Abstract
Autosomal dominant polycystic liver disease (ADPLD) is a distinct clinical and genetic entity that can occur independently from autosomal dominant polycystic kidney disease (ADPKD). We previously studied two large kindreds and reported localization of a gene for ADPLD to an ∼8-Mb region, flanked by markers D19S586/D19S583 and D19S593/D19S579, on chromosome 19p13.2-13.1. Expansion of these kindreds and identification of an additional family allowed us to define flanking markers CA267 and CA048 in an ∼3-Mb region containing >70 candidate genes. We used a combination of denaturing high-performance liquid chromatography (DHPLC) heteroduplex analysis and direct sequencing to screen a panel of 15 unrelated affected individuals for mutations in genes from this interval. We found sequence variations in a known gene, PRKCSH, that were not observed in control individuals, that segregated with the disease haplotype, and that were predicted to be chain-terminating mutations. In contrast to PKD1, PKD2, and PKHD1, PRKCSH encodes a previously described human protein termed “protein kinase C substrate 80K-H” or “noncatalytic beta-subunit of glucosidase II.” This protein is highly conserved, is expressed in all tissues tested, and contains a leader sequence, an LDLa domain, two EF-hand domains, and a conserved C-terminal HDEL sequence. Its function may be dependent on calcium binding, and its putative actions include the regulation of N-glycosylation of proteins and signal transduction via fibroblast growth-factor receptor. In light of the focal nature of liver cysts in ADPLD, the apparent loss-of-function mutations in PRKCSH, and the two-hit mechanism operational in dominant polycystic kidney disease, ADPLD may also occur by a two-hit mechanism.
Introduction
Polycystic liver disease is characterized by the presence of multiple bile duct–derived epithelial cysts scattered in the liver parenchyma (Torres 1996). It often occurs in association with autosomal dominant polycystic kidney disease (ADPKD [MIM 173900 and MIM 173910]), but it also exists as a distinct entity (ADPLD [MIM 174050]). Polycystic kidneys were detected in only one-half of polycystic liver disease cases in old autopsy or surgical series (Comfort et al. 1952; Melnick 1955). A large and more recent retrospective study of medicolegal autopsies in Finland also found that polycystic liver disease often occurs as an entity separate from ADPKD (Karhunen and Tenhu 1986). Further evidence that ADPLD is a novel inherited disease came from three studies in which isolated ADPLD was excluded from genetic linkage to the PKD1 or PKD2 loci for ADPKD (Somlo et al. 1995; Pirson et al. 1996; Iglesias et al. 1999). Finally, we recently identified a locus for ADPLD on chromosome 19p13.2-13.1 (Reynolds et al. 2000).
ADPLD is characterized by an overgrowth of biliary epithelium and supporting connective tissue (Torres 1996). As in polycystic liver disease associated with ADPKD, the cysts derive from focal dilatations of small clusters of intralobular bile ductules surrounded by fibrous tissue known as biliary microhamartomas (Ramos et al. 1990; Qian et al. 2003). Some cysts in polycystic liver disease derive by cystic dilatation of the peribiliary glands that surround and communicate with the large intrahepatic bile ducts (Kida et al. 1992; Qian et al. 2003). Other lesions less consistently found in polycystic livers include dilatation of the intrahepatic and extrahepatic bile ducts and focal biliary fibroadenomatosis (Torres 1996). The latter are characterized by fibrosis and enlargement of the portal tracts and proliferation of the bile ducts.
ADPLD is often asymptomatic (Qian et al. 2003). As a consequence, the disease may go undetected and is likely to be underdiagnosed in the general population. When symptoms occur, they are usually either due to mass effects from the expanding cyst burden or due to hemorrhage, infection, or rupture of cysts. Liver metabolic and synthetic functions remain normal. Symptoms caused by the mass effect of the cysts include abdominal distention, early satiety, dyspnea, and back pain. Rarely, ascites can form because of hepatic venous outflow obstruction by cysts, and lower extremity edema can occur secondary to compression of the inferior vena cava (Torres et al. 1994). Most patients with polycystic liver disease require no treatment. In highly symptomatic patients, percutaneous cyst aspiration and sclerosis, cyst fenestration, partial hepatectomy, and liver transplantation may be indicated. Determining factors regarding therapy include the extent, distribution, and anatomy of the cysts, as well as the nature and severity of the symptoms (Que et al. 1995).
Subsequent to our initial discovery of genetic linkage, we narrowed the ADPLD candidate interval to ∼3 Mb and focused our mutation screening efforts on ∼70 genes in the two-thirds of this interval that is conserved in the mouse. We discovered heterozygous putative loss-of-function mutations in PRKCSH, the gene encoding a protein varyingly called “protein kinase C substrate 80K-H” or “the beta-subunit of glucosidase II.” The protein is widely expressed in tissues and is highly conserved as a single-copy gene in evolution from fission yeast to man. The PRKCSH gene product is predicted to be an endoplasmic reticulum luminal protein that recycles from the Golgi. It is predicted to contain low-density lipoprotein receptor domain class A (LDLa) and EF-hand domains, suggesting that Ca2+ binding may play a role in its function. We report that PRKCSH functions in cyst formation in lumen-forming epithelial tissues.
Subjects and Methods
Subjects
Blood samples for DNA analysis were obtained from subjects belonging to 25 unrelated families with ADPLD after they signed an informed written consent form, in accordance with institutional review board–approved protocols. Subjects from 19 families were specifically recruited for this project and were studied at the General Clinical Research Center of the Mayo Clinic (Qian et al. 2003). Six additional patients were referred from other centers. A panel of DNA samples from 15 of the 25 index cases was used for the initial screening for mutations in candidate genes. All patients were evaluated by abdominal ultrasonography, computed tomography scan, or magnetic resonance scan. No patients—or family members, when available—met the diagnostic criteria for ADPKD (Ravine et al. 1994). Subjects aged ⩽40 years who had any liver cysts and those aged >40 years who had at least four liver cysts were considered affected (Reynolds et al. 2000).
Genotyping and Linkage Analysis
Genomic DNA was extracted from EDTA-treated blood with the PureGene Kit (Gentra Systems). Linkage analysis to the disease interval on chromosome 19 was performed with 25 microsatellite markers between D19S586 (32.94 cM) and D19S593 (45.5 cM) labeled with HEX or FAM dye (table A1; fig. 1). PCR products were separated by electrophoresis, using an ABI 3700 (Applied Biosystems), and were analyzed by Genescan and Genotyper software (version 3.5; Applied Biosystems). Allele calls by Genotyper were checked manually for accuracy. Multipoint LOD scores were computed using GENEHUNTER (version 2.1); two-point linkage analysis used the MLINK (version 5.1) program of the LINKAGE package. The ADPLD allele frequency in the population was set at 0.0002, the phenocopy rate was set to 0.01, and the heterozygous disease penetrance was estimated to be 95%. Marker allele frequencies were set to 1/n, where n is the number of alleles observed in the pedigrees analyzed.
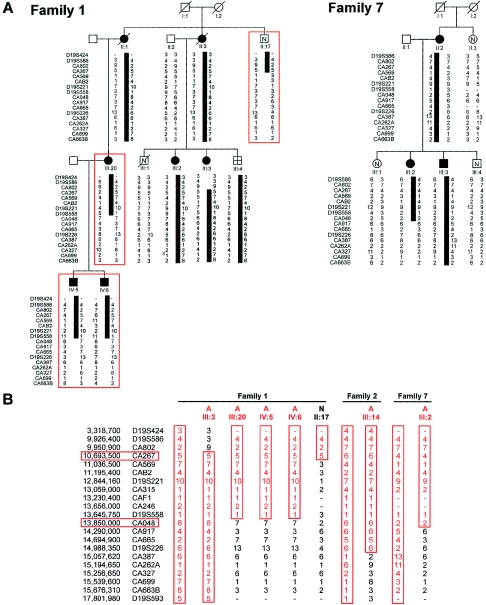
Figure 1.
Refinement of the genetic interval for ADPLD. A, Partial representation of the previously published family 1 (Reynolds et al. 2000), showing the disease-associated haplotype (black bar) and critical recombination events in newly recruited individuals (red boxes). Family 7 is a newly ascertained kindred with possible linkage and a centromeric recombination. B, Schematic representation of the disease-associated haplotypes (all red) in two large kindreds (Reynolds et al. 2000) and family 7, along with critical recombinant chromosomes that define the closest flanking markers (red boxes). Individual identifications correspond to previously published pedigrees for families 1 and 2 (Reynolds et al. 2000).
Identification of Candidate Genes
Genomic sequences were initially retrieved from the Joint Genome Institute database. As genomic sequences became available, each BAC and cosmid clone was analyzed by RepeatMasker followed by gene prediction programs GENSCAN and FGENES. BLAST analyses against the GenBank nonredundant (nr) and EST (dbEST) databases via the National Center for Biotechnology Information (NCBI) Web site were used to identify genes and ESTs mapping to the region of interest. Candidate sequences were further annotated by searches in a series of databases, including UniGene, PubMed, and GenBank (on the NCBI Web site), as well as Celera. Since the completion of the Human Genome Project draft sequence, we have integrated the annotated genome resources from the Human Genome Mapviewer [build 30] and Ensembl Genome Browser into our own gene-prediction data.
Expression of transcripts in liver was confirmed by RT-PCR. Poly-A+ RNA from human adult liver, kidney, brain, and testis was obtained from Clontech and cDNA was synthesized by use of the SUPERSCRIPT Preamplification System for First Strand cDNA Synthesis (Invitrogen). RT-PCR was performed by use of primer-specific reaction conditions.
Mutation Screening
Primers were designed using the program Primer 3, on the basis of the annotated genomic sequence across each exon, including regions of at least 20–40 bp of flanking intervening sequence. A list of candidate genes screened is provided in table A2. The primers for PRKCSH are given in table A3. In case of exons >400 bp in size, overlapping primer pairs were designed to keep amplicons <400 bp in size. PCR was performed in 30-μl reaction volumes using a GeneAmp PCR System 9700 (PE Applied Biosystems). The PCR mixture contained 50 ng DNA, 5 mmol/l of each dNTP (Boehringer Mannheim), 1 U of AmpliTaq DNA Polymerase (PE Applied Biosystems) and 5 pmol of each sense and antisense primer in a reaction buffer (0.5 mmol MgCl2, 10 mmol Tris-HCl pH 8.3, 50 mmol KCl). Reactions were first heated at 95°C for 3 min, followed by 35 cycles of PCR amplifications (at 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s). PCR products were directly analyzed by denaturing high-performance liquid chromatography (DHPLC) on The Wave DNA Fragment Analysis System (Transgenomic). The reference sequences of the amplicons (wild-type DNA) are imported to the WAVEMaker software version 3.3 (and, subsequently, version 4.1), to generate a method that gives sequence-specific separation temperature and separation gradient for the analyzed DNA fragment. Eight to 12 μl of each PCR reaction was injected onto the column and was eluted with a linear gradient at a flow rate of 0.9 ml/min. The mobile phase consisted of a mixture of buffer A (0.1 M TEAA and 1 mM EDTA) and buffer B (25% acetonitrile in 0.1 M TEAA). If the resolution of the DHPLC profiles was not adequate, a second temperature—typically 2°C above or below the first—was used to improve the resolution. Samples displaying altered elution properties were sequenced bidirectionally using Big Dye Terminator Cycle Sequencing Reactions on an ABI 3700 (PE Applied Biosystems). Sequence electropherograms were compared with gene sequence from GenBank and control samples. Variants identified were tested in 20 normal control subjects; if not present, an additional 66 normal control subjects (total 86 samples, 172 chromosomes) were tested, as well as segregation among family members, if available.
Expression Analysis
A 386-bp RT-PCR product from exons 10–13 of PRKCSH was 32P-labeled using the multiprime method and was hybridized to an adult human MTN blot (Clontech), as described elsewhere (Onuchic et al. 2002). The putative splice-site variant in family 1 was examined by RT-PCR from mRNA extracted from Epstein-Barr–transformed lymphoblasts of an affected individual in family 1. Primers spanning exons 2–17 of PRKCSH amplified the entire ORF (table A3). Direct sequencing in the reverse direction showed skipping of exon 16.
Results
We had previously studied two large kindreds and reported localization of a gene for ADPLD without kidney cysts to an ∼8-Mb region flanked by markers D19S586/D19S583 and D19S593/D19S579 on chromosome 19p13.2-13.1 (Reynolds et al. 2000). As the next step in the identification of the underlying gene defect, we refined the genetic interval by fine genetic mapping and by identification of additional disease chromosomes with informative recombination events. For fine mapping, we used the available genomic sequence in the region to identify additional microsatellite markers (table A1). Using the previously reported recombinant chromosomes, we were able to refine the interval to ∼5 Mb flanked by CA802 and CA387 (fig. 1). Recruitment of additional members of family 1 (II:17, III:20, IV:5, and IV:6) permitted further refinement of the ADPLD interval, to ∼3 Mb flanked by CA267 and CA048 (fig. 1). The closest flanking centromeric marker (CA048) was also supported by a recombination event, in the newly identified family 7, that showed possible linkage to chromosome 19 (multipoint Zmax=1.12) (fig. 1).
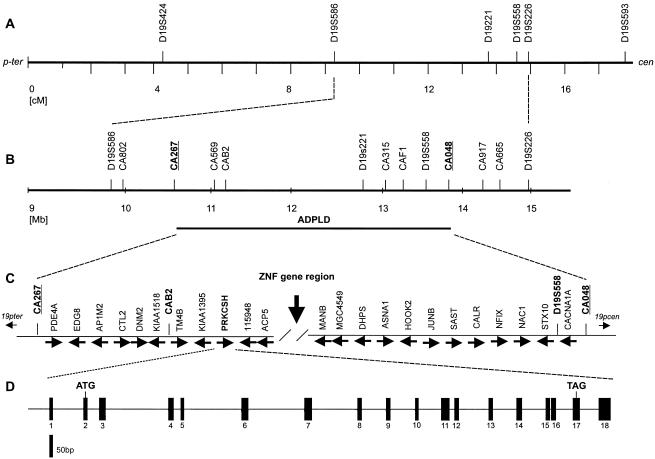
This region of the human genome is very gene rich and did not present any particularly enticing positional candidates on the basis of known structures or functions of polycystin-1 and -2 (fig. 2). To aid in disease-gene identification, we divided the ADPLD genomic region conceptually into three general regions, each ∼1 Mb in length. The telomeric portion, bounded by CA267 and the acid phosphatase 5 gene (ACP5), is syntenic with mouse chromosome 9 and contains ∼25 known genes and ∼10 unknown predicted genes (fig. 2). The centromeric portion, flanked by D19S221 and CA048, is syntenic with mouse chromosome 8 and also contains ∼25 known genes and ∼10 unknown predicted genes (fig. 2). The middle of the interval, ∼1 Mb between ACP5 and D19S221 is not conserved in mouse and contains a large cluster of zinc finger genes (Bellefroid et al. 1995). The latter region was excluded from mutation detection, because we hypothesized that—as is the case with ADPKD (Wu et al. 1998)—the liver cystic pathway in ADPLD will be conserved in mice. In the remainder of the region, we excluded genes from initial mutation detection on the basis of any of the following criteria: (1) the absence of transcript detected by RT-PCR in liver tissue (CNN1, KLF1), (2) that the gene is known to be mutated in another disease (LDLR, EPOR, MAN2B1, CACNA1A), (3) mouse knockout with no liver phenotype (CDKN2D, SMARCA4), (4) genes encoding ribosomal proteins (RPSL30, RPSL18), or (5) enzymes (RNASEH1, TGT, DNASE2, GCDH, FARSL). We sought candidate genes expressed exclusively in liver by screening liver, kidney, brain, and testis cDNA by RT-PCR, but none were found. Although families 1 and 2 were not known to be related (Reynolds et al. 2000), we tested for a possible founder mutation by haplotype segregation analysis, in an effort to extract additional genetic information to narrow the candidate region. Informative sequence variants, SNPs, and small insertion/deletion polymorphisms discovered during mutation detection were analyzed by direct sequencing and were tested by segregation. No common haplotype was observed.
Figure 2.
Positional cloning strategy for ADPLD. A, Genetic map position for microsatellite markers used in linkage mapping. Sex-averaged genetic distances (in cM), from the Marshfield map, are shown. B, Physical map of critical interval, showing newly identified markers including the closest flanking microsatellites CA267 and CA048 (bold). C, Partial representation of known genes and their direction of transcription (arrows). ZNF gene region, ∼1 Mb containing zinc finger genes, that is not conserved in the mouse genome. D, Genomic organization of PRKCSH; exons indicated by vertical bars.
Our mutation screening panel consisted of 15 unrelated affected individuals. Of these, 10 were members of families with at least one other affected member. Although some of the families were large, our ability to determine genetic linkage was hampered by the late onset of the disease—clinical diagnosis of ADPLD cannot be excluded in individuals under age 40 years, particularly not in men (Qian et al. 2003). To compensate for the lack of a large number of individuals with proven linkage to chromosome 19, mutation detection was carried out by a bipartite strategy (table A2). Mutation screening by DHPLC heteroduplex analysis was carried out in 15 unrelated affected individuals. In parallel, mutation detection by direct sequencing was performed in one affected individual from each of the three families with proven or suggestive linkage (families 1, 2 and 7). We chose to screen known genes in the region first (table A2).
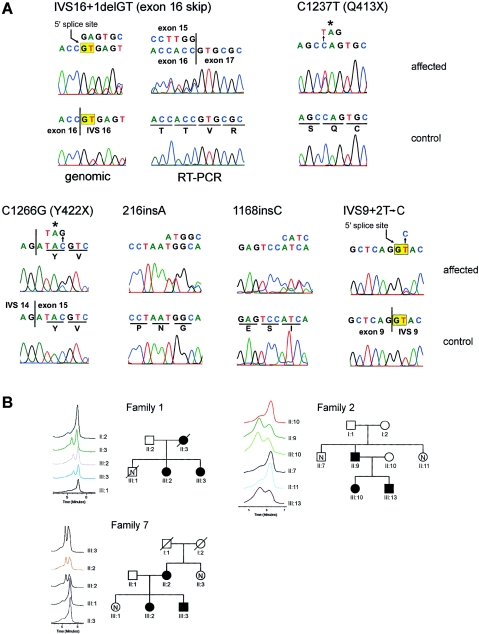
We detected sequence variations in PRKCSH by DHPLC and sequencing that were not observed in 86 normal control individuals (172 chromosomes) and that segregated with the disease haplotype, when it was known (table 1; fig. 3). In addition, we observed several intragenic polymorphisms (table A4). PRKCSH is an ∼15-kb gene encoded in 18 exons, of which the first and last are untranslated (fig. 2). The predicted effect of all of the pathogenic sequence variants we found is premature termination of translation. The mutations occur throughout the gene, from exon 4 to exon 16. The mutation in family 1 (Reynolds et al. 2000) results in deletion of the 5′ splice site in IVS16. We tested the effect of this mutation on the PRKCSH transcript in patient lymphoblasts by RT-PCR. We found skipping of exon 16 in a transcript in which exon 15 is spliced to exon 17, causing a frame shift and premature termination (fig. 3A). This mutation segregates on the affected haplotype in family 1 (fig. 3B). The mutation in family 2 (Reynolds et al. 2000) is a nucleotide substitution 1237C→T resulting in a premature termination codon, Q413X (fig. 3A). This mutation segregates with the affected haplotype (fig. 3B). The mutation in family 7 is also nucleotide substitution producing a stop codon that segregates in all affected individuals (fig. 3). Two families (25 and O-1) have single-nucleotide insertions in exons 4 and 13, respectively, that are predicted to result in premature terminations after frame shifts (fig. 3). The mutation in family 41 is predicted to disrupt the 5′ splice site in IVS9 (table 1; fig. 3). The most likely result of all of these mutations is loss of PRKCSH function.
Table 1.
Mutations in PRKCSH in Patients with ADPLD
| Family | Exon | Nucleotide Changea | ORF Changeb | No. of Affected Individualsb |
| 25 | 4 | 216insA | N72Xfs84 | 2 |
| 41 | 9 | IVS9+2T→C | Splice site | 1 |
| O-1 | 13 | 1168insC | I390Xfs400 | 1 |
| 2 | 14 | 1237C→T | Q413X | 16 |
| 7 | 15 | 1266C→G | Y422X | 3 |
| 1 | IVS16 | IVS16+1delGT | Exon 16 skip; G446Xfs457 | 13 |
Nomenclature for description of sequence variations is from the Leiden Muscular Dystrophy Pages Web site. None of these variants were observed in 172 normal chromosomes.
Number of affected individuals in the respective families—all variants segregated with the disease haplotype where the haplotype could be established.
Figure 3.
Mutations in PRKCSH in ADPLD. A, Sequence electropherograms showing mutant (affected) and wild-type (control) sequences. All template DNA was genomic except as indicated for the IVS16+1delGT mutation where both genomic (left) and RT-PCR (right) was analyzed. Six mutations are shown. B, Segregation of mutations in families 1 (IVS16+1delGT), 2 (C1237T), and 7 (C1266G), as analyzed by DHPLC. Only selected traces are shown, but the entire family was analyzed. Individual identifications correspond to previously published pedigrees (Reynolds et al. 2000) and figure 1.
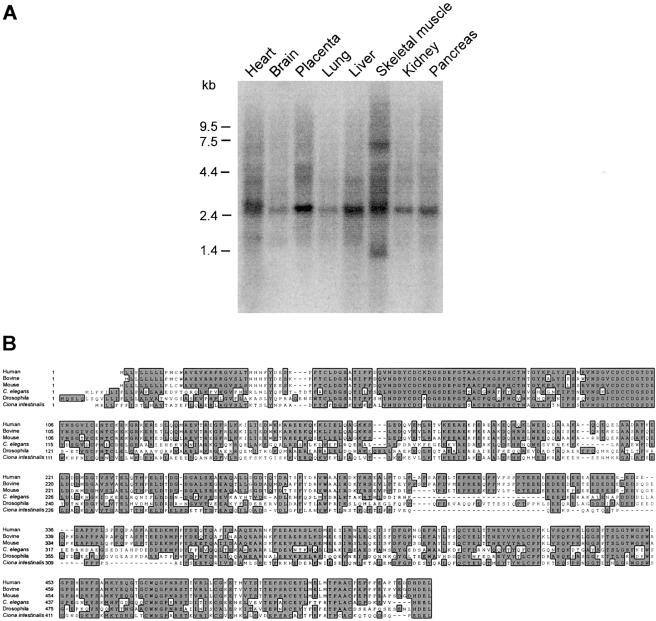
The tissue-expression pattern of PRKCSH has not been reported. A 2.5–3.0-kb transcript for this gene is expressed in all tissues tested (fig. 4). In addition, cardiac muscle seems to have a slightly larger transcript (∼3.0 kb) and skeletal muscle has a 7.0–7.5-kb transcript that is detected even at high stringency (fig. 4). PRKCSH is predicted to encode a 527–amino acid protein that is very highly conserved (fig. 4), even in Schizosaccharomyces pombe and Arabidopsis thaliana (data not shown). Structural features predicted by SMART and PFAM analysis include a leader sequence, and cysteine-rich LDLa type domain and one or two EF-hand domains. The latter two domains likely confer Ca2+ dependent regulation on the PRKCSH product. The terminal HDEL endoplasmic reticulum retention sequence is conserved throughout evolution (fig. 4). Mutations in PRKCSH cause ADPLD linked to chromosome 19p.
Figure 4.
Expression and conservation of PRKCSH. A, Tissue northern showing expression of PRKCSH in all tissues including liver and kidney. The transcript size is ∼2.5-3.0 kb with skeletal muscle showing evidence of a transcript >7.0 kb and cardiac muscle perhaps having a pair of transcripts in the 2.5-3.0 kb range. B, High degree of sequence conservation from C. elegans to humans. S. pombe and A. thaliana also have highly conserved PRKCSH homologs (not shown).
Discussion
We have identified PRKCSH as the gene responsible for ADPLD by positional cloning. In contrast to the polycystic kidney disease genes PKD1, PKD2, and PKHD1, PRKCSH encodes a previously described human protein called either “protein kinase C substrate 80K-H” (80K-H) or “beta-subunit of glucosidase II” (GIIβ). Similar to the other polycystic disease proteins, however, its function remains incompletely understood. The mutations found in patients with ADPLD are all predicted to cause premature chain termination. Although most occur in COOH-terminal exons 13–16, we also found mutations in exons 4 and 9 in the NH2-terminus half of the protein. In light of the nature and distribution of these mutations, it is most likely that they constitute loss-of-function changes, although, in the absence of functional studies, dominant negative or gain-of-function/loss-of-regulation effects in the residual protein products cannot be excluded. A common feature of all truncating mutations in 80K-H/GIIβ is the loss of the highly conserved terminal four amino acids, His-Asp-Glu-Leu (HDEL). This motif constitutes an endoplasmic reticulum (ER) luminal retention sequence (Trombetta et al. 1996; Arendt and Ostergaard 2000), and its loss presumably would result in failure to retain 80K-H/GIIβ in the ER. This would be significant, because the protein has a leader sequence and no membrane spans suggesting that without retention by HDEL-mediated binding to the KDEL receptor, it would enter the secretory pathway and be trafficked out of the cell (Arendt and Ostergaard 2000). There is precedent for reconciling loss-of-function changes with dominant inheritance in polycystic liver and kidney disease. Liver and kidney cyst formation can occur by a two-hit somatic mutation mechanism resulting in homozygous inactivation of PKD1 or PKD2 at the cellular level (Watnick et al. 1998; Wu et al. 1998). This mechanism has been invoked to explain the focal nature of cyst formation in affected organs but may not be the only factor determining the occurrence of cysts. Evidence exists for a potential role of compound heterozygous states and gene dosage (high or low) in cyst formation in ADPKD as well. However, given the similarity of the clinical presentations of liver disease in ADPLD and ADPKD, the two-hit hypothesis is extensible to ADPLD and bears further investigation.
The available functional data do not immediately suggest role for PRKCSH in a common pathway with the other polycystic disease genes, but the similarities in the liver phenotype warrant consideration of such a convergence of pathways. PRKCSH is broadly expressed in tissues including those with lumen-forming epithelia (e.g., liver, kidney, and pancreas). However, cyst formation is confined to the liver, suggesting that there may be tissue-specific factors required for cyst formation due to mutations in this gene. The 80K-H/GIIβ protein was initially identified during a search for protein kinase C substrates (Hirai and Shimizu 1990). Although 80K-H turned out to be a poor substrate for PKC, other possible functions were later suggested by its ability to bind advanced glycation end products (Li et al. 1996; Thornalley 1998) and by its rapid phosphorylation following activation of fibroblast growth-factor receptors (Shaoul et al. 1995; Goh et al. 1996; Kanai et al. 1997). More recently, homologous proteins were identified as the beta subunit of glucosidase II (GIIβ) (Trombetta et al. 1996; Arendt and Ostergaard 1997) and as a vacuolar system-associated protein-60 (VASAP-60) (Brule et al. 2000).
80K-H/GIIβ is an alternatively spliced protein that migrates as a doublet at ∼80 kDa on polyacrylamide gels (Arendt and Ostergaard 2000; Trombetta et al. 2001). It contains an NH2-terminal cysteine-rich LDLa region, two EF-hand domains, a highly acidic domain flanked by proline-rich segments presenting putative Grb2-binding domains, and the aforementioned HDEL endoplasmic reticulum luminal retention sequence (Arendt and Ostergaard 2000; Trombetta et al. 2001). An alternatively spliced region and two interaction domains responsible for the heterodimerization with the alpha catalytic subunit of glucosidase II (GIIα) have been described in the mouse GIIβ (Arendt and Ostergaard 2000). The overall domain structure of 80K-H/GIIβ suggests that it may be regulated by Ca2+ binding—the LDAa, EF-hand, and acidic regions are all potential Ca2+ interacting motifs. Interestingly, among the other polycystic disease gene products, polycystin-1 contains an LDLa motif in its extracellular portion and polycystin-2 is a Ca2+ channel and has an EF-hand in its COOH terminus.
GIIβ does not have enzymatic activity, but it is essential for the maturation of GIIα and its retention in the endoplasmic reticulum (D'Alessio et al. 1999; Treml et al. 2000). Glucosidase II plays a major role in regulation of proper folding and maturation of glycoproteins (Ellgaard et al. 1999). A preliminary step to protein N-glycosylation is the sequential addition of N-acetylglucosamine, mannose, and glucose to a dolichol pyrophosphate lipid carrier to form a mature oligosaccharide composed of two glucosamines, nine mannoses, and three glucoses (Freeze 1998). This preassembled oligosaccharide is transferred to an N-X-S/T sequence in a nascent polypeptide chain in the ER. Cleavage of the terminal glucose by glucosidase I and of the middle glucose by glucosidase II generates a monoglucosylated product (Ellgaard et al. 1999). This product is recognized by the chaperones calnexin and calreticulin and forms a complex with Erp57, a thiol reductase necessary for proper protein folding. By chance, calreticulin also maps to the ADPLD candidate region and was excluded by mutation detection (fig. 2; table A2). It is conceivable that alterations in the folding and maturation of specific glycoproteins results in the development of biliary cysts. Polycystin-1, polycystin-2, and fibrocystin/polyductin are all glycoproteins. It has been suggested that the polycystin-1/2 complex forms in the ER (Newby et al. 2002) and polycystin-2 is largely retained in the ER membrane (Cai et al. 1999) and only selectively trafficked to the primary cilia (Pazour et al. 2002). Improper association and trafficking of polycystins due to defective glycosylation with mutant GIIβ may tie ADPLD into the ADPKD pathway. Direct or indirect interaction of GIIβ with the polycystins can also be considered. Since 80K-H/GIIβ is an ER luminal protein, it could interact with the extracellular domains of either polycystin. Interestingly, autosomal recessive carbohydrate-deficient glycoprotein syndrome type Ib is associated with congenital hepatic fibrosis and results from nearly complete absence of phosphomannose isomerase, the enzyme needed to maintain the dolichol pyrophosphate-oligosaccharide pool (Jaeken et al. 1998; Niehues et al. 1998; Westphal et al. 2001). Understanding how mutations in PRKCSH lead to cyst formation in the liver and why they do not appear to affect the kidney will prove instructive in understanding all human polycystic diseases.
Acknowledgments
We thank the family members for their generous participation in this study. We thank Kristin Simonson and Patricia Urban for help with patient recruitment, Michael Ott and York Pei for referring patients, Joan Steitz and Richard Lifton for helpful discussions, and Asghar Rastegar for timely support. DNA sequencing was performed by the Keck Biotechnology Resource at Yale. This work was supported by National Institutes of Health (NIH) grant DK51041 (to S.S. and V.E.T.), Mayo Clinic General Clinical Research Center grant M01-RR00585, and Yale Liver Center Training Grant T32 DK07356 (to A.L.). A.L., S.D, L.F, X.T., and S.S. are members of the Yale Center for the Study of Polycystic Kidney Disease (NIH grant P50 DK57328).
Appendix A
Table A1.
Primer Sequences for New Microsatellite Markers
|
Primer |
|||||
| Marker | Position in Build 30(bp) | Forward | Reverse | Tm(°C) | Product Size(bp) |
| CA802 | 9950900 | ACTCTGGCAAACAACTTACAGAT | CCTTAATCCTGGCTCCCTTC | 60 | 152–170 |
| CA267 | 10693500 | TGCCCTTGGACACAAACATA | GAGGGTGGATCAAGCATCTG | 60 | 170–186 |
| CA569 | 11036500 | TGCTATGTGCTCATTGTAAACAG | CATGGATGTCTGTCTACAAG | 58 | 160–186 |
| CAB2 | 11195400 | ATTACAGGAATGAGCCACCAC | ATGCAAACCTACATAGGAAGC | 60 | 143–157 |
| CA315 | 13059000 | CGGGTTTCTCCATACTGGTC | GCAAAATAGGAAGTCCCTGTC | 60 | 164–174 |
| CAF1 | 13230400 | TTCTTCCCATTGCAGTTGTG | ACACATCCTCATTCAAAGTTC | 60 | 106–118 |
| CA246 | 13656000 | AGTTCTGCGTGGAATTGGAAG | CAGAGAGCAGTGTGTGGACAA | 58 | 139–149 |
| CA048 | 13850000 | TGTGGACTAGAGATGGAGCT | GCTGATTTATGTGTCATTTCTCC | 60 | 141–155 |
| CA917 | 14290000 | TTGGTCTCAAATTCCTGGCC | GCTCTTACAGGCTGTTCTTC | 60 | 148–170 |
| CA665 | 14694900 | GGTTCATTTTCTGCCTGGGG | GACCAGCAATTCCCCAATTCC | 58 | 191–205 |
| CA387 | 15057620 | CCAGGTCTTGTCCCTCCTAC | TCTTCCGTGTGTGAATCCAA | 60 | 172–198 |
| CA262A | 15194650 | AGCTGTCCCAAAGCTGAGTC | CACACAGACCAGCACACAAA | 60 | 162–178 |
| CA327 | 15256650 | CCAGCAGGAAAGCACAATAA | AGAGCCACATGGTGGAAACT | 60 | 145–169 |
| CA663B | 15676310 | CCCATTCAGAAATAACCTAGTCAC | GCCAAGATTGTGCCACTGTA | 60 | 163–179 |
| CA699 | 16016000 | ATTGAACTTGGCCTTGAGGA | AAGGGAGAGGGAGCGTATGT | 60 | 144–158 |
| CA255 | 17656190 | CGCAAGTCAATGCTTTTTGA | ATGATTGCACCACTGTACGC | 60 | 174–188 |
Table A2.
Candidate Genes Excluded by DHPLC and Direct Sequencing[Note]
| Gene Name | Gene | Locus | DHPLCa | Sequencingb |
| PDE4A | Phosphodiesterase 4A, cAMP-specific | 5141 | ✓ | |
| EDG8 | Endothelial differentiation, sphingolipid G-protein-coupled receptor, 8 | 53637 | ✓ | |
| MGC15906 | Hypothetical protein MGC15906 | 84971 | ✓ | |
| AK023011 | Hypothetical protein FLJ12949 | 65095 | ✓ | |
| AP1M2 | Adaptor-related protein complex 1, mu 2 subunit | 10053 | ✓ | |
| CTL2 | CTL2 | 57153 | ✓ | |
| ILF3 (NF90) | Interleukin enhancer binding factor 3 | 3609 | ✓ | |
| DNM2 | Dynamin 2 | 1785 | ✓ | |
| KIAA1518 | KIAA1518 protein | 25959 | ✓ | |
| AW245557 | EST AW245557 | AW245557 | ✓ | |
| TM4B | Tetraspanin TM4-B | 26526 | ✓ | ✓ |
| KIAA1395 | KIAA1395 protein | 57572 | ✓ | |
| RAB3D | Ras-related small GTP binding protein 3D | 9545 | ✓ | |
| ELAV3 | Hu antigen C | 1995 | ✓ | |
| AK023117 | Hypothetical protein FLJ13055 | 64748 | ✓ | |
| BE885128 | EST BE995128 | 126074 | ✓ | |
| BC011875 | Hypothetical protein DKFZp547J036 | 84241 | ✓ | |
| 115950 | Hypothetical protein BC016816 | 115950 | ✓ | |
| 115948 (MGC20983) | Hypothetical protein MGC20983 | 115948 | ✓ | |
| ECSIT | Signaling intermediate in Toll pathway-evolutionarily conserved | 51295 | ✓ | |
| MGC4549 (AK001171) | Hypothetical protein MGC4549 | 84337 | ✓ | |
| PTD008 | PTD008 protein | 51398 | ✓ | ✓ |
| DHPS | Deoxyhypusine synthase | 1725 | ✓ | |
| MGC4238 | Hypothetical protein MGC4238 | 84292 | ✓ | |
| ASNA1 | ArsA arsenite transporter, ATP-binding, homolog 1 | 439 | ✓ | |
| VMD2L1 (AK000139) | Vitelliform macular dystrophy 2-like protein 1 | 54831 | ✓ | |
| 199695 | LOC199695 | 199695 | ✓ | |
| MGC10870 | Hypothetical protein MGC10870 | 84261 | ✓ | |
| Hook 2 | Hook2 protein | 29911 | ✓ | ✓ |
| JUNB | Jun B proto-oncogene | 3726 | ✓ | |
| SAST | Syntrophin associated serine/threonine kinase | 22983 | ✓ | |
| CALR | Calreticulin | 811 | ✓ | |
| RAD23A | RAD23 homolog A | 5886 | ✓ | |
| NFIX | Nuclear factor I/X (CCAAT-binding transcription factor) | 4784 | ✓ | |
| LYL1 | Lymphoblastic leukemia derived sequence 1 | 4066 | ✓ | |
| FLJ20244 | Hypothetical protein FLJ20244 | 55621 | ✓ | |
| NAC1 | Transcriptional repressor NAC1 | 112939 | ✓ | |
| STX10 | Syntaxin 10 | 8677 | ✓ | ✓ |
| ETR101 | Immediate early protein | 9592 | ✓ |
Note.— ✓ denotes that the gene was excluded by this method.
DHPLC was performed across all exons under two conditions in 15 affected, unrelated individuals.
Direct sequencing of all exons in one affected individual from families 1, 2, and 7.
Table A3.
PCR Primers Used for Mutation Detection in PRKCSH
|
Oligonucleotide |
||||
| Exon | Sense | Antisense | Annealing Temperature(°C) | Product Size(bp) |
| 1a | GTTCGCGGGCATTTTTCAGGAAC | GATCTCCCCAATCCTGGCCA | 60 | 480 |
| 2a | CGGGAAACTGAGTCAAAAGG | GTGAAGACACAGCGCATCTC | 60 | 254 |
| 3a | GGCACTGAGCAGTGTTCAATAA | ATGGGAGGACAGAGGTGGTA | 60 | 244 |
| 4a | GTGATGGGAGGGGTACTGTC | TCTGTGGATGGATGGGACAT | 60 | 349 |
| 5a | GTGATGGGAGGGGTACTGTC | TCTGTGGATGGATGGGACAT | 60 | 349 |
| 6a | TTGCAAGCCCACACTATGAG | GCAGACCTGGTGGATCCTAA | 60 | 244 |
| 7a | GCAGCATGATCAAAAACCTG | AGCTGGTCTCTTGCCTTCTG | 60 | 274 |
| 8a | AAGGAGGATCTGGCTGGTTT | GGGTGACAGAGGTGGCTTCT | 60 | 205 |
| 9a | CTCCCTAGAAGTCCCCAACC | AGGTCCTGAAGCAAGTTCCA | 60 | 240 |
| 10a | CAACCACTCCAGCCCCTGGT | CCAGGTGCCAGAACCAGAGG | 61 | 390 |
| 11a | GCAGGAGGGGCAGAGACACC | CCAAGATACTGGGGCTTGTG | 60 | 500 |
| 12a | GCAGGAGGGGCAGAGACACC | CCAAGATACTGGGGCTTGTG | 60 | 500 |
| 13a | CTGGGAGTCAAGGAGCAGTC | ATGAGGGTATGGGAGCACAC | 59 | 227 |
| 14a | TTCCCCAACCATCAGGAAACTG | AGACCCTCCTGTGTCTGTCG | 60 | 260 |
| 15a | TCCCCTGCCTTGCAGGCCT | GTTCCCCAAC CCATATGTCCC | 60 | 320 |
| 16a | TCCCCTGCCTTGCAGGCCT | GTTCCCCAAC CCATATGTCCC | 60 | 320 |
| 17a | GGTCCATCTTCCTCAGGGCC | CGAGCACCCG TCTGCCCATC | 60 | 320 |
| 18a | CTGGTCAACTCCTGGCCTCA | CACACCCCAGCAAAGCGAGG | 60 | 480 |
| 10–13b | HAF: ACAGACAGACGCCACCTCTT | HAR: TGGACTCCTCCATGTCCTTC | 60 | 386 |
| 2–17c | F2: GTGAAGACACAGCGCATCTC | 17R: GGTCCATCTTCCTCAGGGCC | 60 | 1741 |
| 10–17c | HAF: ACAGACAGACGCCACCTCTT | Ex17R: GGTCCATCTTCCTCAGGGCC | 60 | 893 |
| 1–4d | F1: CTGCTGGACAAGAGGGGTGC | SCHR3: GACCCGGTTGGAGGGGATATA | 60 | 402 |
| 1–8d | F1: CTGCTGGACAAGAGGGGTGC | HBR: TGTCATCATCCAGCTCCTTG | 60 | 800 |
| 1–11d | F1: CTGCTGGACAAGAGGGGTGC | HCR: CTCCTCCTCCTCCTCTGTGG | 60 | 1077 |
| 1–13d | F1: CTGCTGGACAAGAGGGGTGC | HAR : TGGACTCCTCCATGTCCTTC | 60 | 1290 |
| 2–4d | F2: GTGAAGACACAGCGCATCTC | SCHR3: GACCCGGTTGGAGGGGATATA | 60 | 313 |
| 2–8d | F2: GTGAAGACACAGCGCATCTC | HBR: TGTCATCATCCAGCTCCTTG | 60 | 711 |
| 2–11d | F2: GTGAAGACACAGCGCATCTC | HCR: CTCCTCCTCCTCCTCTGTGG | 60 | 988 |
| 2–13d | F2: GTGAAGACACAGCGCATCTC | HAR : TGGACTCCTCCATGTCCTTC | 60 | 1201 |
| 2–17d | F2: GTGAAGACACAGCGCATCTC | 17R: GGTCCATCTTCCTCAGGGCC | 60 | 1741 |
PCR primers and condition for exon-by-exon amplification and mutation detection by DHPLC and direct sequencing.
RT-PCR for Northern probe.
RT-PCR for IVS16+1delGT mutation.
RT-PCR IVS2+6delTCC mutation.
Table A4.
Polymorphisms in PRKCSH
| Exon | Nucleotide Change | ORF Change |
| 2 | IVS2+6delTCC | No |
| 2 | IVS2-75C→T | No |
| 3 | IVS3-7C→G | No |
| 5 | IVS5-32A→C | No |
| 5 | IVS5-36G→C | No |
| 6 | IVS6-5C→T | No |
| 9 | IVS9+76insG | No |
| 11 | G871A | A291T |
| 11 | 956insGGA | 319insE |
| 18 | C1769T | No |
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Celera, http://cds.celera.com/cds/login.cfm
- Ensembl Genome Browser, http://www.ensembl.org/
- FGENES, http://genomic.sanger.ac.uk/gf/gf.shtml
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for PRKCSH homologs PRKCSH [J03075] and Prkcsh [BC009816], Bos taurus [U49178], Caenorhabditis elegans [NM_063672], Drosophila melanogaster [AY058725], Ciona intestinalis [AK112399], S. pombe [D89245], and A. thaliana, [NM_148139])
- GENSCAN, http://genes.mit.edu/GENSCAN.html
- Human Genome Mapviewer, http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/map_search
- Joint Genome Institute, http://www.jgi.doe.gov/
- Leiden Muscular Dystrophy pages, http://www.dmd.nl/mutnomen.html (for nomenclature for the description of sequence variations)
- Marshfield Medical Research Foundation, Center for Medical Genetics, http://research.marshifieldclinic.org/genetics
- NCBI, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ADPKD [MIM 173900, 173910] and ADPLD [MIM 174050])
- PFAM Home Page, http://pfam.wustl.edu/index.html (for EF hand [PF00036])
- Primer 3, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi
- RepeatMasker, http://ftp.genome.washington.edu/cgi-bin/RepeatMasker
- SMART, http://dylan.embl-heidelberg.de/ (for LDLa domain [SMART # SM0192])
- UniGene, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene
References
- Arendt CW, Ostergaard HL (1997) Identification of the CD45-associated 116-kDa and 80-kDa proteins as the alpha- and beta-subunits of alpha-glucosidase II. J Biol Chem 272:13117–13125 [DOI] [PubMed] [Google Scholar]
- ——— (2000) Two distinct domains of the beta-subunit of glucosidase II interact with the catalytic alpha-subunit. Glycobiology 10:487–492 [DOI] [PubMed] [Google Scholar]
- Bellefroid EJ, Marine JC, Matera AG, Bourguignon C, Desai T, Healy KC, Bray-Ward P, Martial JA, Ihle JN, Ward DC (1995) Emergence of the ZNF91 Kruppel-associated box-containing zinc finger gene family in the last common ancestor of anthropoidea. Proc Nat Acad Sci USA 92:10757–10761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brule S, Rabahi F, Faure R, Beckers JF, Silversides DW, Lussier JG (2000) Vacuolar system-associated protein-60: a protein characterized from bovine granulosa and luteal cells that is associated with intracellular vesicles and related to human 80K-H and murine beta-glucosidase II. Biol Reprod 62:642–654 [DOI] [PubMed] [Google Scholar]
- Cai Y, Maeda Y, Cedzich A, Torres VE, Wu G, Hayashi T, Mochizuki T, Park JH, Witzgall R, Somlo S (1999) Identification and characterization of polycystin-2, the PKD2 gene product. J Biol Chem 274:28557–28565 [DOI] [PubMed] [Google Scholar]
- Comfort MW (1952) Polycystic disease of the liver: a study of 24 cases. Gastroenterology 20:60–78 [PubMed] [Google Scholar]
- D'Alessio C, Fernandez F, Trombetta ES, Parodi AJ (1999) Genetic evidence for the heterodimeric structure of glucosidase II: the effect of disrupting the subunit-encoding genes on glycoprotein folding. J Biol Chem 274:25899–25905 [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A (1999) Setting the standards: quality control in the secretory pathway. Science 286:1882–1888 [DOI] [PubMed] [Google Scholar]
- Freeze HH (1998) Disorders in protein glycosylation and potential therapy: tip of an iceberg? J Pediatr 133:593–600 [DOI] [PubMed] [Google Scholar]
- Goh KC, Lim YP, Ong SH, Siak CB, Cao X, Tan YH, Guy GR (1996) Identification of p90, a prominent tyrosine-phosphorylated protein in fibroblast growth factor-stimulated cells, as 80K-H. J Biol Chem 271:5832–5838 [DOI] [PubMed] [Google Scholar]
- Hirai M, Shimizu N (1990) Purification of two distinct proteins of approximate Mr 80,000 from human epithelial cells and identification as proper substrates for protein kinase C. Biochem J 270:583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias DM, Palmitano JA, Arrizurieta E, Kornblihtt AR, Herrera M, Bernath V, Martin RS (1999) Isolated polycystic liver disease not linked to polycystic kidney disease 1 and 2. Dig Dis Sci 44:385–388 [DOI] [PubMed] [Google Scholar]
- Jaeken J, Matthijs G, Saudubray JM, Dionisi-Vici C, Bertini E, de Lonlay P, Henri H, Carchon H, Schollen E, Van Schaftingen E (1998) Phosphomannose isomerase deficiency: a carbohydrate-deficient glycoprotein syndrome with hepatic-intestinal presentation. Am J Hum Genet 62:1535–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M, Goke M, Tsunekawa S, Podolsky DK (1997) Signal transduction pathway of human fibroblast growth factor receptor 3. Identification of a novel 66-kDa phosphoprotein. J Biol Chem 272:6621–6628 [DOI] [PubMed] [Google Scholar]
- Karhunen PJ, Tenhu M (1986) Adult polycystic liver and kidney diseases are separate entities. Clin Genet 30:29–37 [DOI] [PubMed] [Google Scholar]
- Kida T, Nakanuma Y, Terada T (1992) Cystic dilatation of peribiliary glands in livers with adult polycystic disease and livers with solitary nonparasitic cysts: an autopsy study. Hepatology 16:334–340 [DOI] [PubMed] [Google Scholar]
- Li YM, Mitsuhashi T, Wojciechowicz D, Shimizu N, Li J, Stitt A, He C, Banerjee D, Vlassara H (1996) Molecular identity and cellular distribution of advanced glycation endproduct receptors: relationship of p60 to OST-48 and p90 to 80K-H membrane proteins. Proc Natl Acad Sci USA 93:11047–11052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick PJ (1955) Polycystic liver: analysis of seventy cases. Arch Pathol 59:162–172 [PubMed] [Google Scholar]
- Newby LJ, Streets AJ, Zhao Y, Harris PC, Ward CJ, Ong AC (2002) Identification, characterization, and localization of a novel kidney polycystin-1-polycystin-2 complex. J Biol Chem 277:20763–20773 [DOI] [PubMed] [Google Scholar]
- Niehues R, Hasilik M, Alton G, Korner C, Schiebe-Sukumar M, Koch HG, Zimmer KP, Wu R, Harms E, Reiter K, von Figura K, Freeze HH, Harms HK, Marquardt T (1998) Carbohydrate-deficient glycoprotein syndrome type Ib. Phosphomannose isomerase deficiency and mannose therapy. J Clin Invest 101:1414–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuchic LF, Furu L, Nagasawa Y, Hou X, Eggermann T, Ren Z, Bergmann C, Senderek J, Esquivel E, Zeltner R, Rudnik-Schoneborn S, Mrug M, Sweeney W, Avner ED, Zerres K, Guay-Woodford LM, Somlo S, Germino GG (2002) PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel beta-helix 1 repeats. Am J Hum Genet 70:1305–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, San Agustin JT, Follit JA, Rosenbaum JL, Witman GB (2002) Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr Biol 12:R378–R380 [DOI] [PubMed] [Google Scholar]
- Pirson Y, Lannoy N, Peters D, Geubel A, Gigot JF, Breuning M, Verellen-Dumoulin C (1996) Isolated polycystic liver disease as a distinct genetic disease, unlinked to polycystic kidney disease 1 and polycystic kidney disease 2. Hepatology 23:249–252 [DOI] [PubMed] [Google Scholar]
- Qian Q Li A, King BF, Kamath PS, Lager DJ, Huston J III, Shub C, Davila S, Somlo S, Torres VE (2003) Clinical profile of autosomal dominant polycystic liver disease. Hepatology 37:164–171 [DOI] [PubMed] [Google Scholar]
- Que F, Nagorney DM, Gross JB Jr, Torres VE (1995) Liver resection and cyst fenestration in the treatment of severe polycystic liver disease. Gastroenterology 108:487–494 [DOI] [PubMed] [Google Scholar]
- Ramos A, Torres VE, Holley KE, Offord KP, Rakela J, Ludwig J (1990) The liver in autosomal dominant polycystic kidney disease: implications for pathogenesis. Arch Pathol Lab Med 114:180–184 [PubMed] [Google Scholar]
- Ravine D, Gibson RN, Walker RG, Sheffield LJ, Kincaid-Smith P, Danks DM (1994) Evaluation of ultrasonographic diagnostic criteria for autosomal dominant polycystic kidney disease 1. Lancet 343:824–827 [DOI] [PubMed] [Google Scholar]
- Reynolds DM, Falk CT, Li A, King BF, Kamath PS, Huston J 3rd, Shub C, Iglesias DM, Martin RS, Pirson Y, Torres VE, Somlo S (2000) Identification of a locus for autosomal dominant polycystic liver disease, on chromosome 19p13.2-13.1. Am J Hum Genet 67:1598–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaoul E, Reich-Slotky R, Berman B, Ron D (1995) Fibroblast growth factor receptors display both common and distinct signaling pathways. Oncogene 10:1553–1561 [PubMed] [Google Scholar]
- Somlo S, Torres VE, Reynolds D, King BF, Nagorney DM (1995) Autosomal dominant polycystic liver disease without polycystic kidney disease is not linked to either the PKD1 or PKD2 gene loci. J Amer Soc Nephrol 6:727a [Google Scholar]
- Thornalley PJ (1998) Cell activation by glycated proteins: AGE receptors, receptor recognition factors and functional classification of AGEs. Cell Mol Biol 44:1013–1023 [PubMed] [Google Scholar]
- Torres VE (1996) Polycystic liver disease. In: Watson ML, Torres VE (eds) Polycystic kidney disease. Oxford University Press, Oxford [Google Scholar]
- Torres VE, Rastogi S, King BF, Stanson AW, Gross JB Jr, Nogorney DM (1994) Hepatic venous outflow obstruction in autosomal dominant polycystic kidney disease. J Amer Soc Nephrol 5:1186–1192 [DOI] [PubMed] [Google Scholar]
- Treml K, Meimaroglou D, Hentges A, Bause E (2000) The alpha- and beta-subunits are required for expression of catalytic activity in the hetero-dimeric glucosidase II complex from human liver. Glycobiology 10:493–502 [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Fleming KG, Helenius A (2001) Quaternary and domain structure of glycoprotein processing glucosidase II. Biochemistry 40:10717–10722 [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Simons JF, Helenius A (1996) Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J Biol Chem 271:27509–27516 [DOI] [PubMed] [Google Scholar]
- Watnick TJ, Torres VE, Gandolph MA, Qian F, Onuchic LF, Klinger KW, Landes G, Germino GG (1998) Somatic mutation in individual liver cysts supports a two-hit model of cystogenesis in autosomal dominant polycystic kidney disease. Mol Cell 2:247–251 [DOI] [PubMed] [Google Scholar]
- Westphal V, Kjaergaard S, Davis JA, Peterson SM, Skovby F, Freeze HH (2001) Genetic and metabolic analysis of the first adult with congenital disorder of glycosylation type Ib: long-term outcome and effects of mannose supplementation. Mol Genet Metab 73:77–85 [DOI] [PubMed] [Google Scholar]
- Wu G, D'Agati V, Cai Y, Markowitz G, Park JH, Reynolds DM, Maeda Y, Le TC, Hou H Jr, Kucherlapati R, Edelmann W, Somlo S (1998) Somatic inactivation of Pkd2 results in polycystic kidney disease. Cell 93:177–188 [DOI] [PubMed] [Google Scholar]