Abstract
We recently identified a mutation in the I-κB kinase associated protein (IKBKAP) gene as the major cause of familial dysautonomia (FD), a recessive sensory and autonomic neuropathy. This alteration, located at base pair 6 of the intron 20 donor splice site, is present on >99.5% of FD chromosomes and results in tissue-specific skipping of exon 20. A second FD mutation, a missense change in exon 19 (R696P), was seen in only four patients heterozygous for the major mutation. Here, we have further characterized the consequences of the major mutation by examining the ratio of wild-type to mutant (WT:MU) IKBKAP transcript in EBV-transformed lymphoblast lines, primary fibroblasts, freshly collected blood samples, and postmortem tissues from patients with FD. We consistently found that WT IKBKAP transcripts were present, albeit to varying extents, in all cell lines, blood, and postmortem FD tissues. Further, a corresponding decrease in the level of WT protein is seen in FD cell lines and tissues. The WT:MU ratio in cultured lymphoblasts varied with growth phase but not with serum concentration or inclusion of antibiotics. Using both densitometry and real-time quantitative polymerase chain reaction, we found that relative WT:MU IKBKAP RNA levels were highest in cultured patient lymphoblasts and lowest in postmortem central and peripheral nervous tissues. These observations suggest that the relative inefficiency of WT IKBKAP mRNA production from the mutant alleles in the nervous system underlies the selective degeneration of sensory and autonomic neurons in FD.Therefore, exploration of methods to increase the WT:MU IKBKAP transcript ratio in the nervous system offers a promising approach for developing an effective therapy for patients with FD.
Familial dysautonomia (FD, Riley-Day syndrome, hereditary sensory, and autonomic neuropathy type III [MIM 223900]) is an autosomal recessive disease that affects the sensory and autonomic nervous systems. FD has a remarkably high carrier frequency of 1 in 32 in the Ashkenazi Jewish population (Dong et al. 2002). In FD, there is progressive depletion of unmyelinated sensory and autonomic neurons (Pearson and Pytel 1978a, 1978b; Pearson et al. 1978; Pearson 1979; Axelrod 1995). The loss of neuronal function results in gastrointestinal dysfunction, abnormal respiratory responses to hypoxic and hypercarbic states, scoliosis, gastroesophageal reflux, vomiting crises, lack of overflow tears, abnormal sweating, and postural hypotension (Riley et al. 1949; Axelrod et al. 1974; Axelrod 1984). Recently, we and others identified a single noncoding mutation in the I-κB kinase–associated protein (IKBKAP) gene that accounts for >99.5% of all cases of FD (Anderson et al. 2001; Slaugenhaupt et al. 2001). This major IKBKAP mutation is a single-base change at base pair 6 of the donor splice site of intron 20 (IVS20+6T→C). We found that this mutation causes a decrease in splicing efficiency with sporadic skipping of exon 20, thereby reducing the level of WT IKBKAP mRNA in patient cells (Slaugenhaupt et al. 2001). By contrast, data presented by Anderson et al. (2001) suggested complete elimination of WT IKBKAP mRNA in lymphoblast cells of patients with FD. The only other mutation found in patients with FD to date is a rare missense alteration (R696P), existing only in four patients heterozygous for the major splicing mutation. It is located in exon 19 and is predicted to disrupt a phosphorylation site in I-κB–associated protein (IKAP), the IKBKAP gene product.
IKAP is a conserved protein found in mouse, fruitfly, mustard plant, nematode, and yeast (Cuajungco et al. 2001). It was initially reported as a scaffold protein for the I-κB kinase (IKK) complex (Cohen et al. 1998), but this function was subsequently contested by Krappmann et al. (2000), who reported that IKAP was, in fact, a member of a different cellular complex. IKAP is now recognized as the largest member of the three-subunit protein complex called “core Elongator,” which was originally identified in yeast and, more recently, in humans (Otero et al. 1999; Wittschieben et al. 1999; Fellows et al. 2000; Hawkes et al. 2002; Kim et al. 2002). Studies in yeast and human have revealed that core Elongator interacts with another three-subunit protein to form the six-subunit holo-Elongator complex (Krogan and Greenblatt 2001; Li et al. 2001; Winkler et al. 2001, 2002; Hawkes et al. 2002). Initial investigations in yeast have suggested that core Elongator plays a role in transcription elongation because of its observed histone acetyl transferase activity, its observed interaction with naked or nucleosomal DNA, as well as its interaction with the hyperphosphorylated C-terminal domain of elongating RNA polymerase II during transcription in vitro (Otero et al. 1999; Winkler et al. 2001). Using purified human core Elongator complex from HeLa cell extracts, Kim and colleagues (2002) demonstrated that core Elongator indeed facilitates transcription by RNA polymerase II in a chromatin- and acetyl-coA-dependent manner. However, recent findings in yeast have called previous observations into question. For example, when chromatin immunoprecipitation assay was used, no evidence was observed to suggest that yeast Elongator is directly involved in transcription elongation in vivo (Pokholok et al. 2002) or in vitro (Krogan et al. 2002). Likewise, using tandem affinity purification, Krogan et al. (2002) failed to copurify yeast Elongator with RNA polymerase II, even at microgram quantities. It is interesting that immunofluorescence localization studies show that each core Elongator subunit, including IKAP, is primarily detected in the cytoplasm of human (Krappmann et al. 2000; Hawkes et al. 2002; Holmberg et al. 2002; Kim et al. 2002) or yeast cells (Pokholok et al. 2002). More recently, Holmberg et al. (2002) reported a novel scaffolding role for IKAP in the c-Jun N-terminal kinase (JNK) signal transduction pathway, owing to its discerned strong physical interaction with JNK and enhancement of JNK activation. These studies suggest that IKAP may indeed have multiple roles in the cell, and it remains to be determined how a decrease in cellular IKBKAP causes the FD phenotype and why this phenotype displays tissue specificity.
The regulation of gene splicing (reviewed by Nissim-Rafinia and Kerem [2002]) can potentially play an important role in inherited disorders, influencing phenotypic parameters such as organ specificity, disease severity, and age at onset. Splicing mutations that result in exon skipping have been reported for many disorders, including adrenoleukodystrophy, ataxia telangiectasia, Marfan syndrome, cystic fibrosis, retinoblastoma, tuberous sclerosis, and neurofibromatosis (Mayer et al. 2000; Pagani et al. 2000; Alonso et al. 2001; Birrell et al. 2001; Guimaraes et al. 2001; Guo et al. 2001; Nissim-Rafinia and Kerem 2002). Often, these mutations are associated with a milder disease phenotype, suggesting the persistence of residual protein activity due to a low level of correctly spliced mRNA. The remarkable features of FD are that a single splicing mutation is responsible for virtually all cases of the disease and that, in our hands, homozygous mutant cultured cells are capable of producing substantial amounts of WT IKBKAP mRNA and protein (Slaugenhaupt et al. 2001). These observations suggest that manipulation of splicing mechanisms could offer a route to treating FD and other human disorders involving splicing defects.
In view of the differences in the reported effects of the major FD mutation on splicing of exon 20 (Anderson et al. 2001; Slaugenhaupt et al. 2001), we have characterized the defective IKBKAP splicing using both densitometry and real-time quantitative PCR assays in patient cell lines, freshly collected blood, and FD postmortem tissues. We collected patient blood samples through the Dysautonomia Diagnostic and Treatment Center at New York University Medical Center, with approval from by the Institutional Review Boards of New York University Medical Center, Massachusetts General Hospital, and Harvard Medical School. For each patient, total RNA was extracted, and a lymphoblast cell line was initiated. We also used previously generated patient lymphoblast and fibroblast lines, as well as additional FD cell lines, purchased from the Coriell Mutant Cell Repository, for comparative experiments. All cultured cells were propagated in a standard 37°C incubator at 85% humidity and 5% CO2, with lymphoblast lines grown in RPMI-1640 and primary fibroblasts in Minimum Essential Medium with Earle’s balanced salts. Both media were supplemented with 2 mM L-glutamine and either 10% (GUS lines), 15% (GM lines), or 20% (MIN lines) fetal bovine serum (FBS) (Invitrogen). In some experiments, lymphoblast lines were grown with or without 100 U/mL penicillin, and 100 μg/mL streptomycin (Sigma). Postmortem tissues were obtained from the Pathology Department at New York University Medical Center and the University of Miami Brain and Tissue Bank for Developmental Disorder.
Total RNA was extracted using Tri-Reagent for all tissues and cultured cell samples, or Tri-Reagent BD for blood (Molecular Research Center), and quantitated by UV absorbance. Reverse-transcription reactions using oligo-dT15/random hexamer primers (Promega), Superscript II reverse transcriptase (200 U/μL; Invitrogen) and RNase-inhibitor (80 U/μL; Roche) were performed as described (Cuajungco et al. 2001). PCR was done on all samples to evaluate both WT and MU transcripts (fig. 1). The PCR products were fractionated by electrophoresis on a 1.5% agarose gel stained with ethidium bromide. Each gel band was assessed using an Alpha 2000 Image Analyzer and software coupled with automatic background subtraction (Alpha Innotech). Subsequent analysis of the integrated density value (IDV) to calculate the WT:MU transcript ratio enabled us to obtain the relative differences in expression within each patient tissue or cell samples. To validate our densitometry findings, we used real-time quantitative PCR (QPCR). For QPCR, the 18S ribosomal RNA (Applied Biosystems) was used as an internal normalization control to correct for variations in input RNA amount or reverse transcription efficiency. The 18S rRNA is known for its robustness and stable expression across different tissues and cell lines (Schmittgen and Zakrajsek 2000; Goidin et al. 2001). We used a One-Step RT-Platinum Taq kit (Invitrogen), optimized to 5 mM Mg++, with 500 nM forward and reverse primers, 400 nM TaqMan probe, and 200 ng of total RNA sample. QPCR was done on a BioRad thermal ICycler with the following PCR program: 1 cycle at 50°C for 15 min; 1 cycle at 95°C for 5 min; and 45 cycles at 95°C for 15 sec, and at 59 °C for 1 min. All reactions were carried out in triplicate in a 96-well plate format, with a final volume of 50 μL. Trials for each WT, MU, or 18S assay included four duplicated tissue samples and “no RT” controls. For relative gene-expression analysis, we used the standard curve method (generated from the same serial dilutions of MIN9741 lymphoblast sample used in every plate run) to correct for minor differences in PCR efficiency (Heid et al. 1996; Winer et al. 1999; Giulietti et al. 2001). QPCR values were expressed relative to an FD spleen sample that showed virtually equal WT and MU IKBKAP expression and was included as a calibrator in every experimental run. For protein analysis, available FD postmortem tissues and cell lines were homogenized and total protein quantitated by use of bicinchoninic assay (Pierce). For each sample, 400 μg of total protein was run on a 6% denaturing polyacrylamide gel and Western blotted. The blots were probed using an anti-IKAP C-terminal polyclonal antibody (a kind gift from Dr. Jesper Svejstrup and Dr. Claus Schedereit) and the protein detected by ECL (Amersham), as described elsewhere (Slaugenhaupt et al. 2001). The polyvinylidene fluoride membrane was stained for total protein, using 0.2% Ponceau-S (Sigma) to confirm equal loading and transfer.
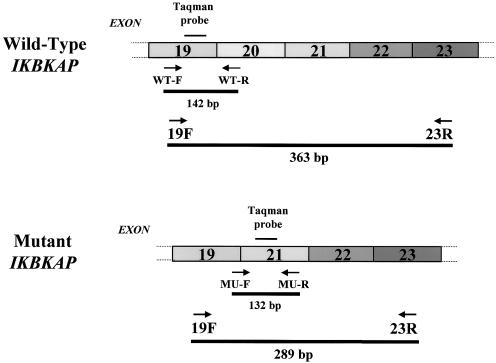
Figure 1.
IKBKAP amplicon sizes and their corresponding PCR products. A schematic representation of the primer sets and Taqman probes used to amplify either the WT or MU IKBKAP transcripts. PCR primers 19F–23R amplify both WT and MU bands. The real-time QPCR primers designated “WT-F/R” (forward/reverse) or “MU-F/R,” with their matching dual-labeled Taqman probes, amplify WT and MU transcripts, respectively. The position of WT-R primer in exon 20 was designed to specifically amplify only the WT message, whereas the MU-F primer located at the junction of exons 19 and 21 will amplify only the MU message. The dual-labeled Taqman probes have a melting temperature (Tm) of 7°C–10°C above the primer Tm for each amplicon. Primer and probe sequences were as follows: 19F: CCT GAG CAG CAA TCA TGT G; 23R: TAC ATG GTC TTC GTG ACA TC; WT-F: GCA GCA ATC ATG TGT CCC A; WT-R: ACC AGG GCT CGA TGA TGA A; WT Taqman probe: FAM-GTT CAC GGA TTG TCA CTG TTG TGC C-BHQ; MU-F: CAC AAA GCT TGT ATT ACA GAC T; MU-R: GAA GGT TTC CAC ATT TCC AAG; and MU Taqman probe: FAM-CTC AAT CTG ATT TAT GAT CAT AAC CCT AAG GTG-BHQ. The housekeeping gene 18S rRNA was used for normalization: 18S-F: GGC CCT GTA ATT GGA ATG AG; 18S-R: GCT ATT GGA GCT GGA ATT AC; and 18S Taqman probe: FAM-TGC TGG CAC CAG ACT TGC CCT C-BHQ.
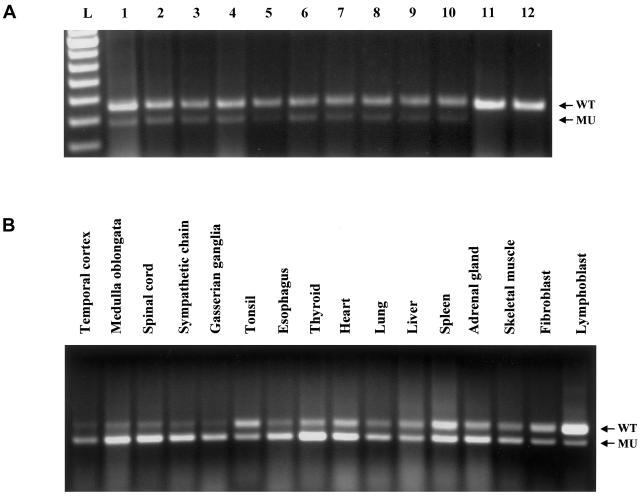
Our original report (Slaugenhaupt et al. 2001) suggested that lymphoblast cell lines from patients with FD were capable of expressing WT IKBKAP transcript from the mutant FD allele. These data contrasted with the absence of WT transcript in FD lymphoblast cells in the report of Anderson et al. (2001). To address this discrepancy, we obtained the GM5106 FD cell line used by Anderson et al. from the Coriell Mutant Cell Repository and compared it with a frozen GM5106 stock already present in our laboratory, as well as with independent lymphoblast cell lines from other patients homozygous for the major mutation. We employed densitometry as a rapid assay to characterize the relative WT:MU transcript ratio of 81 FD lymphoblast cell lines. Of these samples, we consistently found significant WT IKBKAP mRNA in all FD cell lines examined (fig. 2A), including that of the patient sample tested by Anderson et al. (2001) (see lane 6 and lane 7 in fig. 2A).
Figure 2.
A, Expression of WT and MU IKBKAP mRNA in various lymphoblast cell lines of patients with FD. A total of 81 FD lymphoblast cell lines were assayed, using a densitometric assay. Representative samples are shown, including the GM5106 (Coriell) sample used by Anderson et al. (2001). All FD lines consistently expressed both WT IKBKAP (upper band, lane 1–lane 10) and MU (lower band, lane 1–lane 10). Note that MU IKBKAP transcript is never detected in RNA from normal cells or tissues (lane 11–lane 12). L=100-bp ladder; 1=FD (MIN9741, new line); 2=FD (MIN9744, new line); 3=FD (MIN9741, new line); 4=Carrier (GM5105, Coriell); 5=FD (GM5107, Coriell); 6=FD (GM5106, Coriell); 7=FD (GM5106, established line); 8=FD (GM5041, established line); 9=FD (GUS13131, established line); 10=FD (GUS14939, established line); 11=non-FD control (GM5043, Coriell); and 12=non-FD control (GM5110, Coriell). WT amplicon size=363 bp; MU amplicon size=289 bp. B, Expression of WT and MU IKBKAP mRNA in postmortem FD tissue samples and cell lines. All samples were assayed and analyzed as described in our methodology. The relative WT:MU transcript ratio was consistently observed to be highest in FD lymphoblast lines, lowest in FD nervous system tissues, and intermediate in other tissues. The distinctive reduction of relative WT message in most of the CNS and PNS tissues corresponds to the tissue-specific pathology observed among patients with FD.
To assess whether the efficiency of accurate IKBKAP splicing might vary over time, we used four independent lymphoblast cell lines (MIN9741, MIN9744, MIN9745, and MIN9754) from patients with FD who were homozygous for the major mutation. The cells were cultured and assayed weekly for 14 wk (fed every 3 d and harvested on the 7th d at saturation density of ∼20 million cells). The cell lines did not differ significantly in their overall average WT:MU IDV ratio for the entire culture period (table 1). However, the ratio did show significant fluctuation (P<.0001), over a twofold range between harvests, with the relative levels of expression seeming to vary in concert in different cell lines. This suggests that uncontrolled variability in the conditions of culture or harvesting shared by all four cell lines operated to modify the relative efficiency of IKBKAP splicing to some degree.
Table 1.
Quantitative Analysis of WT and MU IKBKAP Expression Ratio in Cultured Cells[Note]
|
Expression Ratio in Lymphoblast Cell Line |
|||||
| Harvest | MIN9741 | MIN9744 | MIN9745 | MIN9754 | Mean±SD |
| 1 | 3.00 | 3.52 | 2.55 | 3.19 | 3.07±0.41 |
| 2 | 2.86 | 2.18 | 3.00 | 2.51 | 2.64±0.37 |
| 3 | 2.70 | 2.15 | 2.17 | 2.60 | 2.41±0.29 |
| 4 | 4.50 | 3.60 | 4.47 | 5.13 | 4.43±0.63 |
| 5 | 4.00 | 4.14 | 3.78 | 4.36 | 4.07±0.24 |
| 6 | 2.09 | 2.22 | 1.90 | 2.28 | 2.12±0.17 |
| 7 | 2.53 | 2.33 | 2.71 | 2.26 | 2.46±0.20 |
| 8 | 3.57 | 3.81 | 3.62 | 3.64 | 3.66±0.10 |
| 9 | 3.69 | 3.88 | 3.33 | 3.19 | 3.52±0.32 |
| 10 | 3.65 | 3.67 | 3.39 | 2.84 | 3.39±0.39 |
| 11 | 3.03 | 3.71 | 2.71 | 2.93 | 3.09±0.43 |
| 12 | 2.21 | 2.97 | 2.53 | 4.18 | 2.97±0.86 |
| 13 | 2.78 | 3.52 | 2.97 | 2.84 | 3.03±0.34 |
| 14 | 2.82 | 3.30 | 3.77 | 3.00 | 3.22±0.41 |
| Mean±SD | 3.10±0.69 | 3.21±0.71 | 3.06±0.70 | 3.21±0.84 | |
Note.— We assayed four independent lymphoblast lines over 14 harvest periods. The cells were harvested at saturation phase (every 7th d) and assayed using densitometry for WT:MU expression ratio, as described. The results showed that, over the cultured time period assessed, no significant difference existed between cell lines (N=4). A significant difference, however, was observed between harvest times (P<.0001, N=14).
We performed several experiments to vary specific aspects of the culture conditions to determine whether they had an impact on the WT:MU ratio. We first tested whether culturing lymphoblast lines with or without the common media antibiotic additives (penicillin/streptomycin) would influence WT:MU IKBKAP ratio, as there was a recent report that pre-mRNA splicing in vitro is inhibited by certain antibiotics (Hertweck et al. 2002). Three different FD lymphoblast lines were grown and harvested every 7th d at saturation density, as above. After assaying for a period of 4 wk, no significant difference was seen in mean WT:MU expression ratios between the respective treated and nontreated cell lines, either at individual harvests or in the average of all harvests (data not shown).
Next, we investigated the effects of changing the percentage of supplemented FBS on lymphoblast lines using 10 independent cell lines. Five of these (MIN lines) were initiated and maintained in 20% FBS, whereas the other five (GUS lines) had been adapted in long-term culture to 10% FBS. Each of the 10 lines was split into two matching cultures, one at 20% FBS and one at 10% FBS. These matching cultures were maintained at their respective FBS levels throughout the experiment and were harvested twice in logarithmic growth and twice at saturation. No significant difference in WT:MU ratio was seen between cultures grown consistently at 10% FBS and their matching cultures increased to 20% FBS (2.32±0.74 vs. 2.29±0.62) between cultures grown consistently at 20% FBS and their matching counterparts decreased to 10% FBS (3.23±0.97 vs. 3.15±1.01), or between all cultures grown at 10% FBS versus 20% FBS (2.73±0.98 vs. 2.76±0.94). However, although the FBS content did not alter the WT:MU ratio, this parameter was significantly affected by the state of the cultures at harvest, as the relative amount of WT IKBKAP RNA was higher in saturated cultures (3.22±0.93) than in logarithmically growing cultures (2.27±0.64) (P<.0005). It is not clear why or how high cell density can apparently influence IKBKAP expression.
Our findings indicate that the WT:MU ratio in a given FD cell line can fluctuate because of culture conditions, implying that the efficiency of producing accurately spliced RNA from the mutant allele can be altered. In these experiments, we have eliminated some obvious potential sources of variation, such as antibiotic treatment and FBS concentration, and implicated an effect of cell density/growth state. However, the fact that the capacity of the mutant allele to be spliced accurately can be modified by as-yet-uncertain external signals provides the proof of principle to search for chemical compounds that can increase the WT:MU IKBKAP ratio and therefore have potential for therapeutic use in FD. The ability to use FD cells to screen for such compounds depends on the degree to which parallel cultures of the test cell line display equivalent WT:MU ratios. We investigated consistency of expression within cell lines, by splitting four independent FD lymphoblast lines into four parallel cultures each and harvesting these four times over a period of 4 wk. Although the independent cell lines showed different WT:MU ratios relative to each other and fluctuated in WT:MU ratio between harvests, replicate cultures of the same line typically yielded very consistent WT:MU IKBKAP expression ratios at any given harvest (table 2). Thus, although the relative efficiency of IKBKAP splicing can vary between independent cell lines and between harvests, multiple replicates of the same culture harvested at the same time yield very small SDs in the mean WT:MU ratios (SD range 0.05–0.62). This suggests that FD mutant cells, assayed simultaneously as multiple parallel cultures diluted from a single source culture, offer a viable route for screening to identify compounds that alter the WT:MU IKBKAP ratio.
Table 2.
Four Parallel Cultures of Four Independent FD Lymphoblast Lines[Note]
|
Expression Ratio in Week |
|||||
| Lymphoblast Culture andHarvest | A | B | C | D | Mean±SD |
| MIN9744: | |||||
| 1 | 2.15 | 1.81 | 200 | 181 | 1.94±0.16 |
| 2 | 2.18 | 2.09 | 2.14 | 1.97 | 2.09±0.09 |
| 3 | 2.93 | 2.94 | 3.00 | 2.89 | 2.94±0.05 |
| 4 | 4.21 | 3.50 | 3.67 | 3.78 | 3.79±0.30 |
| Mean | 2.87 | 2.59 | 2.70 | 2.61 | |
| Overall | 2.69±0.78 | ||||
| GUS14939: | |||||
| 1 | 1.46 | 1.47 | 1.45 | 1.56 | 1.49±0.05 |
| 2 | 2.75 | 271 | 2.90 | 2.80 | 2.79±0.08 |
| 3 | 2.19 | 1.90 | 2.00 | 2.00 | 2.02±0.12 |
| 4 | 2.67 | 2.22 | 2.81 | 2.18 | 2.47±0.32 |
| Mean | 2.27 | 2.08 | 2.29 | 2.14 | |
| Overall | 2.19±0.53 | ||||
| MIN9754: | |||||
| 1 | 3.14 | 2.63 | 3.26 | 3.07 | 3.03±0.27 |
| 2 | 4.10 | 3.72 | 3.56 | 2.39 | 3.89±0.30 |
| 3 | 3.80 | 3.27 | 3.67 | 2.48 | 3.54±0.24 |
| 4 | 3.21 | 2.39 | 3.41 | 2.42 | 2.63±0.39 |
| Mean | 3.56 | 3.00 | 3.24 | 3.27 | |
| Overall | 3.27±0.57 | ||||
| GUS28025: | |||||
| 1 | 2.96 | 1.59 | 1.86 | 1.79 | 2.05±0.62 |
| 2 | 2.49 | 1.94 | 2.50 | 2.53 | 2.37±0.28 |
| 3 | 3.56 | 3.38 | 3.39 | 3.44 | 3.44±0.08 |
| 4 | 2.42 | 1.97 | 2.11 | 2.58 | 2.27±0.28 |
| Mean | 2.86 | 2.22 | 2.47 | 2.59 | |
| Overall | 2.53±0.65 | ||||
Note.— Four independent FD lymphoblast cell lines were cultured over 4 wk to assess the consistency of WT:MU expression ratio in each individual cell line. Each cell line was split into four parallel cultures and assayed using densitometry as described.
To date, we have also assayed three distinct primary fibroblast lines from patients with FD, and we also consistently observed the presence of both WT and MU IKBKAP messages in this cell type. It is interesting that cultured primary fibroblast cells harvested either at high (∼90%–100% confluence) or low cell density (∼50%–60% confluence) give relatively lower WT:MU ratio of 1.21±0.20 and 1.85±0.85, respectively, than that of cultured lymphoblasts (see also table 3), suggesting an inherent cell-type difference in the efficiency of accurate splicing from the mutant allele.
Table 3.
Densitometric and Real-Time QPCR Analyses of Representative FD Postmortem Tissue and Cultured Cell Samples[Note]
| WT:MU Ratio |
||
| FD Postmortem Tissue | Densitometry | Real-Time QPCR |
| Temporal cortex | .35 | .35 |
| Medulla oblongata | .29 | .27 |
| Spinal cord | .25 | .43 |
| Sympathetic chain | .24 | .19 |
| Gasserian ganglia | .23 | .19 |
| Tonsil | 1.21 | 1.14 |
| Esophagus | .33 | .46 |
| Thyroid | .34 | .33 |
| Adrenal gland | .61 | .70 |
| Heart | .55 | .87 |
| Lung | .53 | .64 |
| Liver | .82 | 1.56 |
| Spleen | .94 | 1.00 |
| Skeletal muscle | .68 | .94 |
| Fibroblast | 1.33 | 1.01 |
| Lymphoblast | 3.53 | 3.44 |
Note.— The WT:MU IKBKAP ratio was calculated from data obtained by densitometry and real-time QPCR, using sample tissues represented in figure 2B. Results from the densitometric technique showed very high concordance with real-time QPCR values. The WT:MU data for both CNS and PNS tissues consistently produced a ratio <1.0, whereas cultured cells (lymphoblast and fibroblast) consistently yielded a ratio >1.0. QPCR values were expressed relative to the spleen as calibrator for both WT and MU levels, as described.
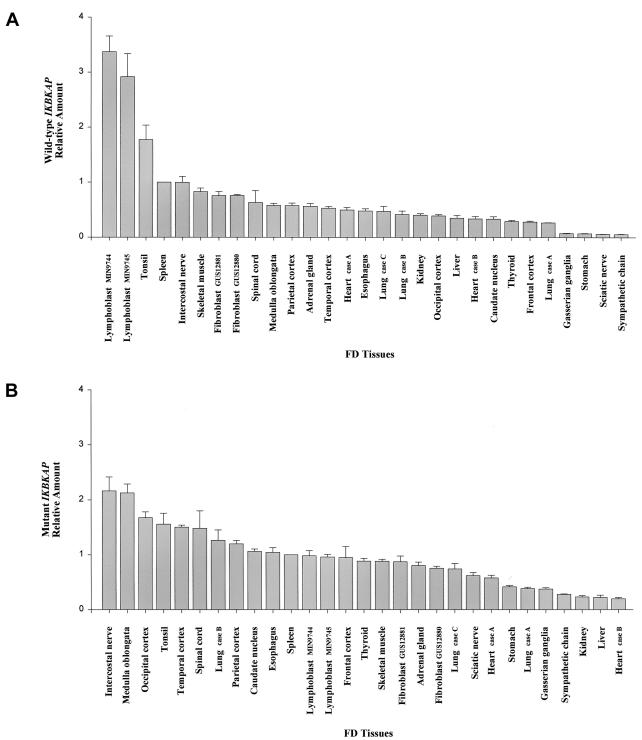
To determine whether the capacity to produce WT transcript from the mutant FD allele reflects the artificial state of cells in culture, we assayed several fresh blood samples taken from patients. We consistently found expression of WT and MU transcripts from the mutant alleles, indicating that FD cells indeed produce WT IKBKAP transcript in vivo (WT:MU=1.46±0.08, N=4). We went on to assay postmortem FD tissues from different CNS and peripheral nervous system (PNS) regions, as well as from other available organs. The results of densitometric assays of these tissues showed that MU IKBKAP transcripts predominate in both the CNS and PNS (fig. 2B). The densitometry data on nervous system tissues gave a WT:MU transcript ratio of <1.0, whereas patient lymphoblast lines gave a consistent ratio >1.0 (table 3). These observations were confirmed using real-time QPCR (table 3; fig. 3). Lymphoblast cell lines from patients with FD displayed relatively high levels of WT IKBKAP transcript, followed by tonsil, spleen, and intercostal nerve, whereas other organs showed relatively intermediate WT levels (fig. 3A). By contrast, relative MU IKBKAP message varied markedly between different CNS and PNS regions, with higher MU levels observed in the brain stem (medulla oblongata), striatum (caudate nucleus), neocortex (frontal, temporal, parietal, and occipital), spinal cord, and intercostal nerve (figs. 2B and 3B). The tonsil, esophagus, and lung tissue samples are among the only nonnervous organs that showed relatively high MU expression. The presence of relatively more MU transcript in various CNS and PNS regions indicates that IKBKAP-splicing efficiency is distinctly affected by the FD gene defect in these tissues. These data also suggest that the identity and relative amounts of cellular splicing factors involved in the correct splicing of IKBKAP may vary from tissue to tissue, with certain neuronal cells, including those in the sensory and autonomic nervous systems, being particularly susceptible to missplicing in FD.
Figure 3.
Graphical representation of relative WT and MU IKBKAP expression levels in various FD postmortem tissues and cultured cell lines, measured by use of real-time QPCR. The QPCR values were normalized using 18S rRNA, and the spleen tissue was used as the calibrator for relative comparisons between patient samples. A, Relative WT IKBKAP levels. The FD lymphoblast cell lines followed by the tonsil tissue showed relatively higher levels of WT message than any other patient tissues examined. B, Relative mutant IKBKAP levels. Several areas of the neocortex, as well as the brain stem medulla, striatum (caudate nucleus), spinal cord, and intercostal nerve tissue samples, showed relatively higher levels of the MU IKBKAP transcript compared with other patient tissues and cell lines investigated. Among the nonnervous tissues tested, only the tonsil, esophagus, and one of the lung samples (case B) showed relatively high MU IKBKAP levels, whereas the others had intermediate levels.
The fact that FD is recessive, with no phenotype in carriers, coupled with the fact that patient cells are capable of producing normal message and protein, presents an exciting possible approach to treatment. By targeting the splicing pathway, it may be possible to increase the level of normal IKAP protein in FD cells. This approach has recently shown promise for the treatment of spinal muscular atrophy (SMA), another neurodegenerative disorder. SMA is caused by loss of function mutations in the SMN1 gene. A nearly identical SMN2 gene encodes transcripts that are missing exon 7 because of a mutation that disrupts an exonic splice enhancer. However, it has been shown that SMN2 encodes a small amount of full-length mRNA, the amount of which is inversely correlated with SMA disease severity, because it compensates for the absent SMN1 protein. Recent studies have shown that SMN2 mRNA processing can be modulated in vitro and in vivo by both splicing factors and drugs, offering a potential for the treatment of SMA in humans (Hofmann et al. 2000; Andreassi et al. 2001; Chang et al. 2001). Although the exon-skipping mechanism in SMA is different from that in FD, these observations clearly demonstrate that certain drugs, or other transacting factors, have the potential to preclude skipping of exon 20 in IKBKAP.
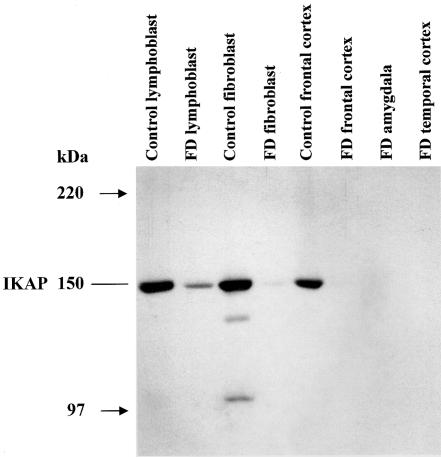
To confirm that the observed change in IKBKAP mRNA expression results in a corresponding decrease in IKAP protein, we performed a Western blot analysis using an anti-IKAP C-terminal antibody (Slaugenhaupt et al. 2001) (fig. 4). As expected, there is a reduction in IKAP protein in both the FD lymphoblast and FD fibroblast samples, and no IKAP was detected in the FD brain postmortem tissues available for study. This observation confirms that the significant reduction of WT IKBKAP mRNA in FD nervous tissue leads to either a complete absence of normal IKAP or reduction of the protein to an undetectable level.
Figure 4.
Western blot showing relative IKAP expression levels in FD postmortem tissues and cultured cell lines. Protein was extracted from several cell lines and postmortem tissues, as shown, and was run on 6% denaturing polyacrylamide gel. The samples were transferred to a PVDF membrane and stained with Ponceau-S to confirm equal protein loading. IKAP was detected using a C-terminal polyclonal antibody. Here, we show that IKAP levels are reduced in the FD lymphoblast and fibroblast samples compared with controls, and IKAP is either absent or below the level of antibody detection in FD brain tissues.
In conclusion, our data demonstrate that all FD cell types examined are capable of producing WT IKBKAP transcripts from the mutant FD alleles, regardless of the tissue source or culture conditions. However, the relative efficiency of accurate splicing varies dramatically, with nervous system tissues displaying the lowest relative amounts of WT IKBKAP transcript. This is consistent with FD being caused not by the absence of IKAP per se but by its reduction below critical levels in sensory and autonomic neurons. Most importantly, the fact that the FD mutation does not fully obliterate splicing, but rather results in a tissue-specific decrease in WT IKBKAP mRNA levels, provides an exciting avenue for therapeutic exploration. This result provides a basis for the identification of cellular splicing modulators, such as drugs and/or splicing factors that enhance the efficiency of inclusion of exon 20 in IKBKAP transcripts from mutant alleles. Indeed, a crucial by-product of our studies is the definition of quantifiable baselines of expression, for both WT and MU IKBKAP transcripts from patient cell lines, that can be employed for future drug screening, to identify compounds with therapeutic potential for patients with FD.
Acknowledgments
We thank Dr. David Friedlander and Ms. Inna Logvinsky for their help in tissue acquisition. Samples used in this study were provided by the Department of Pathology, New York University Medical Center, and by the University of Miami Brain and Tissue Bank for Developmental Disorders through the National Institute of Child Health and Human Development contract NO1-HD-8-3284. We are grateful to Drs. Nicola Hawkes and Jesper Svejstrup, as well as Drs. Daniel Krappmann and Claus Schedereit, for providing the anti-IKAP C-terminal antibody. This study was supported by grants from the Dysautonomia Foundation, the National Institute of Neurological Disorders and Stroke grant RO1 NS36326, and the Massachusetts General Hospital Fund for Medical Discovery (to M.P.C.).
Electronic-Database Information
Accession number and URL for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/Omim/ (for FD [MIM 223900]) [PubMed]
References
- Alonso J, Garcia-Miguel P, Abelairas J, Mendiola M, Sarret E, Vendrell MT, Navajas A, Pestana A (2001) Spectrum of germline RB1 gene mutations in Spanish retinoblastoma patients: phenotypic and molecular epidemiological implications. Hum Mutat 17:412–422 [DOI] [PubMed] [Google Scholar]
- Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, Ekstein J, Rubin BY (2001) Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet 68:753–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C, Jarecki J, Zhou J, Coovert DD, Monani UR, Chen X, Whitney M, Pollok B, Zhang M, Androphy E, Burghes AH (2001) Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum Mol Genet 10:2841–2849 [DOI] [PubMed] [Google Scholar]
- Axelrod FB (1984) Familial dysautonomia and other congenital and sensory autonomic neuropathies. In: Blake IB (ed) Cell and molecular biology of neuronal development. Plenum Press, New York, pp 331–340 [Google Scholar]
- ——— (1995) Familial dysautonomia. In: Robertson D, Biaggioni I (eds) Disorders of the autonomic nervous system. Harwood Academic Publishers, Luxembourg, pp 217–231 [Google Scholar]
- Axelrod FB, Nachtigal R, Dancis J (1974) Familial dysautonomia: diagnosis, pathogenesis and management. Adv Pediatr 21:75–96 [PubMed] [Google Scholar]
- Birrell GW, Ramsay JR, Tung JJ, Lavin MF (2001) Exon skipping in the ATM gene in normal individuals: the effect of blood sample storage on RT-PCR analysis. Hum Mutat 17:75–76 [DOI] [PubMed] [Google Scholar]
- Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H (2001) Treatment of spinal muscular atrophy by sodium butyrate. Proc Natl Acad Sci USA 98:9808–9813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L, Henzel WJ, Baeuerle PA (1998) IKAP is a scaffold protein of the Iκ-B kinase complex. Nature 395:292–297 [DOI] [PubMed] [Google Scholar]
- Cuajungco MP, Leyne M, Mull J, Gill SP, Gusella JF, Slaugenhaupt SA (2001) Cloning, characterization, and genomic structure of the mouse Ikbkap gene. DNA Cell Biol 20:579–586 [DOI] [PubMed] [Google Scholar]
- Dong J, Edelmann L, Bajwa AM, Kornreich R, Desnick RJ (2002) Familial dysautonomia: detection of the IKBKAP IVS20+6T→C and R696P mutations and frequencies among Ashkenazi Jews. Am J Med Genet 110:253–257 [DOI] [PubMed] [Google Scholar]
- Fellows J, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2000) The Elp2 subunit of elongator and elongating RNA polymerase II holoenzyme is a WD40 repeat protein. J Biol Chem 275:12896–12899 [DOI] [PubMed] [Google Scholar]
- Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C (2001) An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25:386–401 [DOI] [PubMed] [Google Scholar]
- Goidin D, Mamessier A, Staquet MJ, Schmitt D, Berthier-Vergnes O (2001) Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and β-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem 295:17–21 [DOI] [PubMed] [Google Scholar]
- Guimaraes CP, Lemos M, Menezes I, Coelho T, Sa-Miranda C, Azevedo JE (2001) Characterisation of two mutations in the ABCD1 gene leading to low levels of normal ALDP. Hum Genet 109:616–622 [DOI] [PubMed] [Google Scholar]
- Guo D, Tan FK, Cantu A, Plon SE, Milewicz DM. (2001) FBN1 exon 2 splicing error in a patient with Marfan syndrome. Am J Med Genet 101:130–134 [DOI] [PubMed] [Google Scholar]
- Hawkes NA, Otero G, Winkler GS, Marshall N, Dahmus ME, Krappmann D, Scheidereit C, Thomas CL, Schiavo G, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2002) Purification and characterization of the human elongator complex. J Biol Chem 277:3047–3052 [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM (1996) Real time quantitative PCR. Genome Res 6:986–994 [DOI] [PubMed] [Google Scholar]
- Hertweck M, Hiller R, Mueller MW (2002) Inhibition of nuclear pre-mRNA splicing by antibiotics in vitro. Eur J Biochem 269:175–183 [DOI] [PubMed] [Google Scholar]
- Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B (2000) Htra2-β 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proc Natl Acad Sci USA 97:9618–9623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C, Katz S, Lerdrup M, Herdegen T, Jaattela M, Aronheim A, Kallunki T (2002) A novel specific role for I-κ B kinase complex-associated protein in cytosolic stress signaling. J Biol Chem 277:31918–31928 [DOI] [PubMed] [Google Scholar]
- Kim JH, Lane WS, Reinberg D (2002) Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc Natl Acad Sci USA 99:1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann D, Hatada EH, Tegethoff S, Li J, Klippel A, Giese K, Baeuerle PA, Scheidereit C (2000) The Ik-B kinase (IKK) complex tripartite and contains IKKγ but not IKAP as a regular component. J Biol Chem 275:29779–29787 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Greenblatt JF (2001) Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol Cell Biol 21:8203–8212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22:6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Takagi Y, Jiang Y, Tokunaga M, Erdjument-Bromage H, Tempst P, Kornberg RD (2001) A multiprotein complex that interacts with RNA polymerase II elongator. J Biol Chem 276:29628–29631 [DOI] [PubMed] [Google Scholar]
- Mayer K, Ballhausen W, Leistner W, Rott H (2000) Three novel types of splicing aberrations in the tuberous sclerosis TSC2 gene caused by mutations apart from splice consensus sequences. Biochim Biophys Acta 1502:495–507 [DOI] [PubMed] [Google Scholar]
- Nissim-Rafinia M, Kerem B (2002) Splicing regulation as a potential genetic modifier. Trends Genet 18:123–127 [DOI] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell 3:109–118 [DOI] [PubMed] [Google Scholar]
- Pagani F, Buratti E, Stuani C, Romano M, Zuccato E, Niksic M, Giglio L, Faraguna D, Baralle FE (2000) Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J Biol Chem 275:21041–21047 [DOI] [PubMed] [Google Scholar]
- Pearson J (1979) Familial dysautonomia (a brief review). J Auton Nerv Syst 1:119–126 [DOI] [PubMed] [Google Scholar]
- Pearson J, Pytel BA (1978a) Quantitative studies of ciliary and sphenopalatine ganglia in familial dysautonomia. J Neurol Sci 39:123–130 [DOI] [PubMed] [Google Scholar]
- ——— (1978b) Quantitative studies of sympathetic ganglia and spinal cord intermedio-lateral gray columns in familial dysautonomia. J Neurol Sci 39:47–59 [DOI] [PubMed] [Google Scholar]
- Pearson J, Pytel BA, Grover-Johnson N, Axelrod FB, Dancis J (1978) Quantitative studies of dorsal root ganglia and neuropathologic observations on spinal cords in familial dysautonomia. J Neurol Sci 35:77–92 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell 9:799–809 [DOI] [PubMed] [Google Scholar]
- Riley CM, Day RL, Greely D, Langford WS (1949) Central autonomic dysfunction with defective lacrimation. Pediatrics 3:468–477 [PubMed] [Google Scholar]
- Schmittgen TD, Zakrajsek BA (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46:69–81 [DOI] [PubMed] [Google Scholar]
- Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, Liebert CB, Chadwick B, Idelson M, Reznik L, Robbins CM, Makalowska I, Brownstein MJ, Krappmann D, Scheidereit C, Maayan C, Axelrod FB, Gusella JF (2001) Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet 68:598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer J, Jung CK, Shackel I, Williams PM (1999) Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem 270:41–49 [DOI] [PubMed] [Google Scholar]
- Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2002) Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci USA 99:3517–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler GS, Petrakis TG, Ethelberg S, Tokunaga M, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2001) RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J Biol Chem 276:32743–32749 [DOI] [PubMed] [Google Scholar]
- Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ (1999) A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell 4:123–128 [DOI] [PubMed] [Google Scholar]