Abstract
The Rho-type GTPase, Cdc42, has been implicated in a variety of functions in the yeast life cycle, including septin organization for cytokinesis, pheromone response, and haploid invasive growth. A group of proteins called GTPase-activating proteins (GAPs) catalyze the hydrolysis of GTP to GDP, thereby inactivating Cdc42. At the time this study began, there was one known GAP, Bem3, and one putative GAP, Rga1, for Cdc42. We identified another putative GAP for Cdc42 and named it Rga2 (Rho GTPase-activating protein 2). We confirmed by genetic and biochemical criteria that Rga1, Rga2, and Bem3 act as GAPs for Cdc42. A detailed characterization of Rga1, Rga2, and Bem3 suggested that they regulate different subsets of Cdc42 function. In particular, deletion of the individual GAPs conferred different phenotypes. For example, deletion of RGA1, but not RGA2 or BEM3, caused hyperinvasive growth. Furthermore, overproduction or loss of Rga1 and Rga2, but not Bem3, affected the two-hybrid interaction of Cdc42 with Ste20, a p21-activated kinase (PAK) kinase required for haploid invasive growth. These results suggest Rga1, and possibly Rga2, facilitate the interaction of Cdc42 with Ste20 to mediate signaling in the haploid invasive growth pathway. Deletion of BEM3 resulted in cells with severe morphological defects not observed in rga1Δ or rga2Δ strains. These data suggest that Bem3 and, to a lesser extent, Rga1 and Rga2 facilitate the role of Cdc42 in septin organization. Thus, it appears that the GAPs play a role in modulating specific aspects of Cdc42 function. Alternatively, the different phenotypes could reflect quantitative rather than qualitative differences in GAP activity in the mutant strains.
In the yeast Saccharomyces cerevisiae, the Cdc42 protein (33) is a member of the Rho subfamily of the Ras superfamily of GTPases, which act as molecular switches and regulate many cellular processes (1, 34). Like all GTPases, Cdc42 can exist in two states, a GTP-bound, active state and a GDP-bound, inactive state. The cycling of Cdc42 between these states is controlled by two sets of proteins: guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). There is one GEF for Cdc42 in yeast, called Cdc24 (62, 67, 69). At the time this study began, there was one protein with demonstrated biochemical GAP activity for Cdc42, Bem3 (68), and one protein with potential GAP activity based on the presence of a 170-amino-acid GAP domain, Rga1 (14, 63). Although Bem3 and Rga1 are fairly similar in their GAP domains, they are divergent over the remainder of their protein sequences. Rga1 contains two tandem Lin-11, Isl-1, Mec-3 (LIM) domains (23, 35, 63, 66). LIM domains bind zinc ions and are thought to mediate protein-protein interactions (2, 18, 45). In contrast, Bem3 contains a Pleckstrin homology (PH) domain. PH domains are thought to play roles in membrane localization and protein-protein interactions (40).
Cdc42 is required for polarization of the actin cytoskeleton, for cytokinesis, and for morphological changes that accompany activation of some signal transduction pathways, specifically the pheromone response pathway and the filamentous growth pathway (for review, see reference 32). Several proteins, aside from Cdc24, Rga1, and Bem3, that bind to Cdc42 have been identified and are presumed to be effectors of various Cdc42 functions. These proteins include Ste20 and Cla4, two protein kinases (17, 61), and Gic1 and Gic2, two proteins of similar sequence but as-yet-undefined biochemical activity (7, 13). Ste20 is required for operation of the pheromone and filamentation pathways (38, 53, 56), Cla4 plays ill-defined roles in septin organization and cytokinesis (4, 9, 17, 41), and together Gic1 and Gic2 are required to polarize the actin cytoskeleton (5).
The finding that there is more than one potential GAP for Cdc42 raises the possibility that each GAP may regulate Cdc42 activity with respect to only a subset of its biological roles. To explore this possibility, we sought to identify the full complement of GAPs for Cdc42 and examine the phenotypic consequences of the absence of one or more of these GAPs. Here we report the identification of a third potential GAP, Rga2, which has strong sequence similarity to Rga1. We demonstrate that Rga1 and Rga2 have biochemical GAP activity for Cdc42. Finally, we provide evidence that strains lacking Rga1, Rga2, and Bem3 display distinct phenotypes, supporting the idea that they influence Cdc42 activity in different biological settings.
MATERIALS AND METHODS
Strains, plasmids, and microbiological techniques.
The yeast strains used are listed in Table 1. Gene deletions were constructed by PCR and confirmed by PCR and phenotyping when applicable. In all cases, the entire coding region was replaced with the indicated marker. Yeast and bacterial strains were propagated using standard methods (8, 57, 59). Yeast extract-peptone-dextrose and synthetic dextrose media were prepared as previously described (58). Yeast transformations were performed using modifications of the LiOAc method (12, 24). Bacterial transformations, DNA preparations, and plasmid constructions were performed by standard methods (59). Growth capabilities at various temperatures were measured by spotting equal volumes of serial dilutions of mid-log phase cultures to agar plates and incubating at the indicated temperatures.
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| DJTD2-16D | MATα cdc42-1ts ura3 trpl leu2 his4 gal2 | 45a |
| YEF24H | MATα cdc24-Hts ura3 trpl leu2 his4 ade2 | 34 |
| EGY40 | MATα ura3-52 his3 trpl leu2 | 26 |
| PJ69-4A | MATaGALI-HIS3 GAL2-ADE8 GAL7-lacZ leu2 ura3 his3 gal4 gal80 | 30 |
| Σ1278B | MATα ura3 | G. Fink |
| YAG512 | Σ1278B except his3::ura3- | A. Goehring |
| YGS45 | YEF24H except rga1::HIS3 | This study |
| YGS46 | YEF24H except rga2::HIS3 | This study |
| YGS47 | YEF24H except bem3::HIS3 | This study |
| YGS156 | PJ69-4A except rga1::URA3 | This study |
| YGS157 | PJ69-4A except rga2::URA3 | This study |
| YGS158 | PJ69-4A except bem3::URA3 | This study |
| SY2002 | MATaFUS1::HIS3 his3 mfa2-Δ1::FUS1-lacZ adel leu2 trpl ura3 | Lab strain |
| YGS2 | SY2002 except rga1::URA3 | This study |
| YGS7 | SY2002 except rga2::TRP1 | This study |
| YGS50 | SY2002 except bem3::TRP1 | This study |
| YGS72 | SY2002 except rga1::URA3 rga2::TRP1 | This study |
| YGS51 | SY2002 except rga1::URA3 bem3::TRP1 | This study |
| YGS56 | SY2002 except rga2::TRP1 bem3::TRP1 | This study |
| YGS57 | SY2002 except rga1::URA3 rga2::TRP1 bem3::TRP1 | This study |
| YGS58 | SY2002 except ste4::LEU2 | This study |
| YGS59 | YGS2 except ste4::LEU2 | This study |
| YGS60 | YGS7 except ste4::LEU2 | This study |
| YGS61 | YGS50 except ste4::LEU2 | This study |
| YGS62 | YGS51 except ste4::LEU2 | This study |
| YGS63 | YGS72 except ste4::LEU2 | This study |
| YGS64 | YGS56 except ste4::LEU2 | This study |
| YGS65 | YGS57 except ste4::LEU2 | This study |
| YGS281 | YAG512 except rga1::HIS3 | This study |
| YGS328 | YAG512 except rga2::HIS3 | This study |
| YGS329 | YAG512 except bem3::HIS3 | This study |
| YGS105 | MATamfa2-Δ1::FUS1-lacZ his3 leu2 trp1 ura3 ade1 | This study |
| YGS286 | YGS105 except ste20::URA3 | This study |
| YGS351 | YGS286 except rga1::HIS3 | This study |
| YGS352 | YGS286 except rga2::HIS3 | This study |
| YGS353 | YGS286 except bem3::HIS3 | This study |
| IDY22 | MATαcla4::TRP1 mfa2-Δ1::FUS1-lacZ leu2 ura3 ade2 ade3 lys2 | D. Mitchell |
| YGS80 | IDY22 except rga1::URA3 | This study |
| YGS81 | IDY22 except rga2::URA3 | This study |
| YGS82 | IDY22 except bem3::LEU2 | This study |
Two-hybrid analysis.
Two-hybrid analysis (22) of interactions between the panel of GTPases and Rga1, Rga2, and Bem3 was performed with strain EGY40 (26). To assess β-galactosidase activity, strain EGY40 containing the LexAop-lacZ reporter plasmid pSH18-34 (26) was transformed with a pEG202-derived LexA DNA-binding domain (DBD) plasmid and a pJG4-5-derived Gal4 activation domain (AD) plasmid. At least three separate isolates were grown to mid-log phase in rich medium containing 2% galactose and quantitated as previously described (63). Most of the LexA DBD fusions in pEG202 and Gal4 AD fusions in pJG4-5 were previously described (63). pJG4-5-RGA2 was constructed using gap repair (42) of the entire RGA2 coding region and EcoRI-cut pJG4-5 (26). pEG202-RGA2 was constructed by ligating an XhoI-EcoRI fragment of pJG4-5-RGA2 into XhoI-EcoRI-cut pEG202 (26). The expression of two-hybrid constructs was confirmed by Western blotting techniques (59) using α-LexA DBD and α-Gal4 AD antibodies, respectively (52). β-Galactosidase assays were used to determine the relative strength of the interactions and were performed essentially as described previously (29).
Additional two-hybrid analysis was performed with the PJ69-4A strain (30). pGS38 and pGS39 were constructed by ligating the EcoRI-XhoI fragments from pEG202-CDC42G12V,C188S and pEG202-CDC42Q61L,C188S (63), respectively, into the Gal4 activation domain fusion vector pGAD-C(2) (30), which was cut with EcoRI and SalI. pSL2682 contains a truncation of STE20 (encoding amino acids 1 to 565) cloned into pOBD. CYH2 (64) was a kind gift from Megan Keniry. The expression of these fusion proteins was confirmed by Western blot analysis using a α-Gal4 DBD monoclonal antibody (52). pGS40 was constructed by ligating a KpnI-HindIII fragment from YEp13-RGA1 containing the coding region and 1,500 nucleotides upstream of RGA1 to KpnI-HindIII-cut YEp352 (28). pGS41 was constructed by ligating a SacI-XhoI fragment from YEp13-RGA2 containing the coding region and 800 nucleotides upstream of RGA2 to SacI-XhoI-cut YEp352. pGS42 was constructed by ligating a SacI-PstI fragment from YEp13-BEM3 containing the coding region and 800 nucleotides upstream of BEM3 to SacI-PstI cut YEp352. β-Galactosidase assays were used to determine the relative strength of the interactions and were performed essentially as described previously (29).
Protein purification.
The GTPases Cdc42 and Rsr1 (Bud1) were purified as glutathione S-transferase (GST) fusion proteins using hybrid genes present in the plasmid pGEX-5X-1 (47). pGEX-5X-1-Cdc42C188S and pGEX-5X-1-Cdc42Q61L,C188S were gifts from David Mitchell, and pGEX-Rsr1 was kindly provided by Hay-Oak Park (49). Escherichia coli cells (DH5α) were grown to mid-log phase, protein expression was induced with 1 mM IPTG (Sigma) for 2 h, and cells were lysed by two passages through a French press. The lysate was centrifuged at 5,000 rpm in an SS-34 Sorvall rotor for 10 min to pellet unbroken cells and cellular debris. Swollen glutathione agarose beads were added to the supernatant, incubated at 4°C for 2 h, washed, and frozen at −80°C in GTPase storage buffer (20 mM Tris-HCl [pH 7.5], 1 mM dithiothreitol [DTT], 100 μM GTP, 1× protease inhibitors cocktail [Roche], and 15% glycerol).
The GAP domains of Rga1, Rga2, and Bem3 were purified as maltose binding protein (MBP) fusions. These fusions were expressed from plasmids based on pMAL-c2 (25). pGS53 was constructed by addition of SalI and HindIII sites to the ends of an RGA1 PCR product using a plasmid containing the RGA1 coding region as a template. This PCR product was digested and ligated to SalI-HindIII-cut pMAL-c2. pGS54 was constructed by addition of SalI and PstI sites to the ends of an RGA2 PCR product by using a plasmid containing the RGA2 coding region as a template. This PCR product was digested and ligated to SalI-PstI-cut pMAL-c2. pGS55 was constructed by addition of EcoRI and SalI sites to the ends of a BEM3 PCR product by using a plasmid containing the BEM3 coding region as a template. This PCR product was digested and ligated to EcoRI-SalI-cut pMAL-c2. Bacterial protein expression and cell lysis were performed essentially as described above for the GST fusions, except proteins were purified using an amylose column (44) and eluted with Column Buffer II (20 mM Tris-HCl [pH 7.4], 1 mM EDTA, 200 mM NaCl, 10 mM maltose). In all cases, standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis separation and Coomasie staining confirmed protein expression and purification (59).
Biochemistry.
Purified GST, GST-Cdc42C188S, GST-Cdc42Q61L,C188S, or GST-Rsr1 was incubated with ∼2 μCi of [γ-32P]GTP or [α-32P]GTP (New England Nuclear Life Sciences) at room temperature for 10 min in reaction buffer (20 mM Tris-HCl [pH 7.5], 1 mM DTT, 100 μM GTP). Then, 2 nmol of GTP was added to each reaction mixture. Each reaction mixture contained one of the following: 25.6 pmol of GST-Cdc42 (C188S), 21.3 pmol of GST-Cdc42 (Q61L, C188S), or 71.6 pmol of GST-Rsr1. As a result, 5.7% of the GST-Cdc42 (C188S) bound [α-32P]GTP, 5.9% of the GST-Cdc42 (C188S) bound [γ-32P]GTP, and 7.6% of the GST-Rsr1 bound [γ-32P]GTP. Glutathione agarose beads bound with a [γ-32P]GTP- or [α-32P]GTP-loaded GTPase were added to micro-spin columns (Bio-Rad) and washed six times with 250 μl of wash buffer (20 mM Tris-HCl [pH 7.5], 1 mM DTT, 5 mM MgCl2). The beads were resuspended, removed from the column, and added to ∼2 μg (28.6 pmol) of MBP, MBP-Rga1, MBP-Rga2, or MBP-Bem3. The mixtures were incubated at room temperature for the indicated time, added to a micro-spin column, spun, and washed. Then, 100 μl of elution buffer (1% sodium dodecyl sulfate-20 mM EDTA) was added to the column and incubated at 65°C for 15 min to denature the protein. The samples were centrifuged to collect the released nucleotides. The reaction buffer, wash buffer, and eluate were counted in a scintillation counter. The ratio of bound to released counts was used to determine GAP activity.
Microscopy.
Standard microscopic techniques were used and cells were examined using a 100× objective. Methods for staining with rhodamine phalloidin to visualize F-actin, Calcofluor to visualize chitin deposits in bud scars, and DAPI (4′,6′-diamidino-2-phenylindole) for visualization of DNA were performed essentially as previously described (51). Septins were visualized by immunofluorescence using a α-Cdc3 antibody (36; purified by April Goehring) followed by fluorescence microscopy.
FUS1-lacZ assays.
Cells containing a FUS1-lacZ reporter construct integrated at mfa2Δ (6) were grown to mid-log phase at 30°C in yeast extract-peptone-dextrose medium. β-Galactosidase assays were performed as described previously (31).
Invasive growth assays.
The plate-washing assay was performed as previously described (56). The single cell assay was also performed essentially as previously described (16). For some experiments, cells were scraped from plates using 4 ml of distilled water, concentrated by centrifugation, resuspended in 20 μl of water, and visualized by microscopy. For other experiments, a coverslip was placed directly on the agar medium and cells were visualized by microscopy at 100×.
RESULTS
Rga2 is putative Rho-GTPase-activating protein for Cdc42.
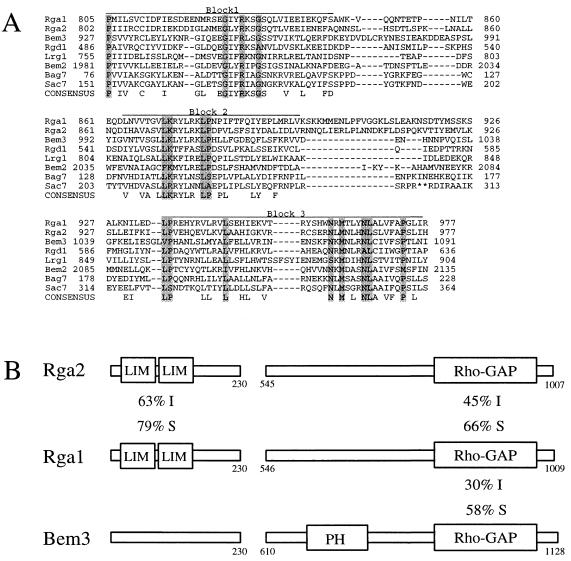
In an effort to find putative GAPs for Cdc42, we performed a homology search of the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/) by using the BLAST algorithm. Searching for proteins similar to Rga1 identified seven proteins that contained a potential Rho-GAP motif: Ydr379w, Bem3, Lrg1, Rgd1, Bem2, Sac7, and Bag7 (Fig. 1A). Many of these proteins have been previously described as GAPs for a variety of Rho-type GTPases. Bem3 was shown to be a GAP for Cdc42 (68). Rgd1 was demonstrated to have GAP activity for Rho3 and Rho4, but not for Cdc42 (3, 20). The putative Rho-GAP, Lrg1, acts as a GAP for Rho1 during 1,3-β-glucan synthesis (65). Bem2 was shown to be a GAP for Rho1, but not for Cdc42 (67, 68). Sac7 is thought to act as a GAP for Rho1 (60). Since Rgd1, Lrg1, Bem2, Sac7, and Bag7 have been at least partially characterized as being involved in other processes, they were not pursued as putative GAPs for Cdc42.
FIG. 1.
Protein sequence alignments of Rho-GAPs in yeast. (A) Protein alignment of putative GAP motifs for Rho-type GTPases in the yeast S. cerevisiae. Consensus residues are shaded for residues shared by seven or eight of the proteins and unshaded for residues shared by four to six of the proteins. ∗∗, 61 omitted residues for Sac7. (B) A graphical representation of the known and putative GAPs for Cdc42. The numbers correspond to amino acid positions in the protein sequence. I, identity; S, similarity; LIM, tandem LIM domains; PH, a pleckstrin homology domain; Rho-GAP, consensus GAP motif as shown in panel A.
One of the open reading frames identified, YDR379W, showed a high degree of similarity to RGA1 across the entire protein sequence and was renamed RGA2 for Rho-GTPase-activating protein 2. RGA2 encodes a 1,009-amino-acid protein that contains two tandem LIM domains and a Rho-GAP domain (Fig. 1B). Rga1 and Rga2 share particularly high levels of identity in these regions. Rga2 also has sequence similarity to Bem3 over the GAP domain, but Bem3 does not contain LIM domains (Fig. 1B). Due to its high sequence similarity to Rga1 and, to a lesser extent, to Bem3, Rga2 was designated a putative GAP for Cdc42p.
Genetic support for Rga2 as a GAP for Cdc42.
To investigate the possibility that Rga2 is indeed a GAP for Cdc42, we first asked whether RGA2 showed the same genetic interactions with CDC42 and CDC24 as do RGA1 and BEM3 (63). We reasoned that an increase in GAP activity for Cdc42 should reduce the amount of active Cdc42 in the cell, thereby lowering the restrictive temperature of a temperature-sensitive CDC42 mutant. Consistent with this reasoning, overexpression of RGA2 lowered the restrictive temperature of a cdc42-1 strain (data not shown), as was observed for RGA1 and BEM3 (63). Conversely, we expected that a reduction in GAP activity would reduce the requirement for Cdc24 (GEF) activity. In accord with this expectation, deletion of RGA2 or BEM3 increased the restrictive temperature of a cdc24-H strain (data not shown), just as previously shown for RGA1 (63). Thus, genetic evidence supports the hypothesis that Rga2, as well as Rga1 and Bem3, are GAPs for Cdc42.
As a second test of the possibility that Rga2 is a GAP for Cdc42, we investigated the two-hybrid interactions between Rga2 and known GTPases of the Rho subfamily. Rga2 interacted with activated forms (G12V and Q61L) but not an inactive form (D118A) of Cdc42, as had previously been seen for Rga1 and Bem3 (63; Table 2). Rga2 failed to interact with Rho1, Rho2, Rho3, or Rho4, including activated versions of these proteins that could not be prenylated (data not shown). In addition, Rga1 and Rga2, but not Bem3, interacted with the GTPase, Rsr1, in both its wild-type and activated (G12V) forms (Table 2). This interaction pattern is distinct from the pattern seen for GTPases and their GAPs: GAPs usually interact preferentially with the active form of the GTPase. Rsr1 is a Ras-type GTPase involved in bud site selection and has been shown to interact with Cdc24 (49). Together, these experiments support the hypothesis that Rga1, Rga2, and Bem3 are all GAPs for Cdc42 and also suggest that Rga1 and Rga2 may have a connection with Rsr1.
TABLE 2.
Two-hybrid interactions of Rga1, Rga2, and Bem3, with Cdc42 and Rsr1a
| Plasmid | Level of interaction with products of:
|
|||
|---|---|---|---|---|
| Original plasmid | Rga1 hybrid | Rga2 hybrid | Bem3 hybrid | |
| pEG202 | 6 | 6 | 4 | 6 |
| pEG202-CDC42 | 1 | 2 | 1 | 1 |
| pEG202-CDC42C188S | 18 | 135 | 4 | 5 |
| pEG202-CDC42G12V,C188S | 143 | 2,576 | 1,327 | 1,303 |
| pEG202-CDC42Q6IL,C188S | 137 | 2,442 | 1,399 | 1,324 |
| pEG202-CDC42D118A,C188S | 1 | 1 | 1 | 1 |
| pEG202-RSR1 | 1 | 96 | 55 | 1 |
| pEG202-RSR1G12V | 1 | 84 | 56 | 1 |
| pJG4-5 | 9 | 1 | 1 | 1 |
| pJG4-5-CDC42 | 6 | 1 | 1 | 2 |
| pJG4-5-CDC42C188S | 3 | 1 | 1 | 1 |
| pJG4-5-CDC42G12V,C188S | 2 | 100 | 10 | 6 |
| pJG4-5-CDC42Q61L,C188S | 2 | 109 | 8 | 6 |
Products of plasmid pEG202 and its constructs interacted with products of pJG4-5 and its constructs and vice versa. β-Galactosidase activity was determined as described in Materials and Methods. The reported values are given in Miller units and are an average of three independent determinations.
Rga1 and Rga2 have in vitro biochemical GAP activity.
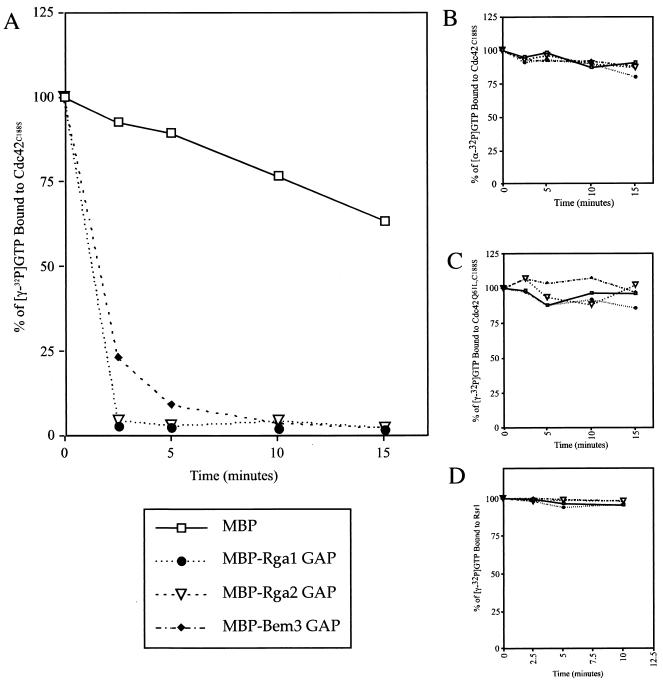
To expand on these genetic studies, we sought to determine whether Rga1 and Rga2 have biochemical GAP activity toward Cdc42; that is, we asked whether they accelerate GTP hydrolysis by Cdc42. A fusion protein containing the Bem3 GAP domain joined to GST was previously shown to catalyze the hydrolysis of GTP to GDP when the nucleotide was bound to Cdc42 (68). GST-Cdc42C188S was purified using glutathione agarose beads and bound to [γ-32P]GTP (see Materials and Methods), and the GAP domains of Rga1, Rga2, and Bem3 were purified as MBP fusions. When mixed with the [γ-32P]GTP-loaded GST-Cdc42C188S, all three GAP domains catalyzed the hydrolysis of GTP to GDP (Fig. 2A). After only 2.5 min, the GAP domains of Rga1 and Rga2 had catalyzed the hydrolysis of over 94% of the GTP on Cdc42 (summary of multiple data sets). In contrast, incubation with MBP alone resulted in hydrolysis of less than 20% of the GTP after 2.5 min. Incubation with the Bem3 GAP domain resulted in hydrolysis of ∼75% of the GTP bound to Cdc42 after 2.5 min. Thus, it appears that Rga1, Rga2, and Bem3 all catalyze GTP hydrolysis on Cdc42. However, Rga1 and Rga2 may do so more efficiently than Bem3.
FIG. 2.
GAP domains of Rga1, Rga2, and Bem3 display biochemical GAP activity for Cdc42, but not for Rsr1. The relative percentage of [γ-32P]GTP bound to GST-Cdc42C188S (A), [α-32P]GTP bound to GST-Cdc42C188S (B), [γ-32P]GTP bound to GST-Cdc42Q61L, C188S (C), and [γ-32P]GTP bound to GST-Rsr1 when incubated with ∼2 μg of purified MBP, MBP-Rga1 GAP, MBP-Rga2 GAP, or MBP-Bem3 GAP (D). The ratio of released counts (32Pi) to counts still bound to protein is plotted as a function of time. (A) Shown is a representative graph from multiple data sets.
To ensure that the loss of counts from the beads was due to GTP hydrolysis and not simply dissociation of the bound nucleoside triphosphate, purified Cdc42C188S was bound to [α-32P]GTP instead of [γ-32P]GTP and then incubated with the GAPs. If Rga1, Rga2, and Bem3 were to cause dissociation of GTP rather than hydrolysis to GDP, [α-32P]GTP would be released in this experiment, resulting in a loss of bound radioactivity. In fact, very little radioactivity was released, indicating that [α-32P]GTP did not dissociate from Cdc42C188S when incubated with the GAP domains of Rga1, Rga2, or Bem3 (Fig. 2B). The GAP assay was also performed with GST-Cdc42Q61L,C188S. Since the Q61L substitution locks Cdc42 in its GTP-bound, active state (70), this mutant should not be capable of GTP hydrolysis. Indeed, incubation with Rga1, Rga2, or Bem3 did not cause hydrolysis of [γ-32P]GTP when bound to Cdc42Q61L, C188S (Fig. 2C). These results show that the GAP domains of Rga1, Rga2, and Bem3 catalyze the hydrolysis of GTP by Cdc42.
Motivated by the two-hybrid results discussed above (Table 2), biochemical GAP assays were performed to ask whether the GAP domain of Rga1, Rga2, and Bem3 could accelerate GTP hydrolysis by the bud-site selection GTPase, Rsr1. The presence of the Rga1, Rga2, or Bem3 GAP domains did not catalyze the hydrolysis of GTP bound to GST-Rsr1 (Fig. 2D). Thus, Rga1, Rga2, and Bem3 are GAPs for Cdc42 and, based on two-hybrid and biochemical evidence, it seems unlikely that they act as GAPs for Rsr1, Rho1, Rho2, Rho3, or Rho4.
Deletion of GAPs for Cdc42 alters cell morphology.
Why are there three GAPs for Cdc42? One possibility is that Rga1, Rga2, and Bem3 are responsible for regulating distinct subsets of Cdc42 function. To explore this possibility, we examined the phenotypes conferred by deletion of RGA1, RGA2, and BEM3, singly and in combination. We first observed cellular morphology, which, for ease of quantification, was divided into five classes: normal, elongated, peanut, fingered, and misshapen. Elongated cells were defined as cells whose length is greater than 1.5 times but less than three times, their width. Peanut cells appeared as two round or elongated cells with a smooth connection between them, reminiscent of two newly fused mating cells. Cells of the fingered class had multiple growth projections or had a length greater than three times their width. Misshapen cells were defined as cells of aberrant morphology that did not fit in any of the other classes. As previously reported (63), some rga1Δ cells displayed an elongated cell morphology (40% versus 9% for wild type; Table 3). No morphological aberrations were observed for rga2Δ cells. Interestingly, a small percentage of the bem3Δ cells (8% versus <3% for the wild type; Table 3) displayed severe morphological defects (peanut, fingered, and misshapen cells). The combination of rga1Δ and bem3Δ deletions resulted in a high percentage (60%) of cells with severe morphological defects. Deletion of rga2Δ from the single and double mutants increased slightly the severity of cell morphology defects in all cases.
TABLE 3.
Quantification of cell morphologies for deletions of the GAPs for Cdc42a
| Strain | Genotype | % Normal | % Elongated | % Fingered | % Peanut | % Misshapen |
|---|---|---|---|---|---|---|
| SY2002 | Wild type | 90 | 9 | <1 | <1 | <1 |
| YGS2 | rga1Δ | 59 | 40 | <1 | <1 | <1 |
| YGS7 | rga2Δ | 90 | 9 | <1 | <1 | <1 |
| YGS50 | bem3Δ | 73 | 18 | 3 | 4 | 1 |
| YGS51 | rga1Δrga2Δ | 49 | 49 | <1 | <1 | <1 |
| YGS72 | rga1Δbem3Δ | 14 | 26 | 21 | 34 | 5 |
| YGS56 | rga2Δbem3Δ | 90 | 21 | 4 | 10 | 3 |
| YGS57 | rga1Δrga2Δbem3Δ | 13 | 14 | 16 | 51 | 4 |
All strains are in the SY2002 background. More than 400 cells were counted for each strain.
Despite the high percentage of severe morphologies observed in rga1Δ rga2Δ bem3Δ mutants, these cells were viable. However, the rga1Δ bem3Δ and rga1Δ rga2Δ bem3Δ deletion strains both showed a growth defect at 30°C when compared to wild-type and single mutant strains (data not shown). The viability of the triple mutant may indicate that there are additional proteins with GAP activity for Cdc42 or that the inherent GTPase activity of Cdc42 is sufficient for viability (27, 67).
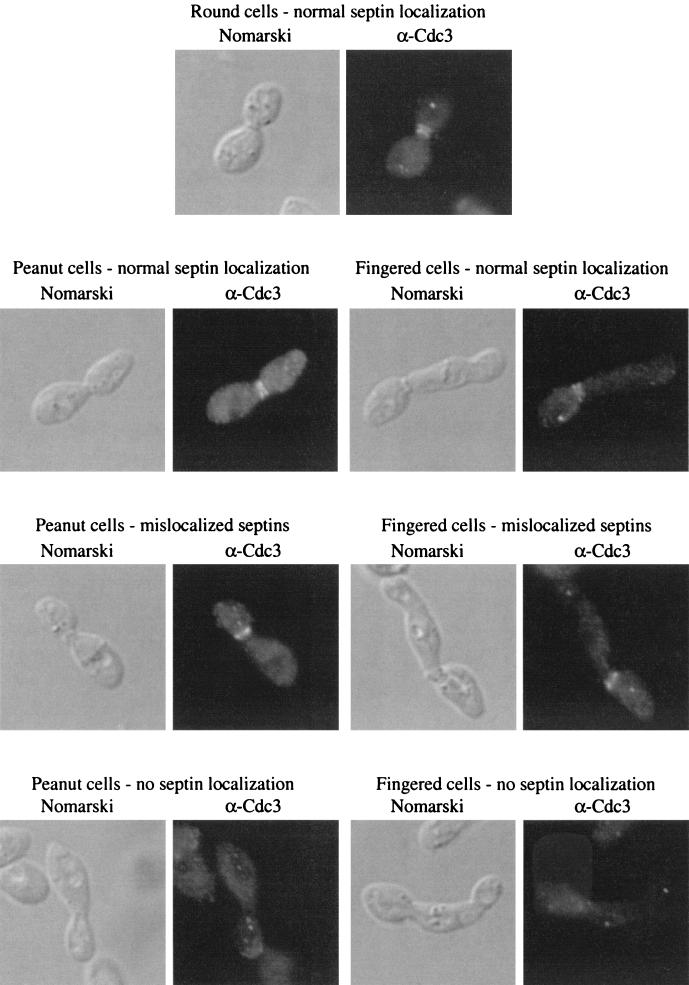
The localization of actin, chitin, and septin rings was examined in peanut and fingered cells. Such cells had defects in chitin localization as revealed by Calcofluor staining. In particular, these cells showed diffuse staining around the periphery in contrast to the rings of staining seen at previous bud sites in normal cells (data not shown). Immunofluorescence microscopy using the α-Cdc3 antibody to highlight septins revealed defects in the peanut and fingered cells (Fig. 3). Septins normally localize as a double ring at the bud neck (19; for review, see reference 21; Fig. 3). However, the septins rings were improperly localized in some peanut and fingered cells (Fig. 3). For cells with large buds, 43% of peanut cells and 68% of fingered cells failed to properly localize their septins rings, compared to only 4% of round cells in the same strain (Table 4). In most of the cells displaying mislocalized septins, a single septin ring had formed on one side of the bud neck. Because many of these cells had not yet undergone nuclear division, the mother cell could be identified by the presence of the only nucleus. In all cases, this single septin ring was located on the daughter side of the bud neck (data not shown). Finally, no defects in actin localization were detected in the GAP mutants (data not shown). Thus, peanut and fingered cells were found to have defects in the organization of septins (Fig. 3; Table 4) and chitin (data not shown), but not actin (data not shown).
FIG. 3.
Septin localization in normal, peanut, and fingered cells. Septins were visualized in the YGS57 (rga1Δ rga2Δ bem3Δ) strain. Nomarski optics was used to examine cell morphology and an α-Cdc3 antibody was used to reveal septins. Localization patterns are shown for round, peanut, and fingered cells. See Table 4 for quantitation.
TABLE 4.
Quantification of septin localizationa
| Strain | Cell type | % with normal localization | % with mislo- calization | % with no localization |
|---|---|---|---|---|
| SY2002 | Round | 99 | <1 | <1 |
| YGS57 | Round | 96 | 2 | 2 |
| YGS57 | Peanut | 57 | 34 | 9 |
| YGS57 | Fingered | 33 | 36 | 32 |
Septin staining was performed as described in the Materials and Methods. Only cells with large buds were counted. More than 200 cells were counted for each class.
Bud site selection defects in GAP deletion strains.
Wild-type haploid cells are round and bud in an axial pattern in which each new bud is adjacent to the site of the previous bud (10, 11). Using Calcofluor to visualize bud scars, we observed nonaxial budding patterns for rga1Δ cells. In toto, 78% of cells in a rga1Δ deletion strain displayed a nonaxial budding pattern (Table 5; see also reference 63): 51% of the rga1Δ cells had a bipolar pattern in which buds were seen at both poles of the cell and 27% of cells showed a random budding pattern with bud scars at either or both poles and in the middle of the cell (Table 5). In contrast, rga2Δ, bem3Δ, and rga2Δ bem3Δ cells exhibited an axial pattern with all of the bud scars at one pole (Table 5). The rga1Δ rga2Δ double mutant cells had a budding pattern similar to that of rga1Δ cells (Table 5). Budding patterns for rga1Δ bem3Δ and rga1Δ rga2Δ bem3Δ cells could not be accurately determined due to the aberrant cell morphologies and diffuse chitin staining (see above). Thus, Rga1 has a distinct function in bud site selection that is not shared by Rga2 or Bem3.
TABLE 5.
Budding patterns of GAP deletion strainsa
| Strain | Genotype | % Axial | % Bipolar | % Random |
|---|---|---|---|---|
| SY2002 | Wild type | 89 | 8 | 3 |
| YGS2 | rga1Δ | 22 | 51 | 27 |
| YGS7 | rga2Δ | 90 | 6 | 3 |
| YGS50 | bem3Δ | 90 | 7 | 3 |
| YGS51 | rga1Δrga2Δ | 14 | 64 | 22 |
| YGS72 | rga1Δbem3Δ | NDb | ND | ND |
| YGS56 | rga2Δbem3Δ | 94 | 4 | 3 |
| YGS57 | rga1Δrga2Δbem3Δ | ND | ND | ND |
Bud scars were visualized using Calcofluor as described in Materials and Methods. Only cells with three of more scars were counted. More than 200 cells were counted for each strain.
ND, not determined. Accurate counts could not be determined due to diffuse chitin staining and abnormal cell morphologies
Activation of FUS1-lacZ reporter in GAP deletion strains.
RGA1 was first identified in a screen for negative regulators of the pheromone response pathway (63). It was shown that ste4Δ rga1Δ cells activated a FUS1-lacZ reporter construct (63). We determined that deletion of RGA2 or BEM3 did not cause activation of the FUS1-lacZ reporter (Table 6). Thus, the rga1Δ deletion appears to be unique in its ability to affect FUS1-lacZ expression. Deletion of RGA2 in a rga1Δ strain caused a slight increase in FUS1-lacZ expression (38.9 for ste4Δ rga1Δ versus 45.5 for ste4Δ rga1Δ rga2Δ). However, deletion of all three GAP genes caused an eightfold increase in expression (Table 6). Thus, although Rga1 has a primary role in controlling the activation of FUS1-lacZ, Rga2, and Bem3 must also have a negative regulatory role, albeit a modest one.
TABLE 6.
Activation of FUS1-lacZ by deletion of GAPs for Cdc42
| Strain | Genotypea | FUS1-lacZ expressionb |
|---|---|---|
| YGS58 | ste4Δ | 1.2 ± 0.1 |
| YGS59 | ste4Δrga1Δ | 38.9 ± 2.5 |
| YGS60 | ste4Δrga2Δ | 1.8 ± 0.5 |
| YGS61 | ste4Δbem3Δ | 1.3 ± 0.1 |
| YGS62 | ste4Δrga1Δrga2Δ | 45.5 ± 2.8 |
| YGS63 | ste4Δrga1Δbem3Δ | 30.3 ± 3.7 |
| YGS64 | ste4Δrga2Δbem3Δ | 3.9 ± 1.2 |
| YGS65 | ste4Δrga1Δrga2Δbem3Δ | 396.1 ± 20.5 |
All strains are in the SY2002 background.
β-Galactosidase activity was determined as described in Materials and Methods. The reported values are given in 100× Miller units and are averages (± standard deviation) of three determinations.
Roles of Rga1, Rga2, and Bem3 in haploid invasive growth.
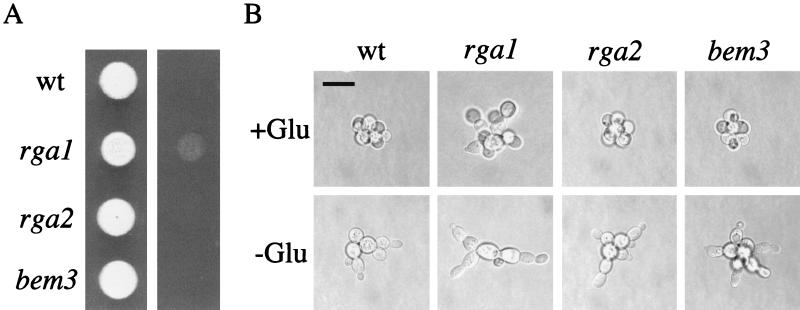
We also examined the roles of Rga1, Rga2, and Bem3 in haploid invasive growth, a process involving Cdc42 (46). These experiments were done with the Σ1278b strain (provided by G. Fink), a strain that exhibits more vigorous agar invasion than other lab strains. Two techniques were used to assess filamentous growth: the plate-washing assay (56), which assesses agar invasion, and the single-cell assay (16), which provides detailed information on cell morphology and budding patterns. By the plate-washing assay, rga1Δ cells displayed hyperinvasive growth, whereas rga2Δ and bem3Δ cells invaded the agar at a level similar to wild-type cells (Fig. 4). On glucose-rich medium, rga1 mutant cells showed a propensity to bud at the distal pole (17%), whereas bem3 (2% distal), rga2 (1%), and wild-type (1%) cells did not (Fig. 4b). The rga1 mutant also had an elongated cell morphology in glucose-rich medium; 45% of the cells were elongated compared to less than 1% for wild-type and the rga2 mutant. The bem3 mutant also had elongated cells (18%) on this medium, but the cells were not as elongated as rga1 cells. On glucose-limited medium, rga1 cells were hyperelongated compared to wild-type cells or rga2 and bem3 mutants, and the rga1 cells displayed a more exaggerated unipolar budding pattern. Thus, among the GAPs, Rga1 appears to play a unique role in regulating haploid invasive growth.
FIG. 4.
Haploid invasive growth phenotypes of strains lacking the Cdc42 GAPs. (A) Cells of the Σ1278b background were grown for 4 days at 30°C on synthetic medium. Plates were washed with water and rubbed with a wet finger. (B) Single-cell invasive growth assay. Equal concentrations of wild-type (wt), rga1, rga2, and bem3 cells were spread onto SCD (+Glu) or SC (−Glu) medium, incubated for 16 h at 25°C, and photographed at 100× by placing a coverslip directly on the agar medium. All images were taken at the same scale. Bar, 30 μm.
Genetic interactions between GAP proteins and Cdc42 effectors.
The data presented above suggest that the GAPs for Cdc42 play unique roles in septin organization (Bem3) and haploid invasive growth (Rga1). The p21-activated kinase (PAK) kinases Ste20 and Cla4 both interact with Cdc42 and are known to be important for these processes (17, 61). Ste20 is required for haploid invasive growth (56) whereas Cla4 influences septin organization (4, 17). Furthermore, ste20Δ and cla4Δ deletions are synthetically lethal, implying that these kinases share an essential activity (4, 17). It seemed plausible that these PAK kinases could link the GAPs for Cdc42 to the processes they mediate.
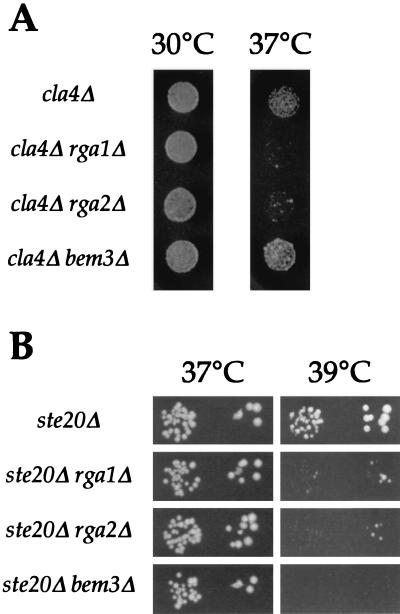
If the GAPs for Cdc42 facilitate the interaction between active Cdc42 and Ste20 or Cla4, the GAPs might exhibit genetic interactions with the protein kinases. We examined the effect of rga1Δ, rga2Δ, and bem3Δ deletions on ste20Δ or cla4Δ strains. The rga1Δ cla4Δ and rga2Δ cla4Δ strains displayed synthetic temperature sensitivity at 37°C (Fig. 5A). The cells arrested with fattened bud necks, and some cells had multiple projections emanating from the cell body (data not shown). In contrast, cells of the cla4Δ strain, the bem3Δ cla4Δ strain, and all GAP single deletions grew well at this temperature (Fig. 5A; data not shown). Since Rga1, Rga2, and Ste20 each displayed synthetic interactions with cla4Δ deletions, we hypothesize that they may play roles in the same cellular function. Moreover, because BEM3 did not show genetic interactions with cla4Δ, we infer that the GAPs can have specific functions with respect to effectors for Cdc42. However, we cannot rule out the possibility that these phenotypic differences reflect quantitative rather than qualitative differences in GAP activity. RGA1, RGA2, and BEM3 also showed genetic interactions with ste20Δ. In this case, loss of any individual GAP for Cdc42 resulted in synthetic temperature sensitivity, though the effect may be more pronounced for bem3Δ (Fig. 5B). These findings suggest that the GAPs may coordinate Cla4 activity, perhaps via Cdc42.
FIG. 5.
Temperature-dependant growth of strains lacking a PAK kinase and one of the Cdc42 GAPs. (A) Aliquots of 10 μl of a 1:100 dilution of IDY22 (cla4Δ), YGS80 (rga1Δ cla4Δ), YGS81 (rga2Δ cla4Δ), or YGS82 (bem3Δ cla4Δ) mid-log phase liquid cultures were spotted onto rich plates and incubated at the indicated temperatures. (B) Aliquots of 10 μl of a 1:1,000 and a 1:10,000 dilution of YGS 286 (ste20Δ), YGS 351 (rga1Δ ste20Δ), YGS 352 (rga2Δ ste20Δ), and YGS 353 (bem3Δ ste20Δ) mid-log phase liquid cultures were spotted onto rich plates and incubated at the indicated temperatures.
Deletion or overexpression of RGA1, RGA2, and BEM3 affects interaction of Cdc42 and Ste20.
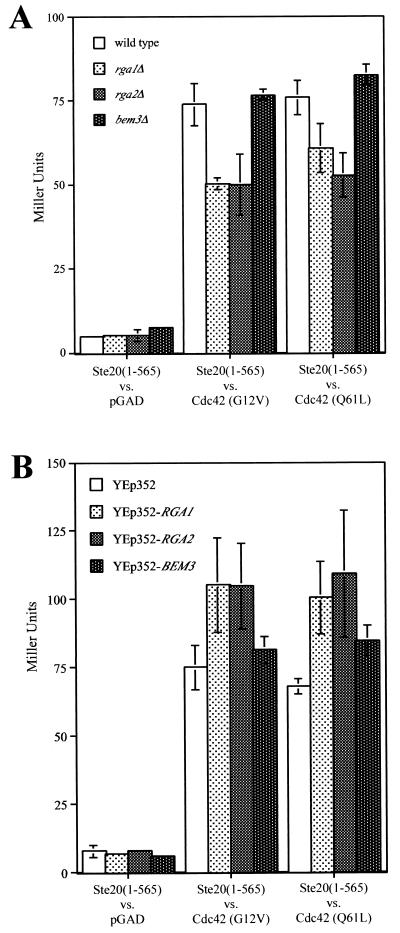
To explore further the hypothesis that Rga1, Rga2, and Bem3 play differential roles in regulating Cdc42 function, we investigated the effects of their deletion or overexpression on the two-hybrid interaction between Cdc42 and Ste20. Specifically, the interaction between two activated forms of Cdc42 (G12V, C188S and Q61L, C188S) and a truncated form of Ste20 (provided by M. Keniry) was compared under a variety of conditions. The interaction was measured for four different strains: PJ69-4A, YGS156 (PJ69-4A rga1Δ), YGS157 (PJ69-4A rga2Δ), and YGS158 (PJ69-4A bem3Δ) (Fig. 6A). Deletion of RGA1 or RGA2 decreased expression of the lacZ reporter, suggesting a decrease in the interaction between Ste20 and Cdc42. Loss of BEM3 had no significant effect. This experiment suggests that Rga1 and Rga2 may facilitate the interaction between Ste20 and active Cdc42. In a complementary experiment, the interaction between Ste20 and active Cdc42 was compared in strains overexpressing the GAPs for Cdc42. The PJ69-4A strain was transformed with YEp352, pGS40 (YEp352-RGA1), pGS41 (YEp352-RGA2), or pGS42 (YEp352-BEM3). Overexpression of RGA1 or RGA2 increased the expression of the lacZ reporter, suggesting an increased interaction between Ste20 and the activated forms of Cdc42 (Fig. 6B). Together, these data suggest that Rga1 and Rga2 facilitate the two-hybrid interaction between Ste20 and Cdc42.
FIG. 6.
Effects of deletion or overexpression of Cdc42 GAPs on two-hybrid interaction between Ste20 and activated versions of Cdc42. (A) The interaction of Ste20 residues 1 to 565 fused to the Gal4 DBD (pSL2682) with three versions of the Gal4 transcription activation domain—GAD itself and GAD fused either to Cdc42G12V,C188S (pGS38) or to Cdc42Q61L,C188S (pGS39)—in four separate strains were compared: PJ69-4A, YGS156 (rga1Δ), YGS157 (rga2Δ), and YGS158 (bem3Δ). Expression of the lacZ reporter in three separate isolates was measured. (B) The same two-hybrid interactions investigated above were compared for strain PJ69-4A transformed with four different plasmids: YEp352, pGS40 (YEp352-RGA1), pGS41 (YEp352-RGA2), and pGS42 (YEp352-BEM3). The expression of the lacZ reporter was measured for three separate isolates in four independent trials. The combined normalized data were graphed.
DISCUSSION
We have presented evidence that there are three distinct GAPs for Cdc42: Rga1, Rga2, and Bem3. Each of these proteins exhibited two-hybrid interactions with Cdc42, and genetic experiments argued that their activity influences the amount of active Cdc42 in the cell. Moreover, each protein was shown to have GAP activity; that is, the ability to accelerate the hydrolysis of GTP bound to Cdc42. The loss of individual GAPs conferred distinct phenotypes, suggesting that each GAP regulates a subset of Cdc42 functions. Hence, these studies complement and extend previous studies that demonstrated that Bem3 has biochemical GAP activity against Cdc42 (68) and that suggested that Rga1 was a Cdc42 GAP (63).
Bem3 and, to a lesser extent, Rga1 and Rga2, play roles in septin organization.
Phenotypic analysis suggests that Bem3 may moderate the function of Cdc42 in morphogenesis. This conclusion is supported by the observation that bem3Δ strains produce peanut and fingered cells whereas rga1Δ and rga2Δ strains do not (Table 3). However, the double and triple deletions, especially the rga1Δ bem3Δ and rga1Δ rga2Δ bem3Δ strains, produce many more peanut and fingered cells than the single deletions, suggesting that Rga1 and Rga2 play a lesser role in the formation of these morphologies. Cells exhibiting the peanut and fingered morphologies were shown to have mislocalized septin rings and occasionally display no septin localization, thus linking loss of BEM3 and, to a lesser extent, RGA1 and RGA2, to septin ring organization.
The finding that ste20Δ rga1Δ, ste20Δ rga2Δ, and ste20Δ bem3Δ cells display synthetic temperature sensitivity (Fig. 5B) also supports the idea that Bem3, Rga1, and Rga2 play roles in septin organization and/or bud neck morphogenesis. This suggestion is based on two pieces of information: Cla4 is involved in the process of bud neck morphogenesis (4, 9, 41) and cla4Δ and ste20Δ are synthetically lethal (17). Thus, proteins that participate in the same process as Cla4 might also display synthetic interactions with ste20Δ, as seen for deletions of RGA1, RGA2, and BEM3. The bem3Δ ste20Δ strain may have a slightly greater defect than the rga1Δ ste20Δ or rga2Δ ste20Δ strains (see Fig. 5B), suggesting that Bem3 may play a more central role in mediating the function of Cdc42 and Cla4 in cytokinesis.
Unpublished results from E. Bi and J. Pringle also support the hypothesis that Rga1, Rga2, and Bem3 play roles in septin organization. They showed the level of expression of Cdc42 GAPs affects the temperature sensitivity of a cdc12-6 strain. Cdc12 is a component of the septin ring (41). Overexpression of CLA4, RGA1, RGA2, or BEM3, but not STE20, raises the restrictive temperature of a cdc12-6 strain. In addition, deletion of any of the GAPs for Cdc42, but particularly bem3Δ, lowers the restrictive temperature of a cdc12-6 strain (E. Bi and J. R. Pringle, unpublished data). Thus, GAP function for Cdc42 appears to play a positive role in septin organization. Furthermore, immunolocalization showed that Rga1, Rga2, and Bem3 localize to the bud neck during late anaphase and early telophase (E. Bi and J. R. Pringle, unpublished data), consistent with a function in septin ring organization.
Finally, mutational analysis of Cdc42 also supports the hypothesis that Bem3 mediates the interaction of Cdc42 with Cla4. Point mutations in the effector domain of Cdc42 lead to a variety of phenotypes, often including abnormal cell morphologies and mislocalization of actin, chitin, and/or septins (37, 54, 55). Furthermore, some of these Cdc42 alleles display altered interactions with Cla4 and Bem3, but interactions with Rga1, Rga2, and Ste20 are not affected (54; T. J. Richman and D. I. Johnson, unpublished data). Thus, mutational analyses provide additional support to link Bem3 and Cla4 in septin ring organization.
Role of Rga1 and Rga2 in haploid invasive growth and in regulating interaction between Cdc42 and Ste20.
The phenotypes of the rga1Δ deletion strain imply a role for Rga1 in haploid invasive growth. In particular, loss of Rga1 causes cells to display hyperinvasion, to elongate, and to exhibit a nonaxial budding pattern, each a characteristic of invasive growth (43). In addition, activation of the FUS1-lacZ reporter, previously thought to be specific for pheromone response activation (63), may actually reflect activation of the Ste12/Tec1 transcription factor complex, another characteristic of haploid invasive growth (15, 48). This explanation is supported by the observation that deletion of the pheromone response pathway scaffold, Ste5, which has no role in invasive growth but is important for pheromone response, does not affect the FUS1-lacZ activation in an rga1Δ strain (63). Moreover, diminished interaction between Ste20 and active Cdc42 had little effect on pheromone response pathway signaling and mating but did result in decreased invasive growth (39, 50). Together, these findings support the idea that Rga1 affects haploid invasive growth, possibly by mediating the interaction of active Cdc42 with Ste20.
A role for Rga1 in mediating the interaction between Cdc42 and Ste20 is also supported by the synthetic temperature sensitivity of rga1Δ and cla4Δ mutations (Fig. 5). Since cla4Δ and ste20Δ are synthetically lethal (17), it would follow that a protein involved in the same process as Ste20 might also display synthetic interactions with cla4Δ. Interestingly, rga2Δ cla4Δ cells, but not bem3Δ cla4Δ cells, also display synthetic temperature sensitivity, suggesting that Rga2 might also play a role in mediating the interaction between Cdc42 and Ste20. These phenotypic differences may reflect different quantitative contributions to overall GAP activity by Rga1, Rga2, and Bem3. However, the phenotypic differences could also indicate that the GAPs have qualitatively different roles in orchestrating Cdc42 activity. Consistent with the proposed role of Rga1 and Rga2 as facilitators of the interaction between Cdc42 and Ste20, deletion of RGA1 or RGA2 decreased the two-hybrid interaction between activated forms of Cdc42 with Ste20, whereas RGA1 or RGA2 overexpression increased this interaction. These experiments suggest that Rga1 and Rga2 facilitate the interaction between Cdc42 and Ste20. This function would appear to be distinct from the GAP activity of Rga1 and Rga2 since these two-hybrid experiments were performed with versions of Cdc42 locked in the GTP-bound, active form (70). How do Rga1 and Rga2 facilitate this interaction? Given that Rga1 and Rga2 have not been observed to interact with Ste20 by two-hybrid analysis (63; data not shown), perhaps these two GAPs do not directly promote an interaction between Ste20 and Cdc42. Rather, perhaps Rga1 and Rga2 prevent Cdc42 from interacting with other effectors or perhaps they recruit some unidentified protein that aids in this process.
Acknowledgments
We thank April Goehring, Megan Keniry, Hay-Oak Park, Phil James, Brian Stevenson, Betsy Ferguson, Dave Mitchell, Erica Golemis, Gerald Fink, and Doug Johnson for providing advice, strains, and/or plasmids. Thanks to Erfei Bi, John Pringle, Tammy Richman, and Doug Johnson for sharing results before publication. We also thank David Rivers, Hilary Kemp, Jesse Dillon, and Phil Kinsey for helpful comments and suggestions.
This work was supported by research (GM-30027 to G.F.S) and training (GM-07413 to G.R.S., HD-07348 to S.A.G., and GM-19888 to P.C.) grants from the U.S. Public Health Service and by a fellowship from the American Heart Association (AHA1206352) to P.C.
REFERENCES
- 1.Adams, A. E., D. I. Johnson, R. M. Longnecker, B. F. Sloat, and J. R. Pringle. 1990. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111:131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, V. E., J. Breton, I. Sanchez-Garcia, H. Osada, A. Forster, A. J. Thomson, and T. H. Rabbitts. 1994. Cysteine-rich LIM domains of LIM-homeodomain and LIM-only proteins contain zinc but not iron. Proc. Natl. Acad. Sci. USA 91:316-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthe, C., G. de Bettignies, O. Louvet, M. F. Peypouquet, C. Morel, F. Doignon, and M. Crouzet. 1998. First characterization of the gene RGD1 in the yeast Saccharomyces cerevisiae. C. R. Acad. Sci. Ser. III 321:453-462. [DOI] [PubMed] [Google Scholar]
- 4.Benton, B. K., A. Tinkelenberg, I. Gonzalez, and F. R. Cross. 1997. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol. Cell. Biol. 17:5067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi, E., J. B. Chiavetta, H. Chen, G. C. Chen, C. S. Chan, and J. R. Pringle. 2000. Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol. Biol. Cell. 11:773-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boone, C., N. G. Davis, and G. F. Sprague, Jr. 1993. Mutations that alter the third cytoplasmic loop of the a-factor receptor lead to a constitutive and hypersensitive phenotype. Proc. Natl. Acad. Sci. USA 90:9921-9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, J. L., M. Jaquenoud, M. P. Gulli, J. Chant, and M. Peter. 1997. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 11:2972-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 9.Carroll, C. W., R. Altman, D. Schieltz, J. R. Yates, and D. Kellogg. 1998. The septins are required for the mitosis-specific activation of the Gin4 kinase. J. Cell Biol. 143:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chant, J., and I. Herskowitz. 1991. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65:1203-1212. [DOI] [PubMed] [Google Scholar]
- 11.Chant, J., M. Mischke, E. Mitchell, I. Herskowitz, and J. R. Pringle. 1995. Role of Bud3p in producing the axial budding pattern of yeast. J. Cell Biol. 129:767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 13.Chen, G. C., Y. J. Kim, and C. S. Chan. 1997. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 11:2958-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, G. C., L. Zheng, and C. S. Chan. 1996. The LIM domain-containing Dbm1 GTPase-activating protein is required for normal cellular morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:1376-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cullen, P. J., J. Schultz, J. Horecka, B. J. Stevenson, Y. Jigami, and G. F. Sprague, Jr. 2000. Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics 155:1005-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cullen, P. J., and G. F. Sprague, Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cvrckova, F., C. De Virgilio, E. Manser, J. R. Pringle, and K. Nasmyth. 1995. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9:1817-1830. [DOI] [PubMed] [Google Scholar]
- 18.Dawid, I. B., J. J. Breen, and R. Toyama. 1998. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14:156-162. [DOI] [PubMed] [Google Scholar]
- 19.DeMarini, D. J., A. E. Adams, H. Fares, C. De Virgilio, G. Valle, J. S. Chuang, and J. R. Pringle. 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doignon, F., C. Weinachter, O. Roumanie, and M. Crouzet. 1999. The yeast Rgd1p is a GTPase activating protein of the Rho3 and Rho4 proteins. FEBS Lett. 459:458-462. [DOI] [PubMed] [Google Scholar]
- 21.Field, C. M., and D. Kellogg. 1999. Septins: cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 9:387-394. [DOI] [PubMed] [Google Scholar]
- 22.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 23.Freyd, G., S. K. Kim, and H. R. Horvitz. 1990. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature 344:876-879. [DOI] [PubMed] [Google Scholar]
- 24.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 25.Guan, C., P. Li, P. D. Riggs, and H. Inouye. 1987. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene 67:21-30. [DOI] [PubMed] [Google Scholar]
- 26.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 27.Hart, M. J., K. Shinjo, A. Hall, T. Evans, and R. A. Cerione. 1991. Identification of the human platelet GTPase activating protein for the CDC42Hs protein. J. Biol. Chem. 266:20840-20848. [PubMed] [Google Scholar]
- 28.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 29.Ijzerman, M. M., J. O. Falkinham III, and C. Hagedorn. 1993. A liquid, colorimetric presence-absence coliphage detection method. J. Virol. Methods 45:229-233. [DOI] [PubMed] [Google Scholar]
- 30.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarvis, E. E., D. C. Hagen, and G. F. Sprague, Jr. 1988. Identification of a DNA segment that is necessary and sufficient for α-specific gene control in Saccharomyces cerevisiae: implications for regulation of α-specific and a-specific genes. Mol. Cell. Biol. 8:309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, D. I. 1999. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63:54-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, D. I., C. W. Jacobs, J. R. Pringle, L. C. Robinson, G. F. Carle, and M. V. Olson. 1987. Mapping of the Saccharomyces cerevisiae CDC3, CDC25, and CDC42 genes to chromosome XII by chromosome blotting and tetrad analysis. Yeast 3:243-253. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, D. I., and J. R. Pringle. 1990. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J. Cell Biol. 111:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karlsson, O., S. Thor, T. Norberg, H. Ohlsson, and T. Edlund. 1990. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature 344:879-882. [DOI] [PubMed] [Google Scholar]
- 36.Kim, H. B., B. K. Haarer, and J. R. Pringle. 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol. 112:535-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozminski, K. G., A. J. Chen, A. A. Rodal, and D. G. Drubin. 2000. Functions and functional domains of the GTPase Cdc42p. Mol. Biol. Cell 11:339-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leberer, E., D. Dignard, D. Harcus, D. Y. Thomas, and M. Whiteway. 1992. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 11:4815-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leberer, E., C. Wu, T. Leeuw, A. Fourest-Lieuvin, J. E. Segall, and D. Y. Thomas. 1997. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 16:83-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, X., H. Wang, M. Eberstadt, A. Schnuchel, E. T. Olejniczak, R. P. Meadows, J. M. Schkeryantz, D. A. Janowick, J. E. Harlan, E. A. Harris, D. E. Staunton, and S. W. Fesik. 1998. NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell 95:269-277. [DOI] [PubMed] [Google Scholar]
- 41.Longtine, M. S., H. Fares, and J. R. Pringle. 1998. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 143:719-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma, H., S. Kunes, P. J. Schatz, and D. Botstein. 1987. Plasmid construction by homologous recombination in yeast. Gene 58:201-216. [DOI] [PubMed] [Google Scholar]
- 43.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 44.Maina, C. V., P. D. Riggs, A. G. I. Grandea, B. E. Slatko, L. S. Moran, J. A. Tagliamonte, L. A. McReynolds, and C. Guan. 1988. A vector to express and purify foreign proteins in Escherichia coli by fusion to, and separation form, maltose-binding protein. Gene 74:365-373. [DOI] [PubMed] [Google Scholar]
- 45.Michelsen, J. W., K. L. Schmeichel, M. C. Beckerle, and D. R. Winge. 1993. The LIM motif defines a specific zinc-binding protein domain. Proc. Natl. Acad. Sci. USA 90:4404-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Miller, P. J., and D. I. Johnson. 1997. Characterization of the Saccharomyces cerevvisiae cdc42-1 allele and new temperature-conditional-lethal cdc42 alleles. Yeast 13:561-572. [DOI] [PubMed] [Google Scholar]
- 46.Mosch, H. U., T. Kohler, and G. H. Braus. 2001. Different domains of the essential GTPase Cdc42p required for growth and development of Saccharomyces cerevisiae. Mol. Cell. Biol. 21:235-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nilsson, B., L. Abrahmsen, and M. Uhlen. 1985. Immobilization and purification of enzymes with staphylococcal protein A gene fusion vectors. EMBO J. 4:1075-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park, H. O., E. Bi, J. R. Pringle, and I. Herskowitz. 1997. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl. Acad. Sci. USA 94:4463-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peter, M., A. M. Neiman, H. O. Park, M. van Lohuizen, and I. Herskowitz. 1996. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 15:7046-7059. [PMC free article] [PubMed] [Google Scholar]
- 51.Pringle, J. R., R. A. Preston, A. E. Adams, T. Stearns, D. G. Drubin, B. K. Haarer, and E. W. Jones. 1989. Fluorescence microscopy methods for yeast. Methods Cell Biol. 31:357-435. [DOI] [PubMed] [Google Scholar]
- 52.Printen, J. A., and G. F. Sprague, Jr. 1994. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138:609-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramer, S. W., and R. W. Davis. 1993. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richman, T. J., and D. I. Johnson. 2000. Saccharomyces cerevisiae Cdc42p GTPase is involved in preventing the recurrence of bud emergence during the cell cycle. Mol. Cell. Biol. 20:8548-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richman, T. J., M. M. Sawyer, and D. I. Johnson. 1999. The Cdc42p GTPase is involved in a G2/M morphogenetic checkpoint regulating the apical-isotropic switch and nuclear division in yeast. J. Biol. Chem. 274:16861-16870. [DOI] [PubMed] [Google Scholar]
- 56.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 57.Rose, A. B., and J. R. Broach. 1990. Propagation and expression of cloned genes in yeast: 2-microns circle-based vectors. Methods Enzymol. 185:234-279. [DOI] [PubMed] [Google Scholar]
- 58.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Schmidt, A., M. Bickle, T. Beck, and M. N. Hall. 1997. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88:531-542. [DOI] [PubMed] [Google Scholar]
- 61.Simon, M. N., C. De Virgilio, B. Souza, J. R. Pringle, A. Abo, and S. I. Reed. 1995. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature 376:702-705. [DOI] [PubMed] [Google Scholar]
- 62.Sloat, B. F., A. Adams, and J. R. Pringle. 1981. Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 89:395-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stevenson, B. J., B. Ferguson, C. De Virgilio, E. Bi, J. R. Pringle, G. Ammerer, and G. F. Sprague, Jr. 1995. Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes Dev. 9:2949-2963. [DOI] [PubMed] [Google Scholar]
- 64.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe, D., M. Abe, and Y. Ohya. 2001. Yeast Lrg1p acts as a specialized RhoGAP regulating 1,3-beta-glucan synthesis. Yeast 18:943-951. [DOI] [PubMed] [Google Scholar]
- 66.Way, J. C., and M. Chalfie. 1988. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell 54:5-16. [DOI] [PubMed] [Google Scholar]
- 67.Zheng, Y., R. Cerione, and A. Bender. 1994. Control of the yeast bud-site assembly GTPase Cdc42. J. Biol. Chem. 269:2369-2372. [PubMed] [Google Scholar]
- 68.Zheng, Y., M. J. Hart, K. Shinjo, T. Evans, A. Bender, and R. A. Cerione. 1993. Biochemical comparisons of the Saccharomyces cerevisiae Bem2 and Bem3 proteins. Delineation of a limit Cdc42 GTPase-activating protein domain. J. Biol. Chem. 268:24629-24634. [PubMed] [Google Scholar]
- 69.Ziman, M., and D. I. Johnson. 1994. Genetic evidence for a functional interaction between Saccharomyces cerevisiae CDC24 and CDC42. Yeast 10:463-474. [DOI] [PubMed] [Google Scholar]
- 70.Ziman, M., J. M. O'Brien, L. A. Ouellette, W. R. Church, and D. I. Johnson. 1991. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol. Cell. Biol. 11:3537-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]