Abstract
Several different environmental signals can induce asexual spore development (conidiation) and expression of developmentally regulated genes in Neurospora crassa. However, under constant conditions, where no environmental cues for conidiation are present, the endogenous circadian clock in N. crassa promotes daily rhythms in expression of known developmental genes and of conidiation. We anticipated that the same pathway of gene regulation would be followed during clock-controlled conidiation and environmental induction of conidiation and that the circadian clock would need only to control the initial developmental switch. Previous experiments showed that high-level developmental induction of the clock-controlled genes eas (ccg-2) and ccg-1 requires the developmental regulatory proteins FL and ACON-2, respectively, and normal developmental induction of fl mRNA expression requires ACON-2. We demonstrate that the circadian clock regulates rhythmic fl gene expression and that fl rhythmicity requires ACON-2. However, we find that clock regulation of eas (ccg-2) is normal in an fl mutant strain and ccg-1 expression is rhythmic in an acon-2 mutant strain. Together, these data point to the endogenous clock and the environment following separate pathways to regulate conidiation-specific gene expression.
Circadian clocks transduce time-of-day information within cells to control a wide range of rhythmic output processes. In the filamentous fungus Neurospora crassa, an organism with one of the most highly described circadian systems (reviewed in references 9 and 37), the best-characterized output of the circadian clock is the daily rhythm in asexual spore (conidiospore) production. Despite significant advances in our understanding of the workings of the clock in N. crassa, we still know very little about how the clock transmits temporal signals to control the components involved in conveying time-of-day information within the complex output pathways.
Conidiation in N. crassa can be induced by several environmental signals, including desiccation, blue light, carbon starvation, and nitrogen starvation (50). However, the only endogenous signal known to induce conidiation is provided by the circadian clock. Conidial development begins with the vegetative hyphae growing away from the growth medium (48; reviewed in references 22 and 47). After several hours of apical aerial growth, the aerial hyphae switch to a budding form of growth that is defined by a series of morphological stages distinguished by the diameters of the constrictions between the incipient conidia. At 4 h after induction of conidiation, the constrictions are subtle; these proconidial chains are called minor constriction chains. As budding continues, the interconidial constrictions become more pronounced, and around 8 h after conidiation is induced, major constrictions are observed. The formation of major constriction chains signals the commitment to the formation of conidia. Around 12 h after induction, crosswalls are evident between proconidia of the major constriction chains. Conidial separation takes place about 16 h after the initial developmental switch.
Mutations in six different genetic loci are known to block specific stages of conidiation. Fluffyoid (fld) and aconidiate-2 (acon-2) mutations block the formation of minor constrictions (48). Fluffy (fl) and aconidiate-3 (acon-3) mutant strains are blocked in the transition from minor to major constriction chain formation (47, 48). Conidial separation-1 (csp-1) and conidial separation-2 (csp-2) mutant strains form chains of conidia, but the crosswalls are not cleaved and the conidia remain adherent to each other. Several alleles of fl have been isolated, including null alleles (5). Only single alleles are available for the other genes, and it is not known if the mutations in these genes completely abolish activity. The acon-2 mutant allele is a temperature- and dark-sensitive mutation (39).
The only key regulator of conidiospore development that has been cloned is the fl gene. fl encodes a C6 zinc cluster protein similar to the Gal4 class of transcription factors, with greatest similarity to the N. crassa NIT-4 protein (5). When development is initiated by environmental cues, fl expression increases significantly after 6 h of induction and remains high before returning to preinduction levels at 9 h (5). This timing of fl induction corresponds to the timing of the transition from minor to major constriction chain formation and is consistent with a role for FL in initiating major constriction chain formation in response to earlier developmental signals.
Several components required for circadian rhythmicity have been identified in N. crassa, including genes involved in input signaling pathways from the environment to the clock, the central oscillator (clock), and output from the clock (reviewed in references 10, 21, 27, 34, and 37). In the well-described FRQ-based oscillator, the light-responsive WHITE COLLAR-1 (WC-1) and WHITE COLLAR-2 (WC-2) proteins form a complex (7, 49) and activate expression of the clock gene frequency (frq) in the morning (16, 19, 31). FRQ protein, produced maximally in the midafternoon from the accumulating frq transcript (3, 24), enters the nucleus (38) and rapidly inhibits its own transcription. Inhibition is proposed to occur through interference of the activity of the WC-1/WC-2 protein complex by FRQ (15, 19, 31, 40). Turnover of FRQ, involving progressive phosphorylation (33), results in the reactivation of frq transcription by WC-1/WC-2 complexes (16, 17, 33). One or more of these proteins, or still-unidentified transcription factors, are predicted to directly regulate the initial components of the output pathways, including the developmental pathway, to control overt rhythmicity (reviewed in reference 9).
Genes that are regulated by the clock and reside in output pathways have been identified by several different methods (reviewed in references 9, 36, and 37). The most highly characterized N. crassa clock-controlled gene (ccg) is the eas (ccg-2) gene. The eas (ccg-2) locus was originally identified through mutation, which resulted in easily wettable (eas) conidiospores (8, 45). The eas (ccg-2) gene was independently cloned on the basis of daily rhythms in the abundance of the transcript as ccg-2 (35) and as a blue-light-inducible gene, bli-7 (46). The abundantly expressed eas (ccg-2) gene encodes a member of a class of low-molecular-weight, cysteine-rich, hydrophobic secreted proteins called hydrophobins (11, 28). The hydrophobins coat the outer cell wall of fungi and maintain the cell surface hydrophobicity essential for air dispersal of the mature conidiospores (reviewed in references 26 and 51). eas (ccg-2) is not only regulated by the circadian clock, it is also induced by the same environmental signals that trigger conidiospore development (4, 12, 28). Developmental induction of eas (ccg-2) occurs about 1 h after the initiation of conidiation and requires the product of the fl gene (28). Similarly, the morning-specific ccg-1 gene is regulated by the circadian clock and is induced by developmental cues (32, 35). Developmental induction of ccg-1 occurs 1 to 2 h after the initiation of conidiation and requires the product of the acon-2 gene (32). Inactivation of ccg-1 has no obvious effect on conidiation, and ccg-1-null strains do not display any discernible phenotypes. In addition, the CCG-1 protein does not have homology to other known proteins (32). Thus, the function of CCG-1 remains a mystery.
We hypothesized that the circadian clock in N. crassa would need only to regulate the first event in the conidiation pathway in order to achieve a daily output rhythm in spore production. To test this hypothesis, we have assayed rhythmic aerial-hypha formation and rhythmic output gene expression in strains harboring mutations of the key developmental regulators. We show that rather than the clock simply triggering rhythmic activation of the earliest event in the developmental pathway, the clock controls several genes in the pathway. We also demonstrate that fl gene expression is rhythmic; however, unlike eas (ccg-2) and ccg-1, the rhythmicity of fl requires the activity of its upstream regulator ACON-2. Together, these data establish that regulation of the developmental output pathway by the clock in N. crassa is complex: some genes are regulated independently of the developmental cascade, whereas other genes require their upstream developmental regulator for normal rhythmicity.
MATERIALS AND METHODS
N. crassa strains and growth conditions.
The strains of N. crassa used in this study include the frq+ strains 30-7 (bd;A) and 87-3 (bd;a) and the long-period mutant strains 695-425 (bd;frq7;A) and 585-70 (bd;frq7;a). The band mutation (bd) enhances the circadian rhythm of conidiation but does not affect the clock itself (43, 44). These strains were obtained from Jay Dunlap (Dartmouth Medical School, Hanover, N.H.). The developmental mutant strains used in this study were obtained from the Fungal Genetic Stock Center (FGSC) (University of Kansas Medical Center). These strains include FGSC 7430 (flL;A), FGSC 7431 (flL;a), FGSC 3263 (acon-2;a), and FGSC 5074 (acon-3;a). The flL null allele was previously shown to contain a 67-bp deletion that spans the start codon and the first segment of the zinc cluster (5). The acon-2 allele is temperature sensitive (conidiating at 25°C but not at 34°C) and dark sensitive (conidiating in the light at 25°C but not in the dark at 25°C) (39). The flL, acon-2, and acon-3 strains were each crossed to the appropriate bd and bd;frq7 strains. Because all strains used in this study contain the bd mutation, they are designated here with only the developmental mutant or clock genotype. The growth media (Vogel's and Fries minimal media), vegetative growth conditions, and crossing protocols were described previously (18).
Race tube assay.
Measurements of circadian period and growth rates were performed on 40-cm-long hollow glass tubes, called race tubes, containing glucose-arginine medium (44). Conidia were inoculated at one end of the tube and grown in constant light for about 24 h. The position of the growth front was marked, and the culture was then transferred into constant darkness at 25°C. The growth front was marked every 24 h with the aid of a red light. The calculation of circadian period length and phase was performed as previously described (20).
Culture harvesting conditions.
For rhythmic RNA analyses, the clock was synchronized by a light-to-dark transition in mycelial mats grown in shaking (100 rpm) liquid culture (Fries minimal medium containing 0.03% glucose and 0.05% arginine) at 25°C (13, 35) or at 34°C for acon-2 mutant strains. Previous studies have shown that under these growth conditions, the clock functions normally but respiration is suppressed (42). Reduced respiration likely results in the arrested growth and development observed. Clock-controlled genes involved in development are expressed under these conditions at specific times of the day (35), but the levels of expression are probably not sufficient to induce development. The light-to-dark transfer sets the clock to circadian time (CT) 12. CT is used to normalize biological time in strains with different endogenous period lengths to 24 circadian hours per cycle. By convention, CT0 represents subjective dawn and CT12 represents subjective dusk. Light-to-dark transfer times were such that the ages of the cultures at harvest were approximately the same but the circadian times varied (13). Tissue for RNA extraction was harvested after the indicated times in the dark for each experiment. Developmental induction was performed as previously described (14, 30). Briefly, conidia were germinated in Vogel's minimal medium for 24 h at room temperature. The cultures were then filtered onto sterile filter paper. The paper and the mycelium were placed onto a layer of glass beads in a petri dish containing sufficient medium (10 ml) to keep the paper moist. The cultures were left uncovered in a sterile hood, and at the designated times after the induction of development by exposure to air, ∼2-cm-square pieces of tissue were removed, placed into microcentrifuge tubes, and immediately frozen. Total RNA was isolated from frozen tissue harvested immediately after filtration of mycelia (time zero) and at 15, 30, 60, and 120 min after exposure of the cultures to air.
Nucleic acid isolation and hybridization.
RNA was isolated by a miniprep method, and Northern blots were processed as described previously (13). Riboprobes were synthesized in the presence of [α32-P]UTP (6,000 Ci/mmol; New England Nuclear). The eas (ccg-2) riboprobes were synthesized from pLW1K (11) using T3 RNA polymerase. The ccg-1 riboprobes were generated from plasmid pKL119 (32) using T3 RNA polymerase. The frq riboprobes were generated from plasmid pKAJ106 (17) using T7 RNA polymerase. The fl riboprobes were generated from plasmid pFLUF3 using T7 RNA polymerase. pFLUF3 was obtained from Daniel Ebbole, Texas A&M University. RNA loading was normalized in each experiment to rRNA, which remains at constant levels under the growth conditions used (35). Densitometric data were acquired and analyzed as described previously (12).
RESULTS
Circadian clock control of aerial-hypha production is unaltered by mutations that block later stages of development.
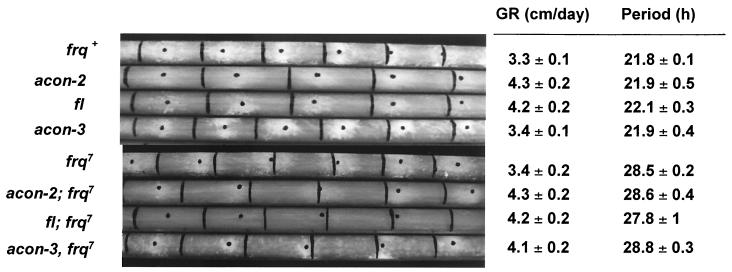
To determine if any of the known mutants that block key stages in conidiospore development in N. crassa affect rhythmic aerial-hypha initiation, we introduced the band (bd) mutation into the aconidial mutant strains and assayed rhythmicity on race tubes (Fig. 1). The bd mutation slows the growth rate of the cultures and allows clear visualization of development in closed culture tubes (44). In the frq+ strain, the period of the rhythm in constant dark and temperature was approximately 22 h. Similar rhythms in aerial-hypha initiation were observed in each of the aconidial mutant strains, despite the differences in their growth rates and the fact that the aerial hyphae do not form conidiospores. These data suggest that the FRQ-based circadian oscillator functions normally in strains that cannot develop conidiospores.
FIG. 1.
The inability to complete conidiation does not affect clock control of the initiation of aerial-hypha formation. Race tube cultures of the indicated strains were grown in constant darkness for 7 days, and the growth fronts were marked every 24 h (vertical black lines). The centers of the developmental bands are indicated by the black dots. The calculated growth rates (GR) and the periods of the rhythms in hours are shown on the right. The values are means ± standard deviations determined from at least three separate experiments for each strain.
We crossed the developmental mutants into the long-period frq7 mutant strain, which has a free-running period of approximately 29 h (23). All of the resulting strains displayed a rhythm in aerial-hypha production of about 29 h (Fig. 1). Examination of the rhythms of the central clock component frq in each of the developmental mutant strains revealed normal frq mRNA rhythms, with peak accumulation occurring around subjective noon (data not shown). Together, these data demonstrated that the inability to complete asexual spore development does not directly affect circadian oscillator components or clock control of the initiation of aerial-hypha formation.
fl gene expression is regulated by the circadian clock.
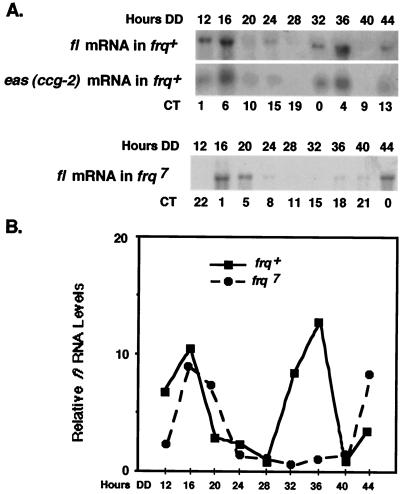
To investigate whether the clock simply triggers a developmental cascade or directly controls individual regulatory elements along this cascade, we examined fl gene expression under conditions in which development is curtailed (see Materials and Methods). Mycelia were grown in constant darkness and temperature and harvested for RNA isolation every 4 h over two consecutive days. fl mRNA accumulated rhythmically, with a period of about 22 h in the frq+ strain (Fig. 2). The mRNA levels peaked in the morning (around CT0 to CT6), at the same time of day as the morning-specific clock-controlled eas (ccg-2) gene. The time of peak expression of fl also corresponds to the times of peak expression of other known clock-controlled genes which are induced at various times during development (13, 29). A rhythm in fl mRNA accumulation was likewise observed in the long-period frq7 mutant strain, and as would be predicted for a gene that is controlled by the frq-based circadian oscillator, the period of fl mRNA accumulation was longer in frq7 than in the frq+ strain. The data revealed that the clock not only controls components required for the initiation of development but also regulates key elements of the subsequent conidiation pathway.
FIG. 2.
The circadian clock regulates fl gene expression. (A) The steady-state levels of fl mRNA were assayed by Northern blot analyses in frq+ (top) and frq7 (bottom) strains. The frq+ membrane was also probed with an eas (ccg-2)-specific probe as a control. Liquid cultures of mycelia were grown in constant darkness and harvested after the indicated number of hours in the dark (Hours DD). The approximate CTs at the times of harvest are shown below the autoradiograms. (B) Using the autoradiograms shown in panel A, fl mRNA was quantitated by densitometry and plotted as relative band intensity normalized to rRNA (not shown) versus time in the dark for both frq+ (solid line) and frq7 (dashed line). Three separate repetitions of these experiments in each strain yielded similar results.
acon-2 is required for rhythmic fl mRNA accumulation.
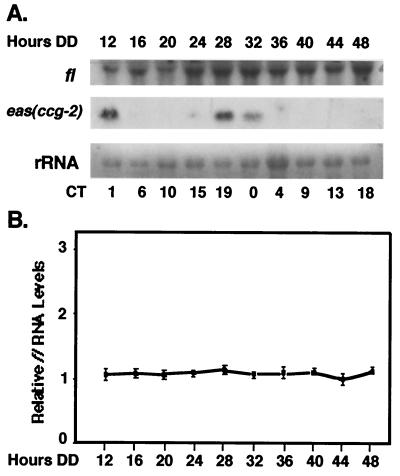
Mutation of fl blocks a later stage in development than mutation of acon-2 (39, 48), and functional ACON-2 is required for fl developmental induction at 6 h after the developmental switch (6; L. Bailey-Shrode and D. J. Ebbole, submitted for publication). To determine if ACON-2 is required for rhythmic fl expression, we investigated whether fl mRNA is rhythmic in the acon-2 mutant strain. The acon-2 mutant strain was grown at 34°C in the dark under nonpermissive conditions and harvested every 4 h over two consecutive days for RNA isolation. The level of fl gene expression was low in the acon-2 mutant strain and remained constant over the course of the day (Fig. 3). As a control, the same blot was probed with eas (ccg-2) to confirm that the cultures were rhythmic. These data suggest that ACON-2 is required for both developmental and rhythmic fl expression.
FIG. 3.
fl mRNA accumulation is arrhythmic in the acon-2 mutant strain. (A) The levels of fl mRNA were assayed by Northern blot analyses in the acon-2 mutant strain. Liquid cultures of mycelia were grown in constant darkness at 34°C and harvested after the indicated number of hours in the dark (Hours DD). The Northern blot was probed with fl and eas (ccg-2) riboprobes. The approximate CTs at the times of harvest are indicated below the autoradiogram. (B) Following autoradiography, fl mRNA was quantitated by densitometry and plotted as relative band intensity normalized to rRNA versus time in the dark. Three separate experiments were used to determine the standard deviation of fl RNA levels.
Distinct output pathways from the clock regulate rhythmicity of developmentally regulated genes.
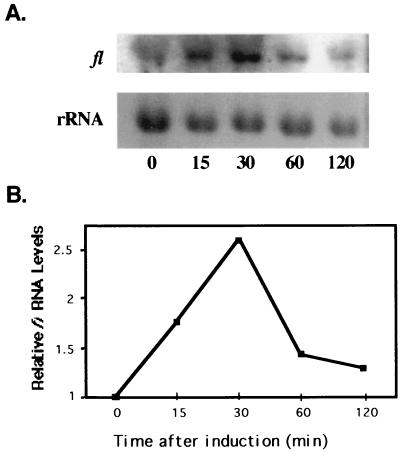
The eas (ccg-2) gene is regulated by both the endogenous circadian clock and environmental cues that trigger conidiation (11, 28). The eas (ccg-2) gene requires FL for developmental induction but is normally induced in acon-3 or fld mutant strains (6, 28; Bailey-Shrode and Ebbole, submitted). High-level expression of eas (ccg-2) also requires ACON-2 (6; Bailey-Shrode and Ebbole, submitted). The requirement for FL in eas (ccg-2) developmental expression was surprising, since high-level eas (ccg-2) induction occurs about 1 h after development is induced by desiccation (13, 28), significantly earlier than fl induction, which peaks transiently about 6 h after development is initiated (5, 6; Bailey-Shrode and Ebbole, submitted). It was suggested that the basal levels of FL in vegetative hyphae are sufficient to induce or permit induction of ccg-2 (eas) expression following developmental cues (6; Bailey-Shrode and Ebbole, submitted). However, the earliest time that fl mRNA induction was investigated in this study was 3 h after the environmental challenge. Therefore, we examined fl mRNA expression at earlier times after the cultures were induced to develop by desiccation. We observed a transient increase in fl mRNA levels 15 min after the cultures were desiccated, with peak levels occurring 30 min after the induction and decreasing by 60 min (Fig. 4). The levels of induction observed at 30 min (2.5-fold) are similar to those observed previously at 6 h (6; Bailey-Shrode and Ebbole, submitted). These data suggest that, along with the role of FL in the switch from minor to major constriction chains, FL functions during aerial-hypha production, and the induced fl expression observed prior to 1 h is consistent with the ability of FL to directly or indirectly induce eas (ccg-2) during the developmental process.
FIG. 4.
fl mRNA is induced early in the conidiation pathway. (A) RNA was isolated from wild-type clock cells at 0, 15, 30, 60, and 120 min after induction of conidiation by desiccation. The RNA was hybridized to an fl-specific RNA probe and subsequently to an rRNA DNA probe. (B) Following autoradiography, fl mRNA was quantitated by densitometry and plotted as relative band intensity normalized to rRNA versus time after induction. Similar results were obtained with two additional repeats of this experiment.
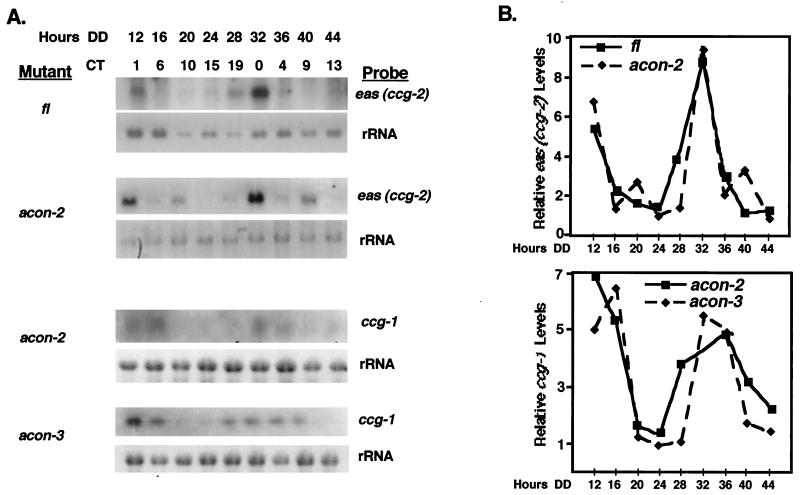
To further investigate whether the clock triggers a cascade from rhythmic aerial-hypha production to downstream genes, we investigated whether eas (ccg-2) mRNA is rhythmic in an fl-null strain. A prediction of the cascade model is that eas (ccg-2) expression would not be rhythmic in the fl mutant strain. However, if the clock regulates expression of developmental genes at multiple points along the pathway, eas (ccg-2) might still be rhythmically expressed in an fl-null strain. While the levels of eas (ccg-2) mRNA were very low in the fl-null strain, they were rhythmic, with a normal periodicity and with a peak in message accumulation occurring at CT0 (Fig. 5).
FIG. 5.
Both eas (ccg-2) and ccg-1 are rhythmically expressed in mutants of key developmental regulators that are required for their induction by exogenous developmental cues. (A) The steady-state levels of eas (ccg-2) mRNA were assayed by Northern blot analyses in fl and acon-2 mutant strains and ccg-1 mRNA in acon-2 and acon-3 mutant strains at 24°C. Liquid cultures of mycelia were grown in constant darkness and harvested after the indicated number of hours in the dark (Hours DD). The approximate CTs at the times of harvest are indicated. (B) Following autoradiography, eas (ccg-2) and ccg-1 mRNA in each mutant background was quantitated by densitometry and plotted as relative band intensity normalized to rRNA versus time in the dark. Similar results were obtained in at least two separate repetitions of these experiments.
To further confirm the independent nature of the clock signal to eas (ccg-2), we examined eas (ccg-2) mRNA rhythmicity in the acon-2 mutant strain in cultures grown in constant darkness at 25 and 34°C. We found that eas (ccg-2) mRNA accumulation is rhythmic in acon-2 (Fig. 5). However, in the acon-2 mutant strain grown at 25°C in the dark, some unusual low-amplitude peaks were consistently observed at about CT10 in each cycle. These low-amplitude peaks disappeared when the cultures were grown at 34°C in the dark (Fig. 3). The unusual eas (ccg-2) mRNA peaks in acon-2 at 25°C may reflect weak activity of ACON-2 at this temperature in dark-grown cultures. The rhythmicity of eas (ccg-2) mRNA in acon-2 strains suggests that the clock can bypass ACON-2 to regulate eas (ccg-2) expression. Rhythmicity of eas (ccg-2) mRNA was found to be normal in an acon-3 mutant strain. This was not surprising, since genetic experiments place acon-3 downstream of fl in the developmental pathway and acon-3 does not affect eas (ccg-2) expression during induced development (6).
The clock-regulated ccg-1 gene requires a functional ACON-2 protein for developmental induction but not FL or ACON-3 (32). We observed that ccg-1 was poorly expressed but normally rhythmic in the acon-2 mutant strain (Fig. 5). Mutations in fl or acon-3 (Fig. 5) did not affect ccg-1 rhythmicity. The above-mentioned results suggest that the clock regulates ccg-1 through a pathway that does not involve FL, ACON-2, or ACON-3.
DISCUSSION
To determine whether the N. crassa circadian clock directly regulates the expression of several components in the developmental output pathway or only controls a key element involved in the initial developmental switch, we examined rhythmic expression of clock- and developmentally regulated genes in strains carrying mutations in genes required for their developmental induction (Fig. 3 and 5). We demonstrated that clock regulation of ccg-1 and eas (ccg-2) can occur independently of the upstream developmental regulators, implying that several output pathways from the clock exist to control rhythmic expression of the clock-controlled genes. Alternatively, we found that fl rhythmicity requires its upstream developmental activator, ACON-2. Therefore, while eas (ccg-2) and ccg-1 are independently regulated by the clock, some aspects of the clock signal can be transduced in a linear fashion from one developmentally regulated gene to another (Fig. 6).
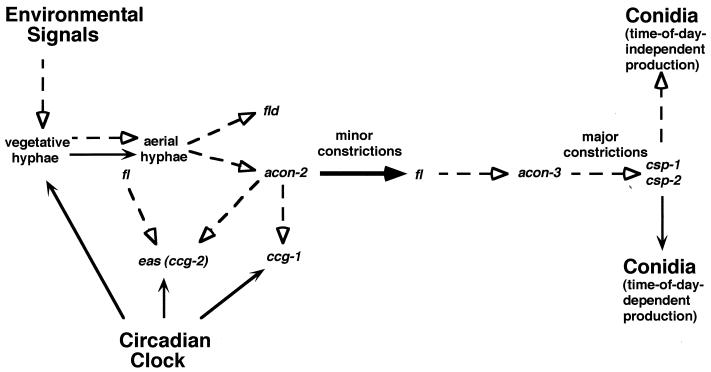
FIG. 6.
Model depicting direct and indirect regulation of clock output genes involved in conidiation. (See the text for a full explanation of the developmental pathway and the clock-regulated events.) Once a day, the endogenous circadian clock signals the production of aerial hyphae from vegetative hyphae (solid arrow). Independent of the time of day, exogenous environmental signals can trigger the same event (dashed arrow). Both pathways eventually lead to the production of conidia. Conidia are formed by budding of the aerial hyphae involving the formation of minor constriction chains, requiring the products of acon-2 and fld, and major constriction chains, requiring the products of fl and acon-3. The csp-1 and csp-2 genes are required for conidial separation. The genes shown to be regulated by the clock independent of the developmental pathway are linked to the clock by solid arrows. The epistatic relationships between the key developmental regulators and developmentally induced genes (6, 22, 47) are depicted by dashed arrows. Rhythmic fl expression requires its upstream developmental regulator, ACON-2 (shown as a heavy solid arrow). A solid line was not drawn to acon-2, since it is not known if acon-2 is rhythmically expressed. The fl gene is shown to act during the production of aerial hyphae and after the formation of minor constriction chains.
During development, there is a transient induction of fl expression at 30 min (Fig. 4) and then another peak of expression at 6 h (5). The early induction of fl mRNA suggests that FL may play a heretofore-undescribed regulatory role in the formation of aerial hyphae in addition to its role in the formation of major constriction chains (Fig. 6). Consistent with this notion, the number of aerial hyphae observed in fl mutant strains is typically lower than that seen in acon-2 or acon-3 mutant strains (Fig. 1). The early induction of fl mRNA also suggests that the basal levels of ACON-2 in the cell prior to aerial-hypha formation are sufficient to induce fl gene expression. This situation is reminiscent of the coordinated activation of gene expression during Aspergillus nidulans conidiospore development (reviewed in reference 2). Here, the brlA, abaA, and wetA genes act centrally to control the temporal aspects of conidiation-specific gene expression. Mutation in any of these three genes blocks conidiation at specific stages and affects the induction of specific classes of developmentally regulated genes. Available evidence suggests that, similar to fl, brlA is required at several different stages during the developmental timeline (41). It is not yet known if a circadian clock in A. nidulans plays a role in the temporal regulation of conidiation.
Examination of the developmental mutant strains on race tubes demonstrated that the inability to complete asexual spore development does not affect clock control of the initiation of aerial-hypha formation (Fig. 1). Thus, while there is evidence for feedback from output pathways back to the clock or input pathways in other organisms (e.g., reference 25), these and other studies (11) suggest that in N. crassa there is no obvious feedback from the developmental output pathway to the clock. In addition, frq expression is rhythmic in all the mutants studied (data not shown), supporting the idea that there is no feedback from the developmental output pathway to regulate frq activity. Interestingly, in the developmental mutant strains, the vegetative growth rate on race tubes is increased compared to that of the bd strain (Fig. 1). These data suggest that in wild-type strains growth energy is redirected from vegetative growth to conidiospore development. Similar observations have been made with A. nidulans, in which forced overexpression of developmental regulators such as brlA in vegetative hyphae results in not only activating development but also inhibiting vegetative growth (1).
The eas (ccg-2) promoter contains distinct elements required for developmental induction, light induction, and clock control (12). However, the developmental element is not required for clock control of eas (ccg-2), and normal developmental induction occurs when the clock element is deleted (12). Neither the clock element nor the developmental element is required for light-induced expression of eas (ccg-2). In support of the molecular data, we observed normal light induction of eas (ccg-2) in the fl mutant strain (data not shown). Furthermore, conidiation can proceed without a functional clock, and environmental conditions that promote rapid development can mask clock regulation of eas (ccg-2) (D. Bell-Pedersen, unpublished data). Together, these findings suggest that under standard environmental conditions, the endogenous clock uses multiple pathways to the developmental program to anticipate light and produce a daily supply of conidia for dispersal, as evidenced by the daily induction of eas (ccg-2) and other developmentally controlled genes. This conclusion is also supported by experiments that show differential regulation of eas (ccg-2) and ccg-1 by the clock; eas (ccg-2) is positively regulated by the clock, whereas ccg-1 is negatively regulated (H. Cho, D. Bell-Pedersen, J. C. Dunlap, and J. J. Loros, unpublished data). However, when stressful conditions are encountered, abundant conidia are produced at all times of day. In this situation, we predict that distinct promoter elements are used to produce high levels of the conidiation-specific transcripts. For high-level eas (ccg-2) developmental expression, this requires a separate promoter element and FL. Alternatively, the genes that appear to follow the same pathway in development and clock regulation (acon-2 and fl) are important regulators of development. This similarity in developmental and clock regulation may be due to the necessity for a specific temporal sequence of the expression of key genes to form conidia irrespective of the time of day.
In summary, we found that rather than the clock simply triggering a rhythmic developmental cascade of gene expression, the clock uniquely regulates genes involved in the developmental process. Thus, similar to what is predicted for higher eukaryotes, clock regulation of output pathways in N. crassa is complex and involves more than one pathway to regulate rhythmic conidium formation. Identification of the regulatory factors that control the clock-controlled genes will eventually help to determine the links between the circadian clock and the output genes.
Acknowledgments
We thank Richard Gomer and Louis Morgan for helpful suggestions on the manuscript. We are also grateful to Lori Bailey-Shrode, Daniel Ebbole, Jennifer Loros, and Jay Dunlap for sharing strains, plasmids, and unpublished data.
This work was supported by the National Institutes of Health (GM58529).
REFERENCES
- 1.Adams, T. H., and W. E. Timberlake. 1990. Developmental repression of growth and gene expression in Aspergillus. Proc. Natl. Acad. Sci. USA 87:5405-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, T. H., J. K. Wieser, and J.-H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson, B. D., K. A. Johnson, J. J. Loros, and J. C. Dunlap. 1994. Negative feedback defining a circadian clock: autoregulation in the clock gene frequency. Science 263:1578-1584. [DOI] [PubMed] [Google Scholar]
- 4.Arpaia, G., J. J. Loros, J. C. Dunlap, G. Morelli, and G. Macino. 1993. The interplay of light and the circadian clock: independent dual regulation of clock-controlled gene ccg-2 (eas). Plant Physiol. 102:1299-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey, L. A., and D. J. Ebbole. 1998. The fluffy gene of Neurospora crassa encodes a Gal4p-type C6 zinc cluster protein required for conidial development. Genetics 148:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey-Shrode, L. 1999. The fluffy gene of Neurospora crassa encodes a C6 zinc cluster protein that is sufficient for conidiation and necessary for conidiophore development. Ph.D. thesis. Texas A&M University, College Station.
- 7.Ballario, P., P. Vittorioso, A. Magrelli, C. Talora, A. Cabibbo, and G. Macino. 1996. white collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 15:1650-1657. [PMC free article] [PubMed] [Google Scholar]
- 8.Beever, R. E., and G. P. Dempsey. 1978. Function of rodlets on the surface of fungal spores. Nature 272:608-610. [DOI] [PubMed] [Google Scholar]
- 9.Bell-Pedersen, D. 2000. Understanding circadian rhythmicity in Neurospora crassa: from behavior to genes and back again. Fungal Genet. Biol. 29:1-18. [DOI] [PubMed] [Google Scholar]
- 10.Bell-Pedersen, D., S. K. Crosthwaite, P. L. Lakin-Thomas, M. Merrow, and M. Vinsjevik. 2001. The Neurospora circadian clock--simple or complex? Phil. Trans. R. Soc. Lond. 356:1697-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell-Pedersen, D., J. C. Dunlap, and J. J. Loros. 1992. The Neurospora circadian clock-controlled gene ccg-2 is allelic to eas and encodes a fungal hydrophobin required for formation of the conidial rodlet layer. Genes Dev. 6:2382-2394. [DOI] [PubMed] [Google Scholar]
- 12.Bell-Pedersen, D., J. C. Dunlap, and J. J. Loros. 1996. Distinct cis-acting elements mediate clock, light, and developmental regulation of the Neurospora crassa eas (ccg-2) gene. Mol. Cell. Biol. 16:513-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell-Pedersen, D., M. Shinohara, J. J. Loros, and J. C. Dunlap. 1996. Clock-controlled genes isolated from Neurospora crassa are late night- to morning-specific. Proc. Natl. Acad. Sci. USA 93:13096-13101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berlin, V., and C. Yanofsky. 1985. Isolation and characterization of genes differentially expressed during conidiation of Neurospora crassa. Mol. Cell. Biol. 5:849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng, P., Y. Yang, C. Heintzen, and Y. Liu. 2001. Coiled-coil domain-mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. EMBO J. 15:101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crosthwaite, S. K., J. C. Dunlap, and J. J. Loros. 1997. Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science 276:763-769. [DOI] [PubMed] [Google Scholar]
- 17.Crosthwaite, S. K., J. J. Loros, and J. C. Dunlap. 1995. Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell 81:1003-1012. [DOI] [PubMed] [Google Scholar]
- 18.Davis, R. L., and D. deSerres. 1970. Genetics and microbial research techniques for Neurospora crassa. Methods Enzymol. 27A:79-143. [Google Scholar]
- 19.Denault, D. L., J. J. Loros, and J. C. Dunlap. 2001. WC-2 mediates the WC-1-FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. EMBO J. 20:109-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dharmananda, S., and J. F. Feldman. 1979. Spatial distribution of circadian clock phase in aging cultures of Neurospora crassa. Plant Physiol. 63:1049-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunlap, J. C. 1999. Molecular bases for circadian clocks. Cell 96:271-290. [DOI] [PubMed] [Google Scholar]
- 22.Ebbole, D. J. 1996. Morphogenesis and vegetative differentiation in filamentous fungi. J. Genet. 75:361-374. [Google Scholar]
- 23.Feldman, J. F., and M. N. Hoyle. 1973. Isolation of circadian clock mutants of Neurospora crassa. Genetics 75:605-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garceau, N. Y., Y. Liu, J. J. Loros, and J. C. Dunlap. 1997. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89:469-476. [DOI] [PubMed] [Google Scholar]
- 25.Herzog, E. D., and G. D. Block. 1999. Keeping an eye on retinal clocks. Chronobiol. Int. 16:229-247. [DOI] [PubMed] [Google Scholar]
- 26.Kershaw, M. J., and N. J. Talbot. 1998. Hydrophobins and repellents: proteins with fundamental roles in fungal morphogenesis. Fungal Genet. Biol. 23:18-33. [DOI] [PubMed] [Google Scholar]
- 27.Lakin-Thomas, P. L. 2000. Circadian rhythms: new functions for old clock genes? Trends Genet . 16:135-142. [DOI] [PubMed] [Google Scholar]
- 28.Lauter, F.-R., V. E. Russo, and C. Yanofsky. 1992. Developmental and light regulation of eas, the structural gene for the rodlet protein of Neurospora. Genes Dev. 6:2373-2381. [DOI] [PubMed] [Google Scholar]
- 29.Lauter, F.-R., and C. Yanofsky. 1993. Day/night and circadian rhythm control of con gene expression in Neurospora. Proc. Natl. Acad. Sci. USA 90:8249-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, K., and D. J. Ebbole. 1998. Analysis of two transcription activation elements in the promoter of the developmentally regulated con-10 gene of Neurospora crassa. Fungal Genet. Biol. 23:259-268. [DOI] [PubMed] [Google Scholar]
- 31.Lee, K., J. J. Loros, and J. C. Dunlap. 2000. Interconnected feedback loops in the Neurospora circadian system. Science 289:107-110. [DOI] [PubMed] [Google Scholar]
- 32.Lindgren, K. M. 1994. Characterization of ccg-1, a clock-controlled gene of Neurospora crassa. Ph.D. thesis. Dartmouth Medical School, Hanover, N.H.
- 33.Liu, Y., J. J. Loros, and J. C. Dunlap. 2000. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc. Natl. Acad. Sci. USA 97:234-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loros, J. J. 1998. Time at the end of the millennium: the Neurospora clock. Curr. Opin. Microbiol. 1:698-706. [DOI] [PubMed] [Google Scholar]
- 35.Loros, J. J., S. A. Denome, and J. C. Dunlap. 1989. Molecular cloning of genes under control of the circadian clock in Neurospora. Science 243:385-388. [DOI] [PubMed] [Google Scholar]
- 36.Loros, J. J., and J. C. Dunlap. 1991. Neurospora clock-controlled genes are regulated at the level of transcription. Mol. Cell. Biol. 11:558-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loros, J. J., and J. C. Dunlap. 2001. Genetic and molecular analysis of circadian rhythms in Neurospora. Annu. Rev. Physiol. 63:757-794. [DOI] [PubMed] [Google Scholar]
- 38.Luo, C., J. J. Loros, and J. C. Dunlap. 1998. Nuclear localization is required for function of the essential clock protein FRQ. EMBO J. 17:1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuyama, S. S., R. E. Nelson, and R. W. Siegel. 1974. Mutations specifically blocking differentiation of macroconidia in Neurospora crassa. Dev. Biol. 41:278-287. [DOI] [PubMed] [Google Scholar]
- 40.Merrow, M., L. Franchi, Z. Dragovic, M. Gorl, J. Johnson, M. Brunner, G. Macino, and T. Roenneberg. 2001. Circadian regulation of the light input pathway in Neurospora crassa. EMBO J. 20:307-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirabito, P. M., T. H. Adams, and W. E. Timberlake. 1989. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell 57:859-868. [DOI] [PubMed] [Google Scholar]
- 42.Nakashima, H. 1981. A liquid culture method for the biochemical analysis of the circadian clock of Neurospora crassa. Plant Cell Physiol. 22:231-238. [Google Scholar]
- 43.Sargent, M. L., and S. H. Kaltenborn. 1972. Effects of medium composition and carbon dioxide on circadian conidiation in Neurospora. Plant Physiol. 50:171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sargent, M. L., W. R. Briggs, and D. O. Woodward. 1966. Circadian nature of a rhythm expressed by an invertaseless strain of Neurospora crassa. Plant Physiol. 41:1343-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selitrennikoff, C. P. 1976. Easily-wettable, a new mutant. Neurospora Newsl. 23:23. [Google Scholar]
- 46.Sommer, T., J. A. A. Chambers, J. Eberle, F.-R. Lauter, and V. E. Russo. 1989. Fast light-regulated genes of Neurospora crassa. Nucleic Acids Res. 17:5713-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Springer, M. L. 1993. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa. BioEssays 15:365-374. [DOI] [PubMed] [Google Scholar]
- 48.Springer, M. L., and C. Yanofsky. 1989. A morphological and genetic analysis of conidiophore development in Neurospora crassa. Genes Dev. 3:559-571. [DOI] [PubMed] [Google Scholar]
- 49.Talora, C., L. Franchi, H. Linden, P. Ballario, and G. Macino. 1999. Role of a WHITE COLLAR-1-WHITE COLLAR-2 complex in blue-light signal transduction. EMBO J. 18:4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turian, G., and D. E. Bianchi. 1972. Conidiation in Neurospora. Bot. Rev. 38:119-154. [Google Scholar]
- 51.Wessels, J. G. J. 1997. Hydrophobins: proteins that change the nature of the fungal surface. Adv. Microb. Physiol. 38:1-45. [DOI] [PubMed] [Google Scholar]