Abstract
In Saccharomyces cerevisiae, OLE1 encodes a Δ9 fatty acid desaturase, an enzyme that plays a critical role in maintaining the correct ratio of saturated to monounsaturated fatty acids in the cell membrane. Previous studies have demonstrated that (i) OLE1 expression is repressed by unsaturated fatty acids (UFAs) and induced by low oxygen tension, (ii) a component of this regulation is mediated through the same low oxygen response element (LORE) in the OLE1 promoter, and (iii) Mga2p is involved in LORE-dependent hypoxic induction of OLE1. We now report that LORE-CYC1 basal promoter-lacZ fusion reporter assays demonstrate that UFAs repress the reporter expression under hypoxic conditions in a dose-dependent manner via LORE. Electrophoretic mobility shift assays show that UFAs repress the hypoxia-induced complex formation with LORE. Studies with a construct encoding a truncated form of Mga2p support the hypothesis that both hypoxia and UFA signals affect the processing of Mga2p and the UFA repression of OLE1 hypoxic induction is mediated through Mga2p. Data from Western blot assays provide evidence that under normoxic conditions, Mga2p processing produces approximately equimolar levels of the membrane-bound and processed forms and is unaffected by UFAs. Hypoxic induction of OLE1, however, is associated with increased processing of the protein, resulting in an approximately fivefold increase in the soluble active form that is counteracted by exposure of the cells to unsaturated fatty acids. Data from this study suggest that the Mga2p-LORE interaction plays an important role in OLE1 expression under both normoxic and hypoxic conditions.
Saccharomyces cerevisiae is a facultative aerobe that can utilize oxygen for energy production and therefore has developed a number of mechanisms by which to sense and respond to changes in oxygen availability. The regulatory pathways that are involved with the response to hypoxia (low oxygen) have been studied for decades, and there has been significant progress in this field (3, 23, 43).
In mammalian cells, the best-studied hypoxia signal transduction pathway is mediated by hypoxia-inducible factor 1 (HIF-1) (3, 32). HIF-1 is a heterodimer comprised of HIF-1α and ARNT, two basic helix-loop-helix proteins in the PAS family (39). ARNT mRNA and protein levels are not significantly affected by ambient oxygen tension. HIF-1α mRNA levels are also not appreciably affected by oxygen tension, but HIF-1α protein is only minimally present in normoxia and is rapidly degraded by the ubiquitin-proteasome pathway under this condition (15, 16, 21, 30). This process is dependent on the von Hippel-Lindau tumor suppressor protein (pVHL) (24), which serves as the recognition component of a ubiquitin ligase that promotes ubiquitin-dependent proteolysis of HIF-1α and directly interacts with a highly conserved region within the HIF-1α oxygen-dependent degradation domain (7, 22, 28, 35). Recent studies have indicated that a proline hydroxylation within that conserved region is responsible for targeting HIF-1α for pVHL-mediated destruction (17, 18). In hypoxic cells, HIF-1α degradation is suppressed, leading to heterodimer formation with ARNT. It can then interact with other DNA-binding proteins, such as HNF4 and p300, which provide tissue and developmental specificity and enable enhanced transcription of target genes in response to hypoxia (3, 10).
The S. cerevisiae OLE1 gene encodes the Δ9 fatty acid desaturase, an enzyme that forms monounsaturated palmitoleic (16:1) and oleic (18:1) fatty acids from palmitoyl (16:0) or stearoyl (18:0) coenzyme A. Unsaturated fatty acids (UFAs) are essential for maintaining optimal cell membrane fluidity and expansion in growing cells and serve as major components of storage lipids in stationary-phase cells. OLE1 is highly regulated in response to various physiologically important signals. It has been shown that OLE1 transcription is regulated by fatty acid pools. Its transcription is only weakly induced (1.6-fold) by exogenous saturated fatty acids, but UFAs strongly repress OLE1 transcription, by as much as 60-fold (6, 25). Systematic OLE1 promoter deletion-lacZ fusion analysis indicates that fatty acid response (FAR) elements located 466 to 576 nucleotides upstream from the translation start ATG within the OLE1 promoter contribute to OLE1 repression by UFAs (6).
Studies also indicate that OLE1 expression can be induced under hypoxic conditions, by cobalt and iron chelation, similar to several mammalian hypoxia-inducible genes (23, 38). We recently identified and characterized a cis transactivation element, LORE (low oxygen response element), in the promoter region of OLE1 (38). A similar, but longer element has also been described by Nakagawa et al. (26). In addition, data from both our laboratory and Nakagawa et al. suggest that the same LORE is involved in the UFA repression of OLE1. Genetic analyses have demonstrated that MGA2 or SPT23 is required for OLE1 transcription (42).
Hoppe et al. (14) have identified a novel processing pathway in S. cerevisiae termed regulated ubiquitin-proteasome-dependent processing. The crucial factors in this pathway are Spt23p and Mga2p, which are initially made as dormant precursors (p120) that are firmly anchored in the endoplasmic reticulum-nuclear envelope membranes by their C-terminal tails. It was demonstrated that this pathway controls the level of UFAs in S. cerevisiae by regulating OLE1 expression (14). A shortage of UFAs leads to the Rsp5p-mediated ubiquitination of Spt23p and Mga2p, the release of their N-terminal transcription factor domains (processed forms, p90) into the cytosol, and enhanced expression of the Ole1p, Δ9 fatty acid desaturase. Addition of UFAs that contained more than one double bond almost completely blocked Spt23p precursor processing.
Recently, we have demonstrated that MGA2 is involved in LORE-dependent hypoxic induction of genes in S. cerevisiae (19). We hypothesized that UFAs also repress LORE-dependent OLE1 hypoxic induction through Mga2p. We further hypothesized that Mga2p processing is regulated by both hypoxia and UFA signals but that the two signals work in opposing manners.
Here we provide both direct and indirect evidence that strongly supports the hypothesis that Mga2p processing is regulated by both hypoxia and UFAs and demonstrate that LORE is involved in the UFA repression of OLE1 expression through MGA2 in S. cerevisiae. We demonstrate that the Mga2p-LORE interaction plays an important role in not only the hypoxic induction but also the UFA repression of OLE1 expression.
MATERIALS AND METHODS
Media, chemicals, and enzymes.
Yeast strains were grown in YPD medium (Bio 101, Inc., Carlsbad, Calif.) or SC dropout medium, depending on the plasmid-selectable markers. Luria-Bertani medium was used to grow bacteria. Ampicillin (U.S. Biochemical Corp., Cleveland, Ohio) was used as necessary at 50 μg/ml unless indicated otherwise. Radiolabeled compounds were purchased from Perkin-Elmer (Boston, Mass.). ULTRAhyb was bought from Ambion (Austin, Tex.). Salmon sperm DNA was from GIBCO BRL (Rockville, Md.). CoCl2, NiCl2, 1,10-phenanthroline, Tween 80 (oleic acid, approximately 70%; balance primarily linoleic, palmitic, and stearic acids), linoleic acid (L.A.), arachidonic acid, oleic acid, γ-linolenic acid (γL.A.), stearic acid, tergitol, ergosterol, and NP-40 were obtained from Sigma Chemical Co. (St. Louis, Mo.). T4 polynucleotide kinase and deoxynucleoside triphosphates were purchased from Promega Corporation (Madison, Wis.). Shrimp alkaline phosphatase and Taq polymerase were purchased from Roche Molecular Biochemicals (Indianapolis, Ind.); other restriction enzymes were from New England BioLabs (Beverly, Mass.). All enzymes were used in accordance with the manufacturers' instructions.
Plasmid and plasmid constructions.
The plasmids used in this study are listed in Table 1. The construction of several of the OLE1 promoter-lacZ fusion deletion series was described previously (6, 38). Plasmid p62::934 is the reporter with the OLE1 promoter sequence (from −934 relative to the ATG translational start codon with the A of the codon designated +1)-lacZ fusion. Plasmid p62::396 is the reporter with the lacZ fusion with the OLE1 promoter sequence from −396. pAM4 is p62::934 with mutations in the LORE of the OLE1 promoter (38). Reporter plasmid pAM6 contains a tandem repeat of OLE1 LORE sequences (−347 to −328 relative to the ATG translational start codon with the A of the codon designated +1) in front of CYC1 basal promoter-lacZ fusion vector pTBA30. A centromeric plasmid, pAM23, containing MGA2 was constructed previously (19). Two truncated forms of Mga2p were constructed by introducing a stop codon into pAM23 at the positions encoding amino acids 941 and 869, respectively, of Mga2p, thereby generating proteins Mga2p-Δ172 (pAM31) and Mga2p-Δ244 (pAM30), which had deletions 172 and 244 amino acids, respectively, from the C terminus of Mga2p. A 2μm plasmid with a LEU2 selection marker, YEplac181-mycMGA2, contains a triple repeat of a myc epitope in front of MGA2. This was a gift from S. Jentsch, Department of Molecular Cell Biology, Max Planck Institute for Biochemistry, Martinsried, Germany (14).
TABLE 1.
S. cerevisiae strains and plasmids used in this study
| Strain or plasmid | Genotype or plasmid description | Source or reference |
|---|---|---|
| RZ53-6 | MATatrp1-289 leu2-3, 112 ura3-52 ade1-100 | R. Zitomer |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | RGa |
| BY4741Δmga2 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ΔMGA2 | RG |
| p62::934 | OLE1 promoter-lacZ fusion | 5 |
| p62::396 | OLE1 promoter-lacZ fusion | 5 |
| pAM4 | LORE in OLE1 promoter contains mutations in p62::934 | 38 |
| pTBA30 | Basal CYC1 promoter-lacZ fusion | A. K. Vershon |
| pAM6 | Tandem repeat LORE-basal CYC1 promoter-lacZ fusion | 38 |
| pAM23 | Centromeric plasmid; contains 5.1-kb fragment that contains MGA2 | 19 |
| pAM30 | Derivative of pAM23 with deletion of 244 aab in MGA2 C terminus | This study |
| pAM31 | Derivative of pAM23 with deletion of 172 aa in MGA2 C terminus | This study |
| pRS315 | Parental centromeric plasmid of pAM23 | ATCCc |
| YEplac181-mycMGA2 | 2μm plasmid; contains triple-myc-tagged MGA2 | 14 |
Strains and growth conditions.
The strains used in this study are listed in Table 1. Yeast cells containing lacZ fusion plasmids were grown at 30°C on uracil dropout medium containing dextrose (37). For UFA repression analysis, yeast cells were grown in medium supplemented with 1% tergitol as described previously (6). Cells were grown in the presence of UFAs for 6 h prior to the β-galactosidase assays. Plasmid amplifications and bacterial transformations were performed with Escherichia coli strain DH5α (Invitrogen Corp., Carlsbad, Calif.). Yeast transformations were performed by the method of Elble (11). Preparative cultures were grown aerobically in a shaker (Innova 4000 incubator shaker; New Brunswick Scientific., Edison, N.J.) at 200 rpm and 30°C to mid-logarithmic phase. For experiments assessing yeast under hypoxic conditions, the procedure was as described previously (38). Growth was monitored by measuring the yeast optical density at 600 nm (OD600) at the completion of each experiment.
β-Galactosidase assays.
Assays of cells containing plasmids derived from the OLE1 promoter-lacZ fusion p62 constructs were performed as described previously (31). Cell densities for these assays were determined by measurement of OD600. The β-galactosidase activities reported here are the results of at least two independent experiments. Each experimental assay was performed in quadruplicate. When appropriate, data from replicate experiments are presented as the mean ± the standard error.
Yeast extract preparation.
Haploid yeast cells were cultured in 1-liter flasks containing 200 ml of YPD (1% yeast extract, 2% peptone, 2% dextrose) under either normoxic or hypoxic conditions, harvested at mid-log phase (OD600 of 0.8), and lysed by vortexing with glass beads as previously described (29). Following addition of ammonium sulfate to 40% and incubation on a rocker table at 4°C for 30 min, the precipitate was collected by centrifugation at 12,000 × g in a microcentrifuge at 4°C for 10 min. The pellet was resuspended in storage buffer (20 mM HEPES [pH 8.0], 5 mM EDTA; 20% [vol/vol] glycerol, 1 mM phenylmethylsulfonyl fluoride, 7 mM β-mercaptoethanol) and stored frozen at −80°C. The soluble protein concentration was determined with a Bradford dye-binding assay (Bio-Rad).
EMSAs. Electrophoretic mobility shift assays (EMSAs) were performed essentially as described previously (38), with paired synthetic oligonucleotides as a LORE probe (−347 to −328 relative to the ATG translational start codon with the A of the codon designated +1). Synthetic paired oligonucleotides were end labeled with polynucleotide kinase and purified with a Sephadex G-25 spin column (Roche Molecular Biochemicals) to remove unincorporated nucleotide. Probes made by PCR were purified away from labeled primers [γ-32P]ATP and Taq polymerase with a QIAquick spin PCR purification kit (Qiagen, Santa Clarita, Calif.). In each reaction mixture, 10 to 20 ng of probe was used. Binding reaction mixtures were in 40 μl of buffer H [25 mM HEPES (pH 7.5) at room temperature, 0.5 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM MgCl2, 1 mM CaCl2, 50 mM NaCl, 7% glycerol, 1% NP-40, 15 ng of poly(dA-dT) per μl] for 20 min at room temperature. Proteins were diluted in binding buffer on ice immediately before use. Reaction mixtures were loaded onto 5% acrylamide gels (acrylamide/bisacrylamide ratio, 29:1), electrophoresed in 0.5× Tris-borate-EDTA, and run for 3 h at 4°C and 15 V/cm. Gels were dried and exposed to X-OMAT AR film (Kodak) to visualize the shifted bands.
RNA isolation and Northern blot analysis.
Total yeast RNA was isolated as described previously (8). Equal amounts (15 μg) of total RNA were analyzed by Northern blots in accordance with standard procedures for separation of RNA with 1% formaldehyde gels. RNA from the gels was transferred to Nytran Plus membranes (Schleicher & Schuell Inc., Keene, N.H.) in 10× SSC (1× SSC is 0.15 M sodium citrate plus 0.015 M NaCl) overnight. Prehybridization, hybridization, and washing of membranes were performed as previously described (2). Northern blots were quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.), and autoradiographs were also prepared on X-OMAT AR film (Kodak).
To make radiolabeled cDNA probes for ACT1 and ATF1, yeast genomic DNA prepared by the rapid isolation of yeast chromosomal DNA protocol (13) was subjected to PCR with appropriate pairs of primers for the particular genes of interest. The PCR products were first purified with a QIAquick spin PCR purification kit (Qiagen), separated by agarose gel electrophoresis in 1× Tris-acetate-EDTA (TAE), and then purified with a Qiagen gel extraction kit (Qiagen) in accordance with the manufacturer's recommendations.
To detect OLE1 mRNA, a radiolabeled DNA probe was made with a 0.5-kb EcoRI fragment from the OLE1 protein coding sequence. DNA fragments were separated by agarose gel electrophoresis in 1×TAE and purified with a Qiagen gel extraction kit (Qiagen) in accordance with the manufacturer's recommendations. The purified DNA fragments were labeled to high specific activity with [32P]dCTP (Perkin-Elmer) by the random primer extension method with Ready to Go DNA labeling beads (Amersham Pharmaceutical Biotech, Piscataway, N.J.) reaction kit. Unincorporated nucleotides were removed from the sample with a Sephadex G-50 spin column (Roche Molecular Biochemicals). The specific activity of labeled probes was determined by liquid scintillation counting.
Biotinylated probe pull-down and Western blot assays.
Both oligonucleotides containing the LORE sequence (5′-CGGACGTTGAACACTCAACAAACCTTATCTAG-3′) and one containing a mutated LORE sequence (5′-CGGACGTTGAACAtaCgACAAACCTTATCTAG-3′; the lowercase letters signify mutations), biotinylated at the 5′ end, were synthesized and purified by Integrated DNA Technologies, Inc., Coralville, Iowa, and equimolar amounts of complementary strands were annealed, respectively. Dynabeads M-280 Streptavidin (Dynal, Inc., Lake Success, N.Y.) were prepared in accordance with the manufacturer's instruction. Probe immobilization was performed by mixing 0.5 ml of Dynabeads M-280 Streptavidin with 1 nmol of the double-stranded oligonucleotides containing LORE or the one containing mutated LORE in accordance with the manufacturer's instructions. Finally, the beads were resuspended in 0.5 ml of binding buffer (see EMSA). For the pull-down experiments, 2 mg of whole-yeast crude extract (CE) from strain BY4741Δmga2 transformed with YEplac181-mycMGA2 in 0.5 ml of binding buffer was incubated with 0.5 ml of beads at room temperature for 30 min with regular mixing to keep the beads in suspension. The beads were then concentrated with a magnetic rack and washed three times with 0.5 ml of binding buffer. The second wash used the binding buffer containing 5 μg of poly(dA-dT). Finally, the beads were eluted with 100 μl of binding buffer containing 1 M NaCl. A 10-μl volume of these final eluates was diluted with 30 μl of binding buffer without NaCl and subjected to EMSA and Western blot analyses.
For Western blot analyses, a total of 30 μg of CE in a 20-μl volume of 1× sodium dodecyl sulfate loading buffer was boiled for 5 min and resolved by sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis. Ready-Gels (Bio-Rad, Hercules, Calif.) were transblotted overnight to Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech) with a Mini Trans-Blot electrophoresis transfer cell (Bio-Rad). A Western blotting kit (SuperSignal West Dura; Pierce, Rockford, Ill.) was used. The membrane was probed with a 1:1,000 monoclonal anti-c-myc antibody (clone 9E10; Sigma). Antigen-antibody complexes were visualized by enhanced chemiluminescence (SuperSignal; Pierce) in accordance with the manufacturer's instructions and detected by Hyperfilm ECL (Amersham Pharmacia Biotech). For quantitation of gels, films were scanned with a Personal Densitometer (Molecular Dynamics) and analyzed with ImageQuant software (Molecular Dynamics).
RESULTS
Tween 80 represses hypoxia-induced OLE1 expression.
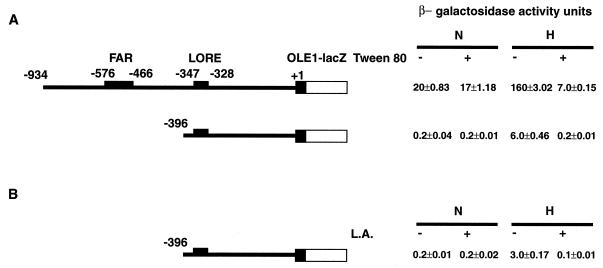
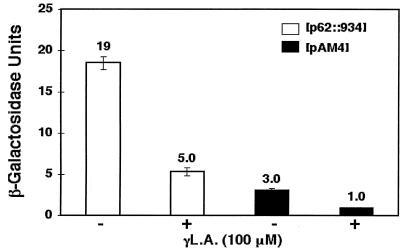
UFA (Tween 80) and ergosterol are required exogenous substrates in yeast cultured under anaerobic conditions (9). Previous studies of the induction of OLE1 expression under low oxygen tension (i.e., hypoxic but not anaerobic conditions) were done with yeast strains grown in medium supplemented with Tween 80 and ergosterol (23). However, when we tested hypoxia-induced reporter expression with an OLE1 promoter-lacZ fusion in yeast in medium containing Tween 80, we found that the robust hypoxic induction of the reporter gene was dramatically repressed (Fig. 1A) whereas the basal reporter expression was not affected. Similar experiments with medium supplemented only with ergosterol showed no effect on OLE1 expression under normoxic and hypoxic conditions (data not shown). This initial observation led us to pursue the identification of the cis elements that may be responsible for the repression. Previous experiments demonstrated FAR elements (−466 to −576) within the OLE1 promoter that contributed to OLE1 repression by UFA (6). To test whether these FAR elements are involved in Tween 80 repression, we performed an OLE1 promoter-lacZ reporter assay with the OLE1 promoter in which the FAR elements were deleted (p62::396). Surprisingly, the hypoxic induction of this reporter construct was still strongly repressed by Tween 80, as shown in Fig. 1A. Because the major components of Tween 80 are various UFAs and it was previously recognized that UFAs exert substantial repression of OLE1 expression, we postulated that the repressive effects of Tween 80 might be mediated by UFAs specifically. As shown in Fig. 1B, when the UFA L.A. was added to the medium, it strongly repressed the expression of the p62::396 reporter as well. These results suggested that there might be another cis element 3′ to −396 of the OLE1 promoter that is critical for OLE1 UFA regulation under hypoxic conditions.
FIG. 1.
Repression effects of UFAs on the β-galactosidase activity of the OLE1 promoter-lacZ fusion. Constructs p62::934 and p62::396 are shown on the left. The narrow line represents the OLE1 promoter sequence. The number above each narrow line indicates the position of the deletion end point with respect to the ATG start codon of the wild-type base sequence with the A of the codon designated +1. The solid black bar represents the amino-terminal 27 amino acids of the OLE1 coding sequence fused to E. coli lacZ (blank bar). The small solid bar above the promoter indicates the FAR and LORE elements, which are also labeled. The values to the right are β-galactosidase activities under normoxic (N) and hypoxic (H) conditions, which were determined as described in Materials and Methods. The values (units of activity) shown are averages of at least three independent experiments that were performed in quadruplicate. (A) Tween 80 repression of β-galactosidase activity of reporters p62::934 and p62::396. (B) L.A. (100 μM) repression of β-galactosidase activity of reporter p62::394.
Dose-dependent repression of OLE1 induction under hypoxic conditions by UFAs.
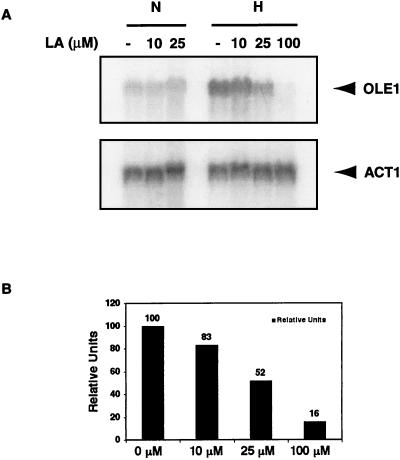
Northern blot analyses were performed to examine the effects of different concentrations of UFAs on OLE1 expression at the mRNA level. Figure 2 shows that the induction of OLE1 expression under hypoxic conditions was repressed by L.A. in a dose-dependent manner. The hypoxia-induced OLE1 mRNA levels were increasingly repressed by the addition of increasing amounts of L.A. to the medium. At 25 μM L.A., the OLE1 mRNA level under hypoxic conditions was about half of that in the control in which no L.A. was added to the medium. At 100 μM L. A., the basal OLE1 mRNA expression under normoxic conditions was repressed to a similar degree as under hypoxic conditions (data not shown). At 1 mM L.A., OLE1 mRNA expression was not detectable by Northern blot analyses, consistent with our previous observation (38). Another UFA, γL.A., was also tested and produced similar results (data not shown).
FIG. 2.
(A) Northern blot analysis under L.A.-treated conditions. Total RNA was extracted from yeast cells grown in increasing concentrations (10, 25, and 100 μM) of L.A. for 6 h. The RNA blot was first hybridized with a specific OLE1 probe, stripped, and rehybridized with a control ACT1 probe. The experiment was done under normoxic (N) and hypoxic (H) conditions. The positions of the OLE1 and ACT1 mRNAs are shown. (B) Quantitation of OLE1 mRNA in the Northern blot under hypoxic conditions in panel A. Signal intensity was quantitated with a Molecular Dynamics PhosphorImager. Transcript levels were normalized to the level of ACT1 mRNA and are presented as 100 relative units for OLE1 mRNA extracted from yeast cells grown without L.A.
Dose-dependent repression of OLE1 hypoxic induction by UFAs via LORE.
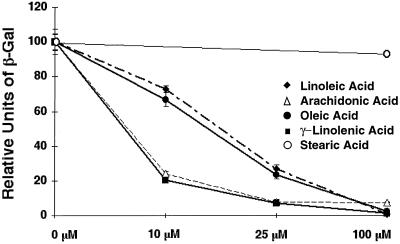
LORE is a previously identified cis transactivation element that plays a critical role in hypoxic induction of OLE1 (38). It is located between −347 and −328 in the OLE1 promoter region. The LORE-CYC1 promoter-lacZ fusion reporter pAM6 was utilized to demonstrate that LORE is involved in L.A. repression in our previous studies (38). Here, we further demonstrate the dose-dependent repression of LORE-mediated hypoxia-induced reporter expression (pAM6) by various UFAs (Fig. 3). Yeast cells were grown under hypoxic conditions in medium supplemented with various concentrations of four different UFAs. All of the UFAs examined demonstrated a clear dose-dependent repression of hypoxia-induced reporter gene expression. However, arachidonic acid and γL.A., which possess four cis double bonds (C-5, C-8, C-11, and C-14) and three cis double bonds (C-6, C-9, and C-12), respectively, exert greater repression of reporter expression (as indicated by the lower β-galactosidase activities), with a 50% inhibitory concentration of ∼5 μM, compared to a 50% inhibitory concentration of ∼20 μM for oleic acid and L.A., which contain one (C-9) and two (C-9 and C-12) double bonds in cis, respectively. Notably, there was almost complete repression of reporter expression with all four UFAs tested at 100 μM. However, stearic acid, a saturated fatty acid, did not have significant effects at this concentration. These results suggest that UFAs specifically repress OLE1 hypoxic induction via LORE and that, at low concentrations, the number of double bonds in the UFAs affects the intensity of UFA repression, regardless of their position and stereochemistry.
FIG. 3.
Dose-dependent repression by UFAs of hypoxia-induced reporter gene expression via LORE. β-Galactosidase assays were performed as described in Materials and Methods. Yeast strains carrying reporter pAM6 were grown in increasing concentrations of fatty acids as indicated in the diagram (0, 10, 25, and 100 μM) for 6 h under hypoxic conditions. The β-galactosidase activity of each strain grown without UFAs under hypoxia was designated 100 U. The values shown are averages of two independent experiments that were performed in quadruplicate.
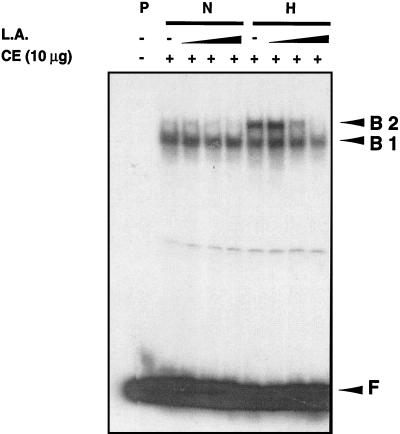
The effects of UFAs on hypoxia-induced complex formation involving LORE were investigated by EMSA. CEs from yeast grown under normoxic and hypoxic conditions with or without increasing amounts of L.A. were utilized for EMSAs with the end-labeled double-stranded LORE oligonucleotides as a probe. As shown in Fig. 4, specific hypoxia-induced complex B2 was increasingly repressed by CEs from yeast cells grown in medium supplemented with increasing amounts of L.A. It appears that complex B2 from normoxic CEs was also repressed. Another UFA, γL.A. (100 μM), was also tested, and it produced similar results (data not shown).
FIG. 4.
L.A. repression of hypoxia-induced complex formation in LORE. EMSAs were performed as described in Materials and Methods. A 10-μg sample of CE from S. cerevisiae grown under normoxic (N) or hypoxic (H) conditions was incubated for 20 min at 25°C with a 32P-labeled LORE and then subjected to electrophoresis at 4°C for ∼3 h. Free probe (F) and bound complexes (B1 and B2) were detected by autoradiography. A minus sign indicates that no CE was added or that the CE was from cells grown without L.A. A plus sign indicates that CE was added. The letter P represents the DNA probe. CEs from yeast cells grown in increasing concentrations of L.A. (10, 25, and 100 μM) were used as indicated.
EMSAs were also performed with hypoxic CEs and addition of increasing amounts of L.A. to the binding reaction mixture in vitro. At increasing concentrations of up to 1 mM, L.A. did not significantly affect the formation of the specific hypoxia-induced complex (B2) (data not shown).
Other cis elements besides LORE are involved in OLE1 expression under normoxic conditions.
OLE1 promoter-lacZ reporter assays were performed to test whether other cis elements besides LORE are involved in OLE1 expression under normoxic conditions. As shown in Fig. 5, γL.A. exhibited repressive effects on reporter gene expression (p62::934) under normoxic conditions. When the reporter pAM4 was used, in which mutations were introduced into the LORE region of the full-length (>900-bp) OLE1 promoter, decreased basal reporter gene expression was observed, consistent with our previous report (38) suggesting that LORE contributes to OLE1 expression under normoxic conditions. However, when γL.A. was added, further repression of reporter gene expression was observed, implying that there are additional cis elements in the OLE1 promoter that play a role in UFA repression. An obvious candidate element is the previously described FAR element (6). These results are consistent with our previous conclusion that UFA repression of OLE1 transcription depends on the FAR element under aerobic conditions (6).
FIG. 5.
Effect of γL.A. on expression of reporter activity under normoxic conditions. β-Galactosidase assays were performed as described in Materials and Methods. Yeast strains carrying reporter p62::934 or pAM4, which possesses mutations in LORE, were grown in medium with or without 100 μM γL.A. for 6 h under normoxic conditions. The values shown are absolute units of β-galactosidase activity, which are averages of two independent experiments that were performed in quadruplicate.
Mga2p, a factor required in LORE-dependent hypoxic induction, is also involved in UFA repression.
Previously, we have shown that MGA2 is involved in the LORE-dependent hypoxic induction of genes in S. cerevisiae (19). CEN-based plasmid pAM23, containing the wild-type MGA2 gene, was transformed into the BY4741Δmga2 strain and was able to fully restore the hypoxic induction of OLE1 in a reporter assay, as well as in Northern and EMSA analyses (19). Dose-dependent repression of OLE1 hypoxic induction by UFAs was also tested in strain BY4741Δmga2 and the parental strain. Medium containing 100 μM L.A. almost completely repressed OLE1 expression under normoxic and hypoxic conditions in reporter and Northern assays with both strains BY4741 and BY4741Δmga2 transformed with pAM23 (data not shown).
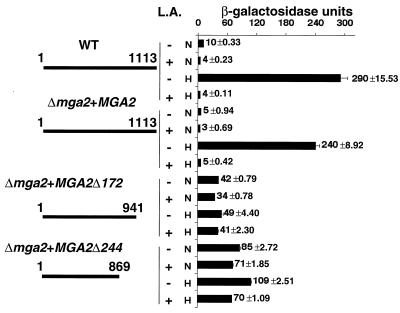
We have previously shown that a soluble, truncated form of Mga2p is regulated by UFA under normoxic conditions when it is independently expressed in an spt23Δ mga2Δ strain (5). To test the hypothesis that Mga2p processing is regulated by hypoxia and UFAs, we constructed two truncated forms of Mga2p by introducing a stop codon at amino acid positions 941 and 869 of MGA2 in pAM23 to generate Mga2-Δ172 (pAM31) and Mga2-Δ244 (pAM30), respectively. The resultant translated Mga2p proteins had deletions of 172 and 244 amino acids, respectively, from the C terminus of wild-type Mga2p. It was hypothesized that these truncated forms of Mga2p might function like endogenously processed Mga2p (p90). If UFA-mediated proteolytic processing of Mga2p is the cause of LORE-dependent UFA regulation, then hypoxia or UFAs should exert little or no additional effect on OLE1 expression in yeast when either truncated form of Mga2p is expressed in BY4741Δmga2. Thus, CEN-based plasmids pAM23, pAM30, and pAM31 were transformed into the BY4741Δmga2 strain carrying the tandem repeat LORE-CYC1 promoter-lacZ fusion, pAM6, and in vivo reporter analyses were performed. As shown in Fig. 6, when the MGA2-containing plasmid (pAM23) was transformed into BY4741Δmga2, the hypoxic induction of β-galactosidase activity was dramatically repressed by the addition of either 100 μM or 1 mM L.A. (data not shown) to the medium. Consistent with previous observations, the basal reporter activity under normoxic conditions was also reduced. However, the reporter activities in the BY4741Δmga2 strain containing either truncated form of Mga2p, Mga2-Δ172 (pAM31) or Mga2-Δ244 (pAM30), increased significantly under normoxic conditions and were similar to the levels found under hypoxic conditions. Additionally, the β-galactosidase activities under both conditions were slightly reduced by addition of 100 μM L.A. to the medium. However, dramatic (>70%) repression of reporter gene expression was observed in the two strains carrying the truncated Mga2p plasmids when 1 mM L.A. was used (data not shown). These data suggest that mechanisms other than Mga2p proteolytic processing are also involved in UFA-mediated OLE1 regulation under normoxic and hypoxic conditions. Additionally, the data suggest that the C-terminally truncated forms of Mga2p, Mga2p-Δ172 and Mga2p-Δ244, do not function as robustly under hypoxic conditions as naturally processed Mga2p.
FIG. 6.
Activities of truncated Mga2p constructs under different conditions. The truncated constructs are shown on the left. The narrow line represents the Mga2p amino acid sequence. The numbers above each narrow line indicate positions in the amino acid sequence with respect to the N terminus on the left and the C terminus on the right. WT indicates that reporter (pAM6) analyses were performed with wild-type strain BY4741. Δmga2+MGA2 indicates that reporter (pAM6) analyses were performed with the BY4741Δmga2 strain transformed with pAM23 containing full-length MGA2. Δmga2+MGA2Δ172 indicates that strain BY4741Δmga2 transformed with pAM31 was used. Δmga2+MGA2Δ244 indicates that strain BY4741Δmga2 transformed with pAM30 was used. L.A indicates growth medium containing 100 μM L.A. dissolved in 1% ethanol. A plus sign indicates that L.A. was added; a minus sign indicates that no L.A. was added. The diagram on the right shows β-galactosidase activities under normoxic (N) and hypoxic (H) conditions. The absolute units of activity indicated are averages of at least three independent experiments that were performed in quadruplicate.
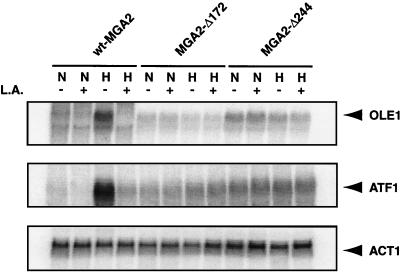
Northern blot analysis was performed as shown in Fig. 7. The OLE1 mRNA levels under normoxia were increased in BY4741Δmga2 transformed with pAM30 compared with those in the wild type, as expected on the basis of the reporter assays. Hypoxia did not further induce OLE1 mRNA levels in BY4741Δmga2 transformed with pAM30. Addition of L.A. had no dramatic effects on the mRNA levels under either normoxic or hypoxic conditions. Experiments with BY4741Δmga2 transformed with pAM31 did not show significantly increased levels of OLE1 mRNA under normoxic conditions. Further, no hypoxic induction or L.A. repression of OLE1 mRNA levels were observed. This is consistent with data from the reporter assays (Fig. 6), in which lower overall reporter activity was observed.
FIG. 7.
Effect of truncated Mga2p on OLE1 gene expression as assessed by Northern blot analysis under various conditions. L.A indicates growth medium containing 100 μM L.A. dissolved in 1% ethanol. A plus sign indicates that L.A. was added; a minus sign indicates that no L.A. was added. The letters N and H indicate that yeast cells were grown under normoxic and hypoxic conditions, respectively, as described in Materials and Methods. The positions of the OLE1, ATF1, and ACT1 mRNAs are shown. wt, wild type.
Another LORE-dependent hypoxic gene, ATF1, was also studied. Previously, we demonstrated that ATF1, encoding an alcohol acetyltransferase, requires MGA2 for hypoxic induction (19, 38). Furthermore, ATF1 expression is repressed by UFAs (12). As shown in Fig. 7, similar to OLE1 expression, the ATF1 mRNA levels of BY4741Δmga2 transformed with either truncated Mga2p plasmid pAM30 or pAM31 were significantly increased under normoxic conditions. Hypoxia had no significant effect compared with normoxia, and addition of 100 μM L. A. had no significant effect under either condition.
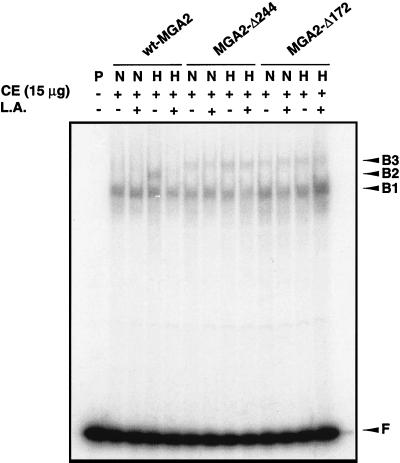
In vitro EMSAs were also performed with CEs from BY4741Δmga2 transformed with plasmids containing wild-type MGA2 or truncated Mga2p and grown under various conditions. As shown in Fig. 8, slower-migrating complex B3 from the CE containing either truncated Mga2p protein was virtually the same under normoxic and hypoxic conditions with or without 100 μM L.A., consistent with the results from the reporter assay and Northern blot analyses. A slightly different migration pattern was observed with CEs containing the two truncated Mga2p proteins. In CE containing Mga2p-Δ244, B3 migrated slightly faster than in CE containing Mga2p-Δ172. This may reflect the 72-amino-acid difference between these two truncated proteins. Additionally, the complexes from CEs with either protein migrated slower than B2, which represents wild-type Mga2p. The migration difference between complexes B2 and B3 may result from the amino acid differences among Mga2p90, Mga2p-Δ244, and Mga2p-Δ172. When a mutated LORE sequence (mLORE) was used as a probe, the complexes from the CEs containing either wild-type or truncated Mga2p disappeared although the nonspecific B1 complex remained intact (data not shown). This suggests that B2 and B3 are functionally equivalent, specific LORE-binding complexes associated with the various Mga2p proteins studied.
FIG. 8.
EMSA analyses of truncated Mga2p CEs. EMSAs were performed as described in Materials and Methods. A 15-μg sample of CE from yeast cells grown under normoxic (N) or hypoxic (H) conditions with (+) or without (−) 100 μM L.A. was incubated for 20 min at 25°C with 32P-labeled LORE and then subjected to electrophoresis at 4°C for ∼3 h. Free probe (F) and bound complexes (B1, B2, and B3) were detected by autoradiography. The letter P represents the DNA probe. wt, wild type.
Mga2p processing is regulated by hypoxia and UFA.
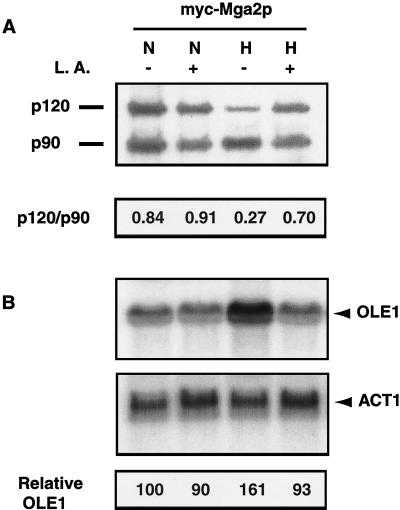
To directly test the hypothesis that Mga2p processing is regulated by hypoxia and UFA, we transformed 2μm-based plasmid YEplac181-mycMGA2 into BY4741Δmga2. CEs and RNA for Western and Northern blot analyses, respectively, were prepared. As shown in Fig. 9A, a Western blot with the anti-myc antibody demonstrated that the Mga2p120/Mga2p90 ratio varies depending upon whether the cells are grown under normoxic or hypoxic conditions. In normoxic cells, levels of membrane-bound Mga2p were about 80% of those of the proteolytically processed p90 form. This was increased to about 90% of the processed form in cells that were exposed to 100 μM L.A. This slight effect of UFAs on the p120/p90 ratio under normoxia is consistent with the previous report of Hoppe et al. (14), who observed that Mga2p processing is only moderately influenced by UFAs.
FIG. 9.
Hypoxia and UFA affect Mga2p processing and OLE1 expression. (A) Western blot assay. BY4741Δmga2 cells transformed with YEplac181-mycMGA2 containing a myc-MGA2 fragment (myc-Mga2p) were grown under different conditions. CEs were prepared as described in Materials and Methods after incubation of the yeast cells under normoxic (N) or hypoxic (H) conditions with (+) or without (−) 100 μM UFA (L.A.). At the bottom are p120/p90 ratios determined by densitometric measurement of p120 and p90 band densities. A Western blot assay was performed as described in Materials and Methods and probed with a monoclonal antibody to c-myc. (B) Northern blot assay. The positions of the OLE1 and ACT1 mRNAs are shown. At the bottom is a quantitative analysis of OLE1 mRNA expression in the Northern blot above. Signal intensity was quantitated with a Molecular Dynamics PhosphorImager. Transcript levels were normalized to the level of ACT1 mRNA and are presented as relative units with respect to the OLE1 mRNA extracted from strain BY4741Δmga2 transformed with YEplac181-mycMGA2 containing the myc-MGA2 fragment (Δmga2+MGA2) and grown without L.A. under normoxic conditions.
Hypoxia significantly increased the relative proportion of p90 (p120/p90 ratio, 0.27 under hypoxia versus 0.84 under normoxia). When UFA was added under hypoxia, a restoration of the p120/p90 ratio toward that observed in normoxia was seen (from 0.27 to 0.70). Northern blot assays were also performed to assess OLE1 mRNA expression. As shown in Fig. 9B, similar hypoxic induction and UFA repression patterns were observed under the same conditions. Again, the UFA repression of mRNA levels under normoxic conditions was not as dramatic.
p90 Mga2p forms a complex with LORE.
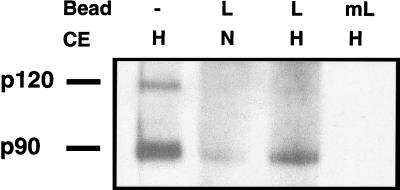
To directly test the hypothesis that the p90 form of Mga2p complexes with LORE, we performed biotinylated probe pull-down assays with probes containing either wild-type LORE or mutated LORE. As shown in Fig. 10, the eluates from beads with LORE primarily contained the p90 form of Mga2p when previously incubated with either hypoxic or normoxic CEs. The lower intensity of the p90 band in normoxic CEs is consistent with the lower levels of processed p90 under normoxic conditions. Importantly, no bands were detected from the eluates obtained from beads with mutated LORE that had been incubated with either hypoxic or normoxic CEs. Similar shifted bands were observed when the same LORE-bead elutes from both hypoxic and normoxic CEs were used in EMSAs (data not shown).
FIG. 10.
p90 Mga2p forms a complex with LORE. A biotinylated probe pull-down assay was done as described in Materials and Methods. Strain BY4741Δmga2 transformed with YEplac181-mycMGA2 containing a myc-MGA2 fragment (myc-Mga2p) was grown under different conditions. CEs were prepared as described in Materials and Methods after incubation of yeast cells under normoxic (N) or hypoxic (H) conditions and incubated with double-stranded, biotinylated LORE (L) or mutated LORE (mL) bound to streptavidin-coated magnetic beads. Protein complexes were purified with a magnet, washed, eluted, and analyzed by Western blotting as described in Materials and Methods, except in lane 1 (no beads), for which the CE from cells grown under hypoxic conditions was used directly in the Western blot. The blot was probed with a monoclonal antibody to c-myc. The positions of the p120 and p90 forms of Mga2p are indicated.
DISCUSSION
The degree of fatty acid unsaturation in cell membrane lipids plays a major role in determining membrane fluidity. Alterations of the ratio of saturated fatty acids to mono-UFAs have been implicated in a variety of diseases, including diabetes, obesity, hypertension, cancer, and neurologic, vascular, and heart diseases, in mammals (27, 33, 36). In yeast, it has been suggested that this ratio is related to the heat shock response, ethanol tolerance, and mitochondrial movement and inheritance (1, 4, 33, 34).
OLE1, which encodes a Δ9 fatty acid desaturase in S. cerevisiae, is an enzyme that plays a critical role in maintaining the correct ratio of saturated fatty acids to mono-UFAs in the cell membrane. Two well-studied signals known to significantly affect OLE1 expression are UFAs and hypoxia, which, respectively, repress and activate OLE1 expression at the transcriptional level (23, 25, 26, 38). We hypothesized that there may be communication between these different signal pathways. Here we provide evidence that the cis element LORE and the trans factor Mga2p are involved in both hypoxic induction and UFA repression of OLE1.
LORE-CYC1 basal promoter-lacZ fusion reporter (pAM6) assays and Northern blot analyses clearly demonstrated that UFAs repress hypoxia-induced expression in a dose-dependent manner (Fig. 2 and 3). The data suggest that, at low concentrations of fatty acids, the number of cis double bonds in a UFA is a major factor in determining the potency of UFA repression. Results of EMSAs further confirmed that UFAs repress LORE-dependent gene expression, as indicated by inhibition of hypoxia-induced complex formation with LORE (Fig. 4). This differential repression might be caused by changes induced in membrane fluidity by the more highly unsaturated species or by differences in the rates of fatty acid import or intracellular movements that produce different concentrations of the various species in the vicinity of the sensors that trigger those regulatory changes. Given the suggestion here that fatty acids affect OLE1 expression by more than one pathway, it is conceivable that Mga2p-mediated expression might respond to changes in membrane lipid composition, as well as internal free fatty acid or fatty acyl coenzyme A pools.
Northern analyses, LORE-dependent reporter assays, and EMSAs with two truncated forms of Mga2p mimicked the processed p90 form of Mga2p and support the hypothesis that both hypoxia and UFA signals affect the processing of Mga2p and that both UFA repression and hypoxic induction of OLE1 are mediated through the same factor, Mga2p, that acts via LORE. One of the truncated Mga2p forms used in this study, Mga2-Δ172, is similar to one studied before (41). Zhang et al. demonstrated that if the C-terminal 173 codons were deleted from MGA2, in either CEN- or 2μm-based plasmids, the truncated gene was able to suppress the Ty-induced mutation lys2-61. Another C-terminally truncated Mga2p protein from a 2μm-based plasmid could suppress ufd1-2 (a temperature-sensitive mutant form of ufd1, an essential gene of unknown function that was identified in a screen for yeast mutants that stabilize an artificial ubiquitin fusion protein [20]), similar to Spt23p and its C-terminally truncated forms (14). It is noteworthy that previous studies demonstrated that Spt23p processing can be strongly regulated by UFAs, which repress OLE1 expression (14). SPT23 could be crucial in general UFA repression under normoxic conditions via a LORE-independent element, for example, the FAR element, which plays an important role in OLE1 normoxic expression.
Finally, Western blot analysis (as shown in Fig. 9) directly demonstrated that both UFAs and hypoxia affect the processing of Mga2p and that the effects of these two signals can counteract one another. These observations regarding Mga2p processing are consistent with the data from Northern blot analyses. Interestingly, results from previous studies have suggested that UFAs have only a modest influence on Mga2p processing (14), and these results are consistent with our findings. We believe that at least part of the reason for this is the much higher levels of expression in the 2μm-based myc-MGA2 plasmid. The results of Northern blot assays in which BY4741Δmga2 was transformed with a CEN-based MGA2 plasmid (Fig. 7), compared to a 2μm-based myc-MGA2 plasmid (Fig. 9B), indicate that the UFA repression of the myc-MGA2 plasmid is less dramatic than that of CEN-based MGA2 plasmid pAM23. The moderate effects of UFA on blocking of Mga2p processing, as assessed by Western analysis, especially under normoxic conditions, support this hypothesis. It is also possible, although unlikely, that the myc tag contributes to these effects. In summary, both direct and indirect evidence from this study strongly supports the hypothesis that both UFA and hypoxia signals affect the processing of Mga2p. The interesting question of exactly how these signals affect the processing of Mga2p remains unsolved.
The results of biotinylated-probe pull-down assays (Fig. 10) provide direct evidence that the processed p90 form of Mga2p, not the precursor p120, is a component of the LORE complex. This could explain the different migration of the two truncated Mga2p forms compared with that of wild-type Mga2p in Fig. 8. The slower migration of the shifted bands from truncated Mga2p may result from the difference in the number of amino acids between the truncated and wild-type forms of Mga2p. Truncated Mga2p may still be bigger than p90.
It is worth noting that the effect of the ubiquitination-proteasomal degradation pathway on a crucial transcription factor of a hypoxia signal transduction pathway provides a potential common theme for hypoxic regulation of gene expression between yeast and mammalian cells. In yeast, the ubiquitin-proteasome-dependent pathway is involved in the processing of Spt23p and Mga2p (14). We have shown that MGA2 is an important transcription factor involved in the LORE-dependent hypoxic induction of genes in S. cerevisiae (19). Although we did not directly address in this study the question of whether the processing of Mga2p is facilitated by hypoxia in a ubiquitin-proteasome-dependent manner, it is quite possible that the mechanism of Mga2p processing under hypoxia is similar to that of UFA repression, in which the processing of Mga2p depends on the ubiquitin-proteasome-dependent pathway. In the case of the mammalian hypoxic response, HIF-1α is usually immediately destroyed by the ubiquitin-proteasome pathway under normoxic conditions and is stabilized by hypoxia (3, 32). Significantly, the proteasomal degradation of Mga2p is only partial and probably occurs under hypoxic conditions to produce higher levels of the active protein form, whereas for HIF-1α, protein degradation is complete under normoxic conditions.
The present study provides another example of how cells may translate different signals to cellular responses, perhaps via the same pathway. The details of the mechanism by which OLE1 expression is regulated by two signals, UFAs and oxygen, are under active investigation.
Acknowledgments
Y.J. and M.J.V. contributed equally to this work.
We thank H. Franklin Bunn and Fred Winston for invaluable support through all of the phases of this project.
This work was supported by National Institutes of Health grants DK45098 to M.A.G., GM45768 to C.E.M., and K08 HL03599 to M.J.V.
REFERENCES
- 1.Alexandre, H., I. Rousseaux, and C. Charpentier. 1994. Relationship between ethanol tolerance, lipid composition and plasma membrane fluidity in Saccharomyces cerevisiae and Kloeckera apiculata. FEMS Microbiol. Lett. 124:17-22. [DOI] [PubMed] [Google Scholar]
- 2.Brown, T., and K. Mackey. 1997. Analysis of RNA by Northern and slot blot hybridization, p. 4.9.1-4.9.14. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Bunn, H. F., and R. O. Poyton. 1996. Oxygen sensing and molecular adaptation to hypoxia. Physiol. Rev. 76:839-885. [DOI] [PubMed] [Google Scholar]
- 4.Carratu, L., S. Franceschelli, C. L. Pardini, G. S. Kobayashi, I. Horvath, L. Vigh, and B. Maresca. 1996. Membrane lipid perturbation modifies the set point of the temperature of heat shock response in yeast. Proc. Natl. Acad. Sci. USA 93:3870-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chellappa, R., P. Kandasamy, C. S. Oh, Y. Jiang, M. Vemula, and C. E. Martin. 2001. The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of Saccharomyces cerevisiae OLE1 gene expression: fatty acid-mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J. Biol. Chem. 276:43548-43556. [DOI] [PubMed] [Google Scholar]
- 6.Choi, J. Y., J. Stukey, S. Y. Hwang, and C. E. Martin. 1996. Regulatory elements that control transcription activation and unsaturated fatty acid-mediated repression of the Saccharomyces cerevisiae OLE1 gene. J. Biol. Chem. 271:3581-3589. [DOI] [PubMed] [Google Scholar]
- 7.Cockman, M. E., N. Masson, D. R. Mole, P. Jaakkola, G. W. Chang, S. C. Clifford, E. R. Maher, C. W. Pugh, P. J. Ratcliffe, and P. H. Maxwell. 2000. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 275:25733-25741. [DOI] [PubMed] [Google Scholar]
- 8.Collart, M. A., and S. Oliviero. 1997. Preparation of yeast RNA, p. 13.12.1-13.12.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 9.Daum, G., N. D. Lees, M. Bard, and R. Dickson. 1998. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast 14:1471-1510. [DOI] [PubMed] [Google Scholar]
- 10.Ebert, B. L., and H. F. Bunn. 1999. Regulation of the erythropoietin gene. Blood 94:1864-1877. [PubMed] [Google Scholar]
- 11.Elble, R. 1992. A simple and efficient procedure for transformation of yeasts. BioTechniques 13:18-20. [PubMed] [Google Scholar]
- 12.Fujiwara, D., H. Yoshimoto, H. Sone, S. Harashima, and Y. Tamai. 1998. Transcriptional co-regulation of Saccharomyces cerevisiae alcohol acetyltransferase gene ATF1 and delta-9 fatty acid desaturase gene OLE1 by unsaturated fatty acids. Yeast 14:711-721. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman, C. S. 1997. Rapid isolation of yeast chromosomal DNA, p. 13.11.2-13.11.4. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 14.Hoppe, T., K. Matuschewski, M. Rape, S. Schlenker, H. D. Ulrich, and S. Jentsch. 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102:577-586. [DOI] [PubMed] [Google Scholar]
- 15.Huang, L. E., Z. Arany, D. M. Livingston, and H. F. Bunn. 1996. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem. 271:32253-32259. [DOI] [PubMed] [Google Scholar]
- 16.Huang, L. E., J. Gu, M. Schau, and H. F. Bunn. 1998. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 95:7987-7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 18.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, Y., M. J. Vasconcelles, S. Wretzel, A. Light, C. E. Martin, and M. A. Goldberg. 2001. MGA2 is involved in the low-oxygen response element-dependent hypoxic induction of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6161-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, E. S., P. C. Ma, I. M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442-17456. [DOI] [PubMed] [Google Scholar]
- 21.Kallio, P. J., I. Pongratz, K. Gradin, J. McGuire, and L. Poellinger. 1997. Activation of hypoxia-inducible factor 1α: posttranscriptional regulation and conformational change by recruitment of the Arnt transcription factor. Proc. Natl. Acad. Sci. USA 94:5667-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamura, T., S. Sato, K. Iwai, M. Czyzyk-Krzeska, R. C. Conaway, and J. W. Conaway. 2000. Activation of HIF1α ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. USA 97:10430-10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwast, K. E., P. V. Burke, B. T. Staahl, and R. O. Poyton. 1999. Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc. Natl. Acad. Sci. USA 96:5446-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 25.McDonough, V. M., J. E. Stukey, and C. E. Martin. 1992. Specificity of unsaturated fatty acid-regulated expression of the Saccharomyces cerevisiae OLE1 gene. J. Biol. Chem. 267:5931-5936. [PubMed] [Google Scholar]
- 26.Nakagawa, Y., S. Sugioka, Y. Kaneko, and S. Harashima. 2001. O2R, a novel regulatory element mediating Rox1p-independent O2 and unsaturated fatty acid repression of OLE1 in Saccharomyces cerevisiae. J. Bacteriol. 183:745-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ntambi, J. M. 1999. Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. J. Lipid Res. 40:1549-1558. [PubMed] [Google Scholar]
- 28.Ohh, M., C. W. Park, M. Ivan, M. A. Hoffman, T. Y. Kim, L. E. Huang, N. Pavletich, V. Chau, and W. G. Kaelin. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2:423-427. [DOI] [PubMed] [Google Scholar]
- 29.Pfeifer, K., B. Arcangioli, and L. Guarente. 1987. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell 49:9-18. [DOI] [PubMed] [Google Scholar]
- 30.Pugh, C. W., J. F. O'Rourke, M. Nagao, J. M. Gleadle, and P. J. Ratcliffe. 1997. Activation of hypoxia-inducible factor-1: definition of regulatory domains within the alpha subunit. J. Biol. Chem. 272:11205-11214. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds, A., V. Lundblad, D. Dorris, and M. Keaveney. 1997. Yeast vectors and assays for expression of cloned genes, p. 13.6.2-13.6.3. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 32.Semenza, G. L. 1999. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 15:551-578. [DOI] [PubMed] [Google Scholar]
- 33.Sherbet, G. V. 1989. Membrane fluidity and cancer metastasis. Exp. Cell Biol. 57:198-205. [DOI] [PubMed] [Google Scholar]
- 34.Stewart, L. C., and M. P. Yaffe. 1991. A role for unsaturated fatty acids in mitochondrial movement and inheritance. J. Cell Biol. 115:1249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanimoto, K., Y. Makino, T. Pereira, and L. Poellinger. 2000. Mechanism of regulation of the hypoxia-inducible factor-1α by the von Hippel-Lindau tumor suppressor protein. EMBO J. 19:4298-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tooke, J. E. 2000. Possible pathophysiological mechanisms for diabetic angiopathy in type 2 diabetes. J. Diabet. Complicat. 14:197-200. [DOI] [PubMed] [Google Scholar]
- 37.Treco, D. A., and F. Winston. 1997. Growth and manipulation of yeast, p. 13.2.1-13.2.12. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 38.Vasconcelles, M. J., Y. Jiang, K. McDaid, L. Gilooly, S. Wretzel, D. L. Porter, C. E. Martin, and M. A. Goldberg. 2001. Identification and characterization of a low oxygen response element involved in the hypoxic induction of a family of Saccharomyces cerevisiae genes. Implications for the conservation of oxygen sensing in eukaryotes. J. Biol. Chem. 276:14374-14384. [DOI] [PubMed] [Google Scholar]
- 39.Wang, G. L., and G. L. Semenza. 1995. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270:1230-1237. [DOI] [PubMed] [Google Scholar]
- 40.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, S., T. J. Burkett, I. Yamashita, and D. J. Garfinkel. 1997. Genetic redundancy between SPT23 and MGA2: regulators of Ty-induced mutations and Ty1 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4718-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, S., Y. Skalsky, and D. J. Garfinkel. 1999. MGA2 or SPT23 is required for transcription of the δ9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics 151:473-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zitomer, R. S., and C. V. Lowry. 1992. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 56:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]