Abstract
The genomic evolution and causes of phenotypic variation among humans and great apes remain largely unknown, although the phylogenetic relationships among them have been extensively explored. Previous studies that focus on differences at the amino acid and nucleotide sequence levels have revealed a high degree of similarity between humans and chimpanzees, suggesting that other types of genomic change may have contributed to the relatively large phenotypic differences between them. For example, the activity of long interspersed element 1 (LINE-1) retrotransposons may impose significant changes on genomic structure and function and, consequently, on phenotype. Here we investigate the relative rates of LINE-1 amplification in the lineages leading to humans, bonobos (Pan paniscus), and chimpanzees (P. troglodytes). Our data indicate that LINE-1 insertions have accumulated at significantly greater rates in bonobos and chimpanzees than in humans, provide insights into the timing of major LINE-1 amplification events during great ape evolution, and identify a Pan-specific LINE-1 subfamily.
Non–long-terminal repeat transposable elements are thought to be ubiquitous in sexually reproducing eukaryotes (Arkhipova and Meselson 2000) and have served as an important driving force in genomic evolution for >600 million years (Malik et al. 1999). Long interspersed elements (LINEs) constitute >20% of the human genome and, through the mobilization of short interspersed elements (SINEs) and 3′ flanking DNAs, have created >40% of human DNA (Holmes et al. 1994; Goodier et al. 2000; Pickeral et al. 2000; Lander et al. 2001). LINE-1 (L1) retrotransposons are the youngest LINEs in mammalian genomes and include the only actively mobile LINEs in modern humans (Lander et al. 2001). Previous investigations into the role of L1 biology in shaping the human genome have been restricted to studies of human genetic sequence.
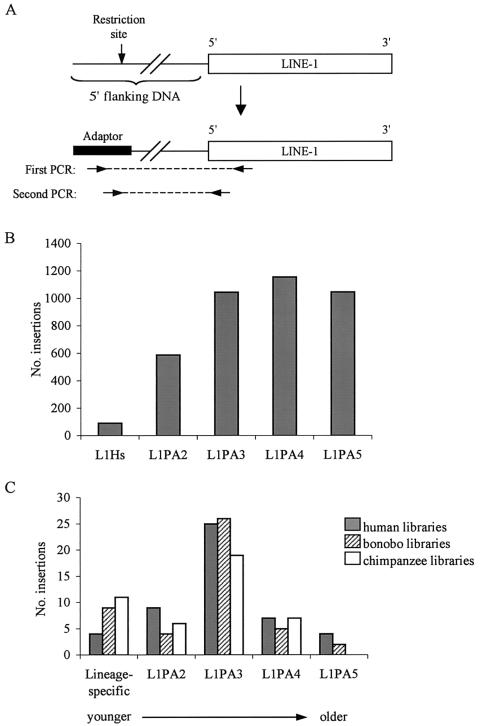
To gain broader insights into the role of L1s in great ape genomic evolution, we developed a molecular procedure to sample L1s that amplified during the period of great ape speciation from the genomes of humans (Homo sapiens), common chimpanzees (Pan troglodytes), and bonobos (P. paniscus). The L1 subfamilies of primary interest in this study are designated “L1Hs” and “L1PA2–L1PA5,” because these are the subfamilies that amplified during the period of ape evolution. L1Hs elements inserted after the divergence of the Homo and Pan lineages and therefore are present only in the genomes of humans. L1PA2–L1PA5 elements are older, with increasing subfamily number reflecting increasing age of the subfamily (Smit et al. 1995). The method, which is described in figure 1A, is designed to construct unbiased libraries of recent L1s, each possessing an intact 5′ end and a variable length of 5′ flanking DNA. The sequences of the flanking DNAs serve as tags for the identification of the insertion target sites in the draft human genome sequence, as determined by BLAST (Altschul et al. 1990) or BLAT (Kent 2002) searches. Two methods were used to ensure that the libraries sampled recent L1 insertions. First, we designed the primers to take advantage of an A→T mutation present in all L1 insertions younger than L1PA4 at position 95 of L1 retrotransposable element 1 (LRE1) (authors' unpublished observation), an active human retrotransposon (Dombroski et al. 1991). Second, insertions that amplified during recent human evolution have a higher fraction of full-length elements than do older L1s (Boissinot et al. 2000; Ovchinnikov et al. 2002). Consequently, libraries containing L1s with intact 5′ ends are enriched for younger insertions. We hereafter refer to these as “LOAF” libraries (LINE one and flanking DNA).
Figure 1.
LOAF library construction and characterization. A, Previously described method of PCR genome walking (Siebert et al. 1995), adapted for the construction of the LOAF libraries. Genomic DNA was digested and ligated overnight to the Genome Walker adaptor (Clontech). Internal L1 primers were designed by aligning bases 1–500 of 32 full-length primate-specific L1 insertions (not shown). Regions near the 5′ end that were highly conserved among the sequences were selected for primer hybridization. First-round PCR analyses were performed with adaptor primer 1 (Clontech) and an internal L1 primer. Second round PCR analyses were performed with adaptor primer 2 (Clontech) and a nested internal L1 primer. Primer sequences are available from the authors upon request. One human library was made by use of the EcoRV-ligated human DNA supplied by the manufacturer, and four libraries each (DraI, EcoRV, PvuII, and StuI) were constructed from a human Pygmy, a chimpanzee, and a bonobo. Products of the secondary PCRs were ligated into the pCR 2.1 Topo vector (Invitrogen); individual colonies were cloned and sequenced. Arrows denote PCR primers, and dashed lines indicate the PCR products. B, Distribution of near–full-length L1 insertions among the L1Hs–L1PA5 subfamilies on five human chromosomes. RepeatMasker data for the L1 insertions on human chromosomes 1, 5, 12, 15, and 22 were downloaded from the UCSC database. The number of L1s belonging to subfamilies L1Hs–L1PA5 was determined for insertions that were >5,500 bp and contained a complete 3′ UTR. Elements that were 5′ truncated beyond bp 67 of the LRE1 sequence were eliminated from consideration, because these insertions would not be amplified by the LOAF primers and therefore would be absent from the libraries. C, Subfamily distribution of near–full-length L1s isolated from the human, bonobo, and chimpanzee LOAF libraries. “Lineage-specific” refers to L1 elements that inserted after the divergence of the Homo (i.e., insertion present only in humans) and the Pan lineages (i.e., insertion present only in bonobos and/or chimpanzees).
Characterization of the LOAF libraries confirmed that they lack both age and position bias and therefore accurately represent the recent L1 insertions found in the Homo and Pan genomes. All of the 56 human, 58 bonobo, and 65 chimpanzee clones that were isolated from the libraries and sequenced contained an L1 5′ end, as predicted. For most of the clones (53 human, 49 bonobo, and 49 chimpanzee), the insertion target sites could be identified in the human genome databases. We completed the database searches at a time when 98% of the euchromatic human genome was represented in GenBank. Thus, the fraction of human clones for which a target site could be identified in GenBank (53/56 [95%]) corresponded well with expectations. The insertion sites for the human, bonobo, and chimpanzee clones were distributed on a broad range of human chromosomes (table 1), indicating no position bias in the libraries. The complete sequence of the target site and 5′ and 3′ flanking DNA was available from GenBank for 49 human clones. RepeatMasker was used to determine the subfamily designation of the insertions. For comparison, we determined the subfamily classifications of all of the near–full-length L1 insertions on five randomly chosen human chromosomes (representing ∼24% of human euchromatic DNA) as a representation of the whole genome (fig. 1B). For the L1Hs–L1PA3 subfamilies, the distributions of the human LOAF library and genomic L1s were similar, indicating a lack of age bias in the libraries (fig. 1C). Older L1 insertions (L1PA4+) are underrepresented in the libraries, as expected, because most of these insertions have the older A nucleotide at the priming site of the first of two nested primers used in the construction of the libraries (fig. 1A). We conclude that the LOAF libraries represent the first substantial and unbiased source of data on the distribution and activity of L1 retrotransposons during the late great ape radiation.
Table 1.
Distribution of LOAF Library Clone Loci by Human Chromosome Number
|
No. of Clone Locifrom Genome of |
|||
| HumanChromosome | Humans | Bonobos | Chimpanzees |
| 1 | 4 | 2 | 3 |
| 2 | 4 | 6 | 4 |
| 3 | 6 | 1 | 5 |
| 4 | 7 | 6 | 5 |
| 5 | 2 | 5 | 6 |
| 6 | 6 | 6 | 5 |
| 7 | 2 | 3 | 2 |
| 8 | 4 | 2 | … |
| 9 | 1 | 2 | 1 |
| 10 | 2 | … | 3 |
| 11 | 3 | 6 | … |
| 12 | 5 | … | 2 |
| 13 | 2 | … | … |
| 14 | 2 | 2 | 2 |
| 15 | 2 | 1 | 1 |
| 16 | … | 2 | 1 |
| 17 | 1 | … | 1 |
| 18 | … | … | 2 |
| 19 | … | 1 | 3 |
| 20 | … | 1 | 3 |
| 21 | … | … | … |
| 22 | 1 | … | … |
| X | … | 1 | … |
| Y | 1 | … | 2 |
The relative rates of L1 accrual in two lineages with a common ancestor can be derived from a reciprocal comparison of the fraction of lineage-specific insertions present in their genomes. For example, if 80% of the randomly sampled L1s from species A are also present in species B, then the remaining 20% of the insertions in A have occurred after the separation of the two species. Likewise, if only 60% of the insertions in B are found in A, then 40% of the insertions in B occurred subsequent to the divergence of A and B. According to this example, the rate of L1 accrual in species B has been two times the rate in species A. The rate of L1 accrual is equal to the rate of L1 amplification (which itself is a function of the rates of transposition and fixation) minus the rate of L1 loss (Boissinot et al. 2001). When the rates of L1 loss have been low, this method yields an estimate of the relative rates of L1 amplification. We next applied this approach to a comparison of the accumulation of L1s in the Homo and Pan lineages.
Three of the 49 human LOAF clones for which complete genomic information was available from GenBank contained human-specific (L1Hs-Ta) insertions. An additional locus had no L1 insertion. We PCR amplified the target site for this insertion from 11 unrelated and ethnically diverse human DNA samples and were unable to detect the presence of the occupied allele (data not shown). We were unable to confirm the presence of this insertion in the DNA of the individual from whom the LOAF library was prepared, because unmodified genomic DNA was not available. Nevertheless, this clone was counted as human specific, because rare polymorphic L1 insertions have been observed (Sheen et al. 2000; Myers et al. 2002). Therefore, in total, 4 of the 49 human LOAF clones contained human-specific L1 insertions. We compared the fraction of human-specific L1s among the LOAF library clones to the fraction of human-specific L1s among the recent insertions in the human genome as a whole (fig. 1B). The differences between these proportions were not statistically significant (P=.348, χ2 test; fig. 1B and 1C), again confirming the lack of age bias in the LOAF libraries.
Surprisingly, a much larger proportion of the L1 insertions isolated from the bonobo and chimpanzee LOAF libraries (each derived from a single individual) were lineage specific (fig. 1C). Complete genomic information was available from the human sequence databases for 47 bonobo and 46 chimpanzee clones, and L1 elements were identified at the insertion target sites of 38 and 35 of them respectively. Thus, 9 of 47 bonobo and 11 of 46 chimpanzee clones were lineage specific. We compared the fractions of Pan-specific LIs in the chimpanzee and bonobo libraries to the fraction of lineage-specific LIs in the human genome sequence (figs. 1B and 1C). The differences in the proportions of lineage-specific insertions were statistically significant for both bonobos (P<.001, χ2 test) and chimpanzees (P<.001, χ2 test). These data indicate that, since the divergence of the Pan and Homo lineages, L1s have accumulated at rates between 2.3- and 3-fold faster in Pan lineages than in the Homo lineage.
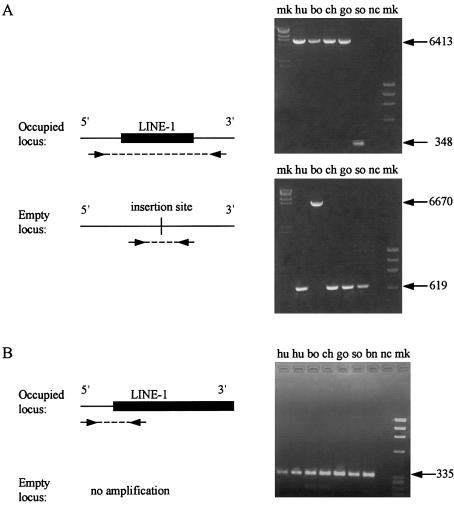
Inferences drawn from the in silico analysis about the species distribution of the LOAF library L1 insertions were confirmed by PCR genotyping. Each of the lineage-specific insertions (n=24) was genotyped in at least two individuals from each species. In most cases, reactions performed with primers that hybridized to the DNA flanking both sides of the target sites (Ovchinnikov et al. 2002) confirmed the presence of the occupied and empty alleles in the different species (fig. 2A). In several cases, PCR amplification of the empty alleles was achieved, but amplification of the full-length L1 insertion was not. For these loci, the presence of the L1 element was confirmed using an internal primer matching the L1 5′ end and the same 5′ flanking primer that had successfully amplified the empty allele (fig. 2B). In total, PCR genotyping successfully confirmed the distribution of 21 of 24 lineage-specific insertions. In the three additional cases, genotyping was unsuccessful. In all cases, the bioinformatic conclusions and PCR genotyping results were concordant.
Figure 2.
PCR genotyping of LOAF library L1 insertion sites. L1 insertion sites were identified by searching the online human genome sequence databases with the sequences of the LOAF clone 5′ flanking DNAs. The presence or absence of L1 elements at the insertion target sites in the various species was determined by PCR amplification of genomic DNA derived from human (hu), bonobo (bo), chimpanzee (ch), lowland gorilla (go), and Sumatran orangutan (so). mk = DNA marker; nc = negative control. Genotyping was successfully performed for 34 human, 36 bonobo, and 19 chimpanzee loci (conditions available upon request). A, PCR amplifications using flanking sequence primers. Primers were designed to hybridize to the 5′ and 3′ flanking sequences surrounding the insertion target sites. Both the occupied and empty alleles could be positively identified using a single reaction. An insertion present in humans, bonobos, chimpanzees, and gorillas but absent from Sumatran orangutans is shown (top). An insertion present only in bonobos is also shown (bottom). B, PCR amplifications across the 5′ junction of the L1 insertion. When the full-length L1 insertion was difficult to amplify, the presence of an insertion was confirmed by amplifying the 5′ junction of the L1, using an internal primer matching nt 96–114 of LRE1 and the same 5′ flanking primer that was used to amplify the empty insertion target site. The panel depicts an L1 insertion present in all species, including a Bornean orangutan. For two loci for which PCR analyses were inconclusive, the presence or absence of the L1 insertion was confirmed by sequencing the cloned PCR products. bo = bonobo; bn = Bornean orangutan; ch = chimpanzee, hu = human; go = lowland gorilla; mk = DNA marker; nc = negative control; and so = Sumatran orangutan.
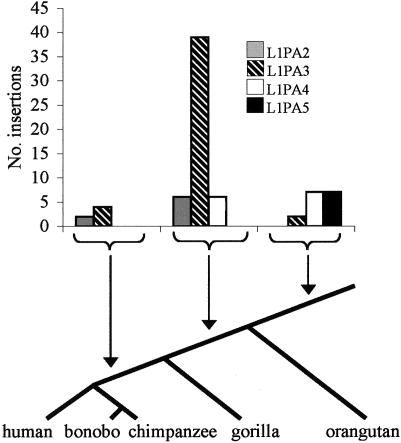
The genotyping data also indicate the evolutionary periods during which the great ape L1 subfamilies appeared and amplified. All five of the tested L1PA5 insertions were present in orangutans, suggesting that the amplification of the L1PA5 subfamily may have concluded before the divergence of the Asian (orangutans) and African apes (fig. 3). In contrast, some of the L1PA4 insertions isolated from the libraries were absent from orangutans, indicating that this subfamily amplified both before and after the Asian-African divergence. L1PA2 elements first appeared before the gorilla divergence and ceased to amplify before the Homo-Pan divergence, whereas L1PA3 elements appear to have amplified over an extended period of great ape evolution. These data are consistent with the accepted phylogenetic relationships among the great apes and with a monophyletic relationship between humans and chimpanzees (Caccone and Powell 1989; Arnason et al. 1996; Ruvolo 1997; Takahata and Satta 1997; Satta et al. 2000; Chen and Li 2001). They also indicate that multiple L1 subfamilies were synchronously retrotransposing during great ape evolution (Smit et al. 1995). None of the tested L1 insertions were present in a species distribution that was inconsistent with the most commonly accepted phylogeny of the great apes (fig. 3). Therefore, these data provide no evidence for the loss of an L1 insertion at these loci in any great ape lineage (Boissinot et al. 2001). The low rate (or absence) of L1 loss also suggests that the large difference in the proportion of lineage-specific L1 insertions in the Homo and Pan genomes is likely due to a difference in the rates of L1 amplification (as opposed to a difference in the rate of L1 loss) in these lineages. Alternatively, these data may reflect a difference in the accumulation of full-length insertions only, rather than a difference in overall amplification rates. We consider this less likely, because it implies that selection against full-length L1s has been greater in the Homo than in the Pan lineages (Boissinot et al. 2001). An alternative explanation—that the processivity of the L1 reverse transcriptase is greater in chimpanzees and bonobos than in humans—is also unlikely. The proportion of full-length L1s among recent human insertions is much higher than among older insertions (Boissinot et al. 2001; Ovchinnikov et al. 2002). If this were the result of an increase in L1 processivity, then an even faster increase in Pan than in Homo lineages would be required to account for our results.
Figure 3.
Species distribution of L1 insertions by subfamily class. The presence or absence of an L1 at LOAF library clone insertion target sites in the different species was determined by PCR genotyping. In nearly all cases, the distribution of the insertions was determined by genotyping at least two individuals from each species. The arrows point to the period of great ape evolution during which the enumerated elements inserted.
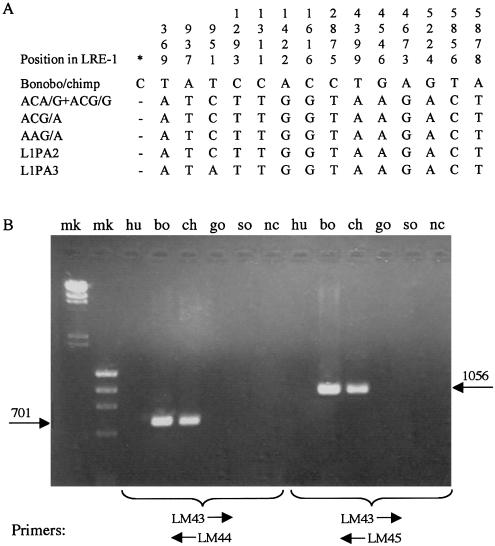
To further characterize Pan-specific L1 insertions, we cloned and entirely sequenced two bonobo-specific L1s, one chimpanzee-specific L1, and one insertion shared by both bonobos and chimpanzees but absent from all other great apes. These represent the first complete sequences of L1s that inserted into the genomes of our closest ape relatives subsequent to the divergence of the Homo and Pan lineages. Previously, a single near–full-length gorilla insertion had been sequenced (DeBerardinis and Kazazian 1998). Alignment of these sequences with 40 L1s belonging to the L1Hs–L1PA5 subfamilies revealed 15 shared sequence variants (SSVs) specific for Pan L1s (fig. 4A). We confirmed that these SSVs were specific for Pan insertions by performing PCR amplifications with primers designed to anneal to L1 regions with SSVs (fig. 4B). Primer LM43 (5′-CACCTGGAAAACTCGGGTCACTCC-3′) hybridizes to a sequence containing the insertion of a C between nt 268 and 269 of LRE1; primer LM44 (5′-GGCGCTCTGCATTTTAGAGTTTCCTGT-3′) hybridizes to a region containing a T→A mutation at position 937 and a C→T mutation at position 951; and primer LM45 (5′-CTCTAGACTTCCCTTCTCGCTTCGTTTC-3′) hybridizes to a sequence containing two T→C mutations at positions 1293 and 1311. Amplification of genomic DNA from the various great apes yielded products of the expected size in bonobos and chimpanzees but no products in any of the other samples (fig. 4B). These results indicate that the SSVs correctly identify a Pan-specific L1 subfamily.
Figure 4.
Identification of a Pan-specific L1 subfamily. A, Shared sequence variants specific for Pan L1s. The sequences of four Pan-specific L1s, obtained by cloning and entirely sequencing L1 elements identified from LOAF library clones, and 40 L1Hs–L1PA5 insertions, downloaded from GenBank, were aligned with the MacVector 7.0 implementation of the ClustalW algorithm (see fig. 5 for a list of the sequences). The sequences have been deposited in GenBank under accession numbers AY189988–AY190012. Only positions that contained nucleotides specific for Pan L1s are depicted. Coordinates (listed in top row) refer to nucleotide positions in the actively transposing element LRE1 (Dombroski et al. 1991). The asterisk denotes a single-base insertion in Pan-specific L1s between nucleotides 268 and 269 of LRE1. B, Results of PCR amplification using great ape genomic DNA and Pan-specific L1 primers. Primers were designed to anneal to L1 sequence at regions near the 5′ end in which one or more Pan-specific SSVs were present (see text). PCR conditions will be supplied upon request. bo = bonobo; ch = common chimpanzee; go = lowland gorilla; hu = human; mk = DNA marker; nc = negative control; so = Sumatran orangutan.
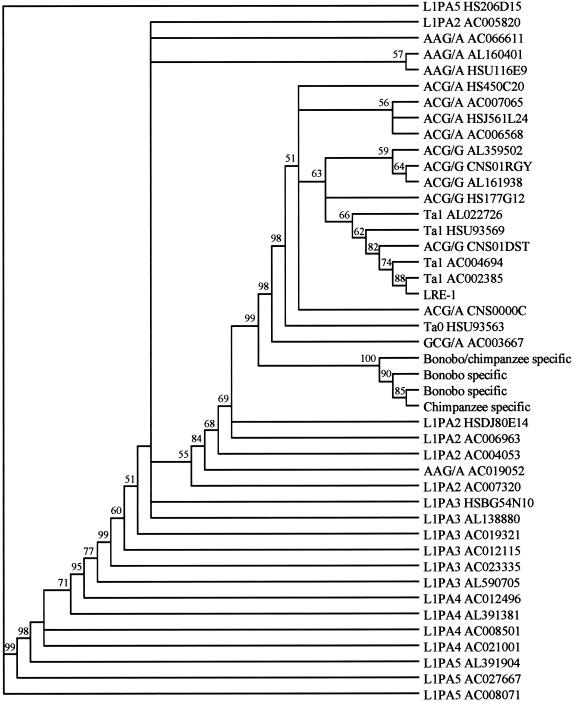
The evolutionary relationships among the L1 insertions were further explored by performing phylogenetic analyses. As shown in figure 5, a neighbor-joining tree of the 4 Pan-specific and 40 L1Hs–L1PA5 L1s displays a well-supported separation between a clade including the Pan-specific L1s and a clade including all human-specific L1 groups (Ta, ACG/G, and ACG/A). The tree also supports a separation between older (ACG/A) and younger (ACG/G and Ta) human-specific insertions, as reported elsewhere (Ovchinnikov et al. 2002). Young, lineage-specific insertions (both Homo and Pan) show greater resolution than do older (L1PA2+) insertions, presumably because the former have had less time to accumulate random mutations. Maximum-parsimony analyses performed in PAUP, version 4.0b10, yielded a tree with similar topology, which is not shown (available upon request).
Figure 5.
Phylogenetic analysis of Pan and human L1 sequences. The figure shows a neighbor-joining tree (Kimura two-parameter, 2,000 bootstrap replications) generated from a ClustalW alignment of full-length L1 sequences. Included are the 4 Pan-specific L1s identified from the LOAF libraries and the 40 L1Hs–L1PA5 human genome sequences downloaded from GenBank. Amplification of the L1PA2 and older subfamilies ceased prior to the Homo-Pan divergence. Human-specific elements in the Ta, ACG/A, ACG/G, and GCG/A groups amplified only after the split between the Homo and Pan lineages, whereas AAG/A elements amplified both before and after the split (Ovchinnikov et al. 2002).
We pose three possible explanations for differential L1 amplification rates in humans and great apes: (1) a change in the frequency of retrotransposition, which requires a differential change in the biochemistry of L1 amplification between the lineages; (2) selection, which requires closely related species to have different responses to the evolutionary dynamics between L1 parasites and host genomes (Yoder et al. 1997; Bestor 1999); and (3) population dynamics. Several studies indicate that the evolution of humans was associated with one or more population bottlenecks (Rogers and Harpending 1992; Rogers 1995; Kimmel et al. 1998). Population constrictions are associated with a reduced genetic diversity, mostly as a result of the loss of low-frequency alleles. Because most active L1s are present in the population at low gene frequencies, the rate of L1 amplification may have been reduced by a contraction in the number of actively mobile L1s during human evolution. To our knowledge, no evidence has been reported for similar bottlenecks in the Pan lineage. In addition, the presence of higher levels of genetic diversity in chimpanzees than in humans (Kaessmann et al. 2001) suggests that any population bottlenecks that might have occurred in the Pan lineage was less severe than those that occurred in humans.
The presence and activity of L1 elements represent substantial sources of genomic instability. Aside from insertional mutagenesis, L1 retrotransposition events may be associated with major changes in genomic structure (Ostertag and Kazazian 2001; Gilbert et al. 2002; Morrish et al. 2002; Symer et al. 2002), and L1 elements may participate in both nonhomologous and unequal homologous recombination events, resulting in large genomic deletions (Drechsler and Royer-Pokora 1996; Burwinkel and Kilimann 1998; Kumatori et al. 1998; Zhang and Zhang 1998; Segal et al. 1999; Suminaga et al. 2000). The 5′ UTR of L1 elements contain transcription regulatory sequences (Swergold 1990); thus, the insertion of full-length L1s may result in the altered expression of nearby genes (Britten 1996; Yang et al. 1998). Our data suggest that the rates of L1 amplification differ substantially between the Homo and Pan lineages, indicating that L1 amplification may change rapidly during primate evolution. What causes L1 amplification rates to change during evolution is not known. However, it is interesting to note that L1 amplification in plants has been reported to be sensitive to evolutionary and environmental stress (Kalendar et al. 2000). Our in silico analysis of near–full-length L1s on five human chromosomes revealed 92 L1Hs insertions (fig. 2A). By extrapolating from this result, we estimate that there are ∼380 near–full-length human-specific L1Hs elements in the modern human genome. Thus, the chimpanzee and bonobo genomes may have as many as 760–1,140 full-length insertions that are absent from the human genome. It will be important to determine the impact of these insertions on genomic evolution and the extent to which they have been involved in the genetic and phenotypic differentiation of the Homo and Pan lineages.
Acknowledgments
We thank R. Jaeger, for helpful discussions, and R. Ramakrishnan, for assistance with the statistical analysis. We are also grateful to the Zoological Society of San Diego for providing DNA samples. This research was supported in part by National Institutes of Health grant R21CA87356-01 (to G.D.S.).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ [Google Scholar]
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for sequences of the L1 insertions [accession numbers AY189988–AY190012])
- RepeatMasker Web Server, http://ftp.genome.washington.edu/cgi-bin/RepeatMasker
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/ (for BLAT search)
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- Arkhipova I, Meselson M (2000) Transposable elements in sexual and ancient asexual taxa. Proc Natl Acad Sci USA 97:14473–14477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnason U, Gullberg A, Janke A, Xu X (1996) Pattern and timing of evolutionary divergences among hominoids based on analyses of complete mtDNAs. J Mol Evol 43:650–661 [DOI] [PubMed] [Google Scholar]
- Bestor TH (1999) Sex brings transposons and genomes into conflict. Genetica 107:289–295 [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV (2000) L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17:915–928 [DOI] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Furano AV (2001) Selection against deleterious LINE-1–containing loci in the human lineage. Mol Biol Evol 18:926–935 [DOI] [PubMed] [Google Scholar]
- Britten RJ (1996) DNA sequence insertion and evolutionary variation in gene regulation. Proc Natl Acad Sci USA 93:9374–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwinkel B, Kilimann MW (1998) Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J Mol Biol 277:513–517 [DOI] [PubMed] [Google Scholar]
- Caccone A, Powell JR (1989) DNA divergence among hominoids. Evolution 43:925–942 [DOI] [PubMed] [Google Scholar]
- Chen FC, Li WH (2001) Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet 68:444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Kazazian HH Jr (1998) Full-length L1 elements have arisen recently in the same 1-kb region of the gorilla and human genomes. J Mol Evol 47:292–301 [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH Jr (1991) Isolation of an active human transposable element. Science 254:1805–1808 [DOI] [PubMed] [Google Scholar]
- Drechsler M, Royer-Pokora B (1996) A LINE element is present at the site of a 300-kb deletion starting in intron 10 of the PAX6 gene in a case of familial aniridia. Hum Genet 98:297–303 [DOI] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran J (2002) Genomic deletions created upon LINE-1 retrotransposition. Cell 110:315–325 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH Jr (2000) Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet 9:653–657 [DOI] [PubMed] [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH Jr (1994) A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet 7:143–148 [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Wiebe V, Weiss G, Pääbo S (2001) Great ape DNA sequences reveal a reduced diversity and an expansion in humans. Nat Genet 27:155–156 [DOI] [PubMed] [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH (2000) Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA 97:6603–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ (2002) BLAT: the BLAST-like alignment tool. Genome Res 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel M, Chakraborty R, King JP, Bamshad M, Watkins WS, Jorde LB (1998) Signatures of population expansion in microsatellite repeat data. Genetics 148:1921–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumatori A, Faizunnessa NN, Suzuki S, Moriuchi T, Kurozumi H, Nakamura M (1998) Nonhomologous recombination between the cytochrome b558 heavy chain gene (CYBB) and LINE-1 causes an X-linked chronic granulomatous disease. Genomics 53:123–128 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH (1999) The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol 16:793–805 [DOI] [PubMed] [Google Scholar]
- Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV (2002) DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet 31:159–165 [DOI] [PubMed] [Google Scholar]
- Myers J, Vincent BJ, Udall H, Watkins WS, Morrish TA, Kilroy GE, Swergold GD, Henke J, Henke L, Moran JV, Jorde LB, Batzer MA (2002) A comprehensive analysis of recently integrated human Ta L1 elements. Am J Hum Genet 71:312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH Jr (2001) Biology of mammalian L1 retrotransposons. Annu Rev Genet 35:501–538 [DOI] [PubMed] [Google Scholar]
- Ovchinnikov I, Rubin A, Swergold GD (2002) Tracing the LINEs of human evolution. Proc Natl Acad Sci USA 99:10522–10527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickeral OK, Makaowski W, Boguski MS, Boeke JD (2000) Frequent human genomic DNA transduction driven by LINE-1 retrotransvposition. Genome Res 10:411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AR (1995) Genetic evidence for a Pleistocene population explosion. Evolution 49:608–615 [DOI] [PubMed] [Google Scholar]
- Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569 [DOI] [PubMed] [Google Scholar]
- Ruvolo M (1997) Molecular phylogeny of the hominoids: inferences from multiple independent DNA sequence data sets. Mol Biol Evol 14:248–265 [DOI] [PubMed] [Google Scholar]
- Satta Y, Klein J, Takahata N (2000) DNA archives and our nearest relative: the trichotomy problem revisited. Mol Phylogenet Evol 14:259–275 [DOI] [PubMed] [Google Scholar]
- Segal Y, Peissel B, Renieri A, de Marchi M, Ballabio A, Pei Y, Zhou J (1999) LINE-1 elements at the sites of molecular rearrangements in Alport syndrome–diffuse leiomyomatosis. Am J Hum Genet 64:62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen F, Sherry ST, Risch GM, Robichaux M, Nasidze I, Stoneking M, Batzer MA, Swergold GD (2000) Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res 10:1496–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23:1087–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Toth G, Riggs AD, Jurka J (1995) Ancestral mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol 246:401–417 [DOI] [PubMed] [Google Scholar]
- Suminaga R, Takeshima Y, Yasuda K, Shiga N, Nakamura H, Matsuo M (2000) Non-homologous recombination between Alu and LINE-1 repeats caused a 430-kb deletion in the dystrophin gene: a novel source of genomic instability. J Hum Genet 45:331–336 [DOI] [PubMed] [Google Scholar]
- Swergold GD (1990) Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol 10:6718–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symer D, Connelly C, Szak S, Caputo E, Cost G, Parmigiani G, Boeke J (2002) Human L1 retrotransposition is associated with genetic instability in vivo. Cell 110:327–337 [DOI] [PubMed] [Google Scholar]
- Takahata N, Satta Y (1997) Evolution of the primate lineage leading to modern humans: phylogenetic and demographic inferences from DNA sequences. Proc Natl Acad Sci USA 94:4811–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Boffelli D, Boonmark N, Schwartz K, Lawn R (1998) Apolipoprotein(a) gene enhancer resides within a LINE element. J Biol Chem 273:891–897 [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13:335–340 [DOI] [PubMed] [Google Scholar]
- Zhang XQ, Zhang JW (1998) The 3′ breakpoint of the Yunnanese (Aγδβ)0-thalassemia deletion lies in an L1 family sequence: implications for the mechanism of deletion and the reactivation of the Gγ-globin gene. Hum Genet 103:90–95 [DOI] [PubMed] [Google Scholar]