. Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells 1:391-408. [DOI] [PubMed] [Google Scholar]
- 28.Kataoka, T., S. Powers, S. Cameron, O. Fasano, M. Goldfarb, J. Broach, and M. Wigler. 1985. Functional homology of mammalian and yeast RAS genes. Cell 40:19-26. [DOI] [PubMed] [Google Scholar]
- 29.Kitagawa, K., D. Skowyra, S. J. Elledge, J. W. Harper, and P. Hieter. 1999. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4:21-33. [DOI] [PubMed] [Google Scholar]
- 30.Kraakman, L., K. Lemaire, P. Ma, A. W. Teunissen, M. C. Donaton, P. Van Dijck, J. Winderickx, J. H. de Winde, and J. M. Thevelein. 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32:1002-1012. [DOI] [PubMed] [Google Scholar]
- 31.Landry, S., and C. S. Hoffman. 2001. The git5 Gβ and git11 Gγ form an atypical Gβγ dimer acting in the fission yeast glucose/cAMP pathway. Genetics 157:1159-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landry, S., M. T. Pettit, E. Apolinario, and C. S. Hoffman. 2000. The fission yeast git5 gene encodes a Gβ subunit required for glucose-triggered adenylate cyclase activation. Genetics 154:1463-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeda, T., N. Mochizuki, and M. Yamamoto. 1990. Adenylyl cyclase is dispensable for vegetative cell growth in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 87:7814-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda, T., Y. Watanabe, H. Kunitomo, and M. Yamamoto. 1994. Cloning of the pka1 gene encoding the catalytic subunit of the cAMP-dependent protein kinase in Schizosaccharomyces pombe. J. Biol. Chem. 269:9632-9637. [PubMed] [Google Scholar]
- 36.Neely, L., and C. S. Hoffman. 2000. PKA and MAPK pathways antagonistically regulate fission yeast fbp1 transcription by using different modes of action at two upstream activation sites. Mol. Cell. Biol. 20:6426-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nocero, M., T. Isshiki, M. Yamamoto, and C. S. Hoffman. 1994. Glucose repression of fbp1 transcription of Schizosaccharomyces pombe is partially regulated by adenylate cyclase activation by a G protein alpha subunit encoded by gpa2 (git8). Genetics 138:39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohi, R., A. Feoktistova, and K. L. Gould. 1996. Construction of vectors and a genomic library for use with his3-deficient strains of Schizosaccharomyces pombe. Gene 174:315-318. [DOI] [PubMed] [Google Scholar]
- 39.Orr-Weaver, T. L., J. W. Szostak, and R. J. Rothstein. 1983. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 101:228-245. [DOI] [PubMed] [Google Scholar]
- 40.Pan, X., T. Harashima, and J. Heitman. 2000. Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 3:567-572. [DOI] [PubMed] [Google Scholar]
- 41.Patton, E. E., A. R. Willems, and M. Tyers. 1998. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 14:236-243. [DOI] [PubMed] [Google Scholar]
- 42.Shirasu, K., T. Lahaye, M. W. Tan, F. Zhou, C. Azevedo, and P. Schulze-Lefert. 1999. A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99:355-366. [DOI] [PubMed] [Google Scholar]
- 43.Southern, J. A., D. F. Young, F. Heaney, W. K. Baumgartner, and R. E. Randall. 1991. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol. 72:1551-1557. [DOI] [PubMed] [Google Scholar]
- 44.Stettler, S., E. Warbrick, S. Prochnik, S. Mackie, and P. Fantes. 1996. The wis1 signal transduction pathway is required for expression of cAMP-repressed genes in fission yeast. J. Cell Sci. 109:1927-1935. [DOI] [PubMed] [Google Scholar]
- 45.Takeda, T., T. Toda, K. Kominami, A. Kohnosu, M. Yanagida, and N. Jones. 1995. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 14:6193-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang, W. J., and A. G. Gilman. 1992. Adenylyl cyclases. Cell 70:869-872. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vassarotti, A., and J. D. Friesen. 1985. Isolation of the fructose-1,6-bisphosphatase gene of the yeast Schizosaccharomyces pombe. Evidence for transcriptional regulation. J. Biol. Chem. 260:6348-6353. [PubMed] [Google Scholar]
- 49.Volland, C., D. Urban-Grimal, G. Geraud, and R. Haguenauer-Tsapis. 1994. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J. Biol. Chem. 269:9833-9841. [PubMed] [Google Scholar]
- 50.Watanabe, Y., Y. Lino, K. Furuhata, C. Shimoda, and M. Yamamoto. 1988. The S. pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 7:761-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe, Y., and M. Yamamoto. 1996. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol. Cell. Biol. 16:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welton, R. M., and C. S. Hoffman. 2000. Glucose monitoring in fission yeast via the gpa2 Gα, the git5 Gβ, and the git3 putative glucose receptor. Genetics 156:513-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yun, C. W., H. Tamaki, R. Nakayama, K. Yamamoto, and H. Kumagai. 1997. G-protein coupled receptor from yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 240:287-292. [DOI] [PubMed] [Google Scholar]