Abstract
The adenomatous polyposis coli (APC) I1307K allele is found in 6% of the Ashkenazi Jewish population and in 1%–2% of Sephardi Jews; it confers a relative risk of 1.5–2.0 for colorectal cancer (CRC) on all carriers. Within the Ashkenazim, the existence of numerous high-prevalence mutations, including I1307K, has sparked controversy over whether genetic drift or selection is the underlying cause. For the present population-based case-control study of CRC in Israel, we tested whether selection has operated at I1307K. We also estimated the age of the I1307K allele, to understand its origin in the context of the Jewish diasporas and subsequent founder events. We genotyped 83 matched pairs, in which one or both members of the pair carried I1307K, at three microsatellites and two SNPs. Haplotypes were statistically constructed using PHASE software. Single-marker age estimates for I1307K were calculated using the approach described by Risch et al. A common progenitor haplotype spanned across APC I1307K from the centromeric marker D5S135 to the telomeric marker D5S346 and was observed in individuals of Ashkenazi, Sephardi, and Arab descent. The ancestor of modern I1307K alleles existed 87.9–118 generations ago (∼2,200–2,950 years ago). This age estimate indicates that I1307K existed at about the time of the beginning of the Jewish diaspora, explaining its presence in non-Ashkenazi populations. Our data do not indicate that selection operated at I1307K (D5S346, P=.114; D5S135, P=.373), providing compelling evidence that the high frequency of disease-susceptibility alleles in the Ashkenazim is due to genetic drift, not selection. This research underscores the importance of the migratory patterns of ancestral populations in the ethnic and geographic distribution of APC I1307K.
Introduction
Colorectal cancer (CRC) ranks third in the United States in cancer incidence and second in Israel in cancer incidence (Greenlee et al. 2000). Low-penetrance susceptibility alleles may contribute significantly to the incidence of CRC not explained by other known risk factors. The low-penetrance susceptibility adenomatous polyposis coli (APC [MIM 175100]) I1307K allele can be found in 6.1% of the Ashkenazi Jewish population, and it confers a relative risk of 1.5–2.0 for CRC (Laken et al. 1997). The APC allele with lysine (K) at codon 1307, commonly referred to in the literature as I1307K, results from the T→A transition at nt 3920 in APC, which causes an extended mononucleotide tract (A8). This mononucleotide repeat impairs replication fidelity, forming a mutational hotspot (Laken et al. 1997).
Several studies have investigated the ethnic distribution of I1307K outside the Ashkenazim. An I1307K carrier frequency of 1.3% was measured in 239 Israeli Jews of non-European origin, which includes Oriental Jews, North African Jews, and Sephardi Jews (Rozen et al. 1999). Within this same study, 261 Ashkenazi Jews—defined as Israeli Jews who primarily immigrated from Russia, Poland, and Romania—had a carrier frequency of 7.7%, which was significantly greater than the 1.3% identified in Israeli Jews of non-European origin (P<.01) (Rozen et al. 1999). Prior et al. subsequently studied 345 non-Ashkenazi Jewish individuals, including black Americans, Italians, Finns, and Hawaiian-Japanese; no I1307K carriers were identified (Prior et al. 1999). In addition, Drucker et al. determined that the I1307K carrier frequency in Yemenite Jews is ∼4.7% (Drucker et al. 2000).
Within the Ashkenazim, the existence of numerous disease-susceptibility alleles in addition to I1307K—including those for Tay-Sachs disease, Gaucher disease, and torsion dystonia—has sparked controversy over whether genetic drift or selection is the underlying cause (Risch et al. 2003). Initially, evidence suggested that the high frequency of lysosomal-storage disorders (LSDs), such as Tay-Sachs disease and Gaucher disease, in the Ashkenazim was likely to reflect selection in favor of heterozygous carriers. However, recent data demonstrated that the increased prevalence of torsion dystonia in the Ashkenazim is due to recent founder events, since this autosomal dominant disorder is unlikely to be subjected to heterozygote advantage (Risch et al. 1995, 2003). In the absence of selection, certain other deleterious alleles, such as APC I1307K, likely became highly prevalent in the Ashkenazim because of the occurrence of one or more founder events during this population’s history. Within the Molecular Epidemiology of Colorectal Cancer (MECC) study, a large population-based case-control investigation of incident CRC, we tested whether selective advantage has operated at I1307K. Furthermore, we estimated the age of the I1307K allele, tracing its exposure to founder events throughout history and framing its current ethnic and geographic distribution in the context of the Jewish diasporas.
Material and Methods
Study Population
The MECC study is a population-based case-control investigation of incident CRC cases collected in northern Israel from March 31, 1998, to December 31, 2002. Incident CRCs were identified via rapid case ascertainment in five hospitals in northern Israel, and all CRC cases for these analyses have histologically confirmed cancer of the colon or rectum. The controls were identified from the Kupat Holim Clalit database and were individually matched for exact year of birth, sex, clinic, and Jewish versus non-Jewish heritage. Controls were required to have no prior diagnosis of CRC. The study was approved by the Institutional Review Boards at the University of Michigan and Technion University. Written, informed consent was required for eligibility. For each MECC participant, blood, frozen tumor specimens (if available), and paraffin-embedded tumor specimens (if available) were collected, and an extensive interview was performed. The interview includes information about religion, self-reported ethnic heritage, and country of birth, as well as similar data for parents and grandparents. Eligible participants from the MECC study included all 83 case-control pairs, in which the case, the control, or both members of the pair carried APC I1307K. Two homozygous I1307K-positive cases, eight positive cases without matched controls, and two positive controls without matched cases were also genotyped.
Genotyping
Genomic DNA was extracted from whole blood, using a commercially available kit, according to the manufacturer’s protocol (Puregene DNA extraction kit; Gentra Systems). Samples were amplified in a 20-μl I1307K PCR volume consisting of 40 ng genomic DNA, 2.0 mM MgCl2, 200 μM dNTPs, 0.75 U AmpliTaq DNA polymerase, 165 nM I1307K-F forward primer, and 165 nM I1307K-R reverse primer (table A). Thermal cycling included an initial denaturation step of 95°C for 5 min, then 35 cycles of 1 min denaturing at 95°C, 1 min annealing at 53°C, and 1 min extension at 72°C, with a final 10-min extension step at 72°C. Allele-specific oligonucleotide (ASO) hybridization for I1307K was performed as described elsewhere (Gruber 2001), with two differences: (1) the hybridization and wash steps were performed at 44°C; and (2) the wild-type probe was 5′-CTTTTCTTTTATTCTGC-3′, and the mutant probe was 5′-CTTTTCTTTTTTTTCTGC-3′ (Gruber 2001). For D1822V genotyping, samples were amplified in a 20-μl PCR volume that contained 100 ng genomic DNA, 2.0 mM MgCl2, 200 μM dNTPs, 1.0 U AmpliTaq DNA polymerase, 491 nM primer D1822V-F, and 490 nM primer D1822V-R (table A [online only]). Thermal cycling was similar to that described above for I1307K, except the annealing step was performed at 54°C. ASO hybridization for D1822V was adapted from the above protocol for APC I1307K, with two differences: (1) the hybridization and wash steps were performed at 51.5°C; and (2) the wild-type probe was 5′-ATTCCAAGGACTTCAATGAT-3′, and the mutant probe was 5′-ATTCCAAGGTCTTCAATGAT-3′. For D5S135 genotyping, the 20-μl PCR analysis was performed as described above for the D1822V PCR analysis, except for the following: (1) 482 nM D5S135-F primer and 500 nM D5S135-R primer were used; and (2) the annealing step was performed at 60°C (table A [online only]). ASO hybridization was performed, with three differences: (1) the hybridization was done at 52°C; (2) the wash step was done at 51.5°C; and (3) different probes were used (the wild-type probe was 5′-TATGGAGAAGGCTCACTG-3′, and the mutant probe was 5′-TATGGAGAAGCCTCACTG-3′) (Gruber 2001). For D5S82, D5S346, and D5S122 genotyping, a 20-μl multiplex PCR volume contained 40 ng genomic DNA, 2.0 mM MgCl2, 200 μM dNTPs, 1.0 U AmpliTaq DNA polymerase, 11 nM 32P end-labeled primer D5S82-R, 12 nM 32P end-labeled primer D5S122-F, 11 nM 32P end-labeled primer D5S346-F, 164 nM primer D5S82-F, 160 nM primer D5S122-R, and 130 nM primer D5S346-R (table A [online only]). Thermal cycling was similar to that for I1307K above except the annealing step was performed at 55°C. The radioactively labeled PCR products were then run on a denaturing acrylamide gel electrophoretic system and were analyzed by autoradiography.
Table A.
PCR Primer Pairs For Genotyping Markers
| Marker | Primer Sequence |
| APC I1307K-F | 5′-TCCACACCTTCATCTAATGCC-3′ |
| APC I1307K-R | 5′-TAAACTAGAACCCTGCAGTCTGC-3′ |
| APC D1822V-F | 5′-GAGTTTTCTCAGACAACAAA-3′ |
| APC D1822V-R | 5′-CTAGAGAACTCAAAGAATCA-3′ |
| D5S135-F | 5′-ACTAGGAAGCAGGAGGAACT-3′ |
| D5S135-R | 5′-CCCTGTCTCAAACCAAAAAC-3′ |
| D5S82-F | 5′-ATCAGAGTATCAGAATTTCT-3′ |
| D5S82-R | 5′-CCCAATTGTATAGATTTAGAAGTC-3′ |
| D5S346-F | 5′-ACTCACTCTAGTGATAAATCGGG-3′ |
| D5S346-R | 5′-AGCAGATAAGACAGTATTACTAGTT-3′ |
| D5S122-F | 5′-GTAGGGGTAAATTAGCCTCTC-3′ |
| D5S122-R | 5′-GATAATGGAGGAGGCTATGC-3′ |
Haplotype Determination
Haplotypes were estimated for each member of all 83 matched pairs, using PHASE, version 1.0 (Stephens et al. 2001). PHASE, a Bayesian Markov chain–Monte Carlo algorithm, incorporates coalescent theory to develop haplotype prior probabilities (Stephens et al. 2001). Because the I1307K-positive chromosomes shared a region surrounding I1307K, a common “progenitor haplotype” was identified from the marker loci that showed conserved marker alleles within the shared region. Genotypes of the four homozygous I1307K carriers were examined to identify a putative progenitor haplotype for comparison with PHASE output.
Statistical Analyses
χ2 tests were performed for each marker locus with one or more putative progenitor alleles. Putative progenitor alleles were compared with all other alleles at the marker locus in I1307K-negative versus I1307K-positive chromosomes and were classified in a 2 × 2 table, and a χ2 test was performed (Risch et al. 1995). The present analysis examines each haplotype regardless of whether it is observed in cases or controls, and therefore an unmatched analysis is appropriate. χ2 tests were conducted using SAS, version 8.02 (SAS Institute).
Linkage Disequilibrium
Methods for calculating linkage disequilibrium (LD) have been described elsewhere (Risch et al. 1995). The LD statistic δ was used for each marker, since δ estimates the proportion of I1307K chromosomes with the progenitor marker allele (Devlin and Risch 1995; Risch et al. 1995). Let pD be the proportion of I1307K chromosomes carrying the progenitor allele at the marker locus, and let pN be the proportion of non-I1307K chromosomes carrying the progenitor allele at the marker locus, then δ=(pD-pN)/(1-pN).
Estimation of Allele Age by Use of a Single-Marker Locus
Allele age can be estimated by analyzing the sequence variability surrounding the allele of interest. Intra-allelic variability refers to the variations in sequence at closely linked polymorphic markers among alleles containing the mutation or polymorphism whose age is in question. Starting at the time of the mutation that created the polymorphism itself, recombination and mutation in subsequent generations break down the initially perfect LD between the polymorphism and the progenitor alleles at nearby markers, thereby creating intra-allelic variability (Slatkin and Rannala 2000). This analytical approach to aging has been described elsewhere (Risch et al. 1995). In brief, allele age in number of generations is related to the observed frequency of I1307K-bearing chromosomes not carrying the progenitor marker allele, the frequency (pN) of the progenitor marker allele on I1307K noncarrier chromosomes, and the recombination fraction θ between the marker allele and I1307K:
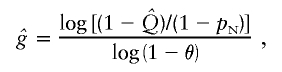 |
where g is the number of generations, and 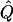 is the observed frequency of I1307K. The marker(s) used in this calculation must demonstrate at least one ancestral allele in strong association with I1307K, but the marker allele cannot be in perfect linkage equilibrium with I1307K (i.e., some recombinations must exist). Because of the small genetic distances involved, the calculated recombination fractions of the Kosambi and Haldane map functions differed by <0.5%, so we report age estimates that incorporate recombination fractions calculated from the Kosambi map function (Haldane 1919; Kosambi 1944). In addition, recombination fraction estimates for D5S346 and D5S135 with respect to APC have been estimated elsewhere, so we report age estimates calculated from these estimates (Nakamura et al. 1988; Olschwang et al. 1995).
is the observed frequency of I1307K. The marker(s) used in this calculation must demonstrate at least one ancestral allele in strong association with I1307K, but the marker allele cannot be in perfect linkage equilibrium with I1307K (i.e., some recombinations must exist). Because of the small genetic distances involved, the calculated recombination fractions of the Kosambi and Haldane map functions differed by <0.5%, so we report age estimates that incorporate recombination fractions calculated from the Kosambi map function (Haldane 1919; Kosambi 1944). In addition, recombination fraction estimates for D5S346 and D5S135 with respect to APC have been estimated elsewhere, so we report age estimates calculated from these estimates (Nakamura et al. 1988; Olschwang et al. 1995).
Because the above approach accounts primarily for recombination, we also estimated the age of the most recent common ancestor of I1307K by using BATWING (Bayesian analysis of trees with internal node generation) software, which incorporates the effects of demographic and mutation processes (Wilson and Balding 1998). BATWING is a Markov chain–Monte Carlo algorithm that uses coalescent theory to generate genealogical trees underlying the sample that are consistent with the sample haplotypes and specified demographic model (Wilson and Balding 1998). The tree height from the generated genealogical trees translates to the time of the most recent common ancestor of I1307K. Two-locus haplotypes, consisting of D5S346 and I1307K, were input into the program, and the stepwise mutation rate for D5S346 was allowed to vary between 0.00028 and 0.0005, according to microsatellite mutation rate data reported by Chakraborty et al. (1997). The specified demographic model assumed exponential growth at a rate of 0.4055, beginning 350 years (14 generations) ago, from a constant-sized ancestral population (Risch et al. 1995). A generation time of 25 years was assumed.
Labuda Correction Factor
A simple approximation for the intra-allelic genealogy assumes a star genealogy, in which all lineages descend independently from the same ancestral allele, so all coalescent times are identical (Slatkin and Rannala 2000). The star genealogy is a reasonable simplification in a very rapidly growing population, such as the Jewish population for the 200 years after 200 bc (Slatkin and Hudson 1991; Barnavi 1992; Reich and Goldstein 1999). However, even slight deviations from a starlike genealogy can introduce a downwardly biased allele age estimate (Slatkin and Rannala 2000). Labuda et al. (1996) present a correction factor for the allele age point estimate, assuming a synchronously bifurcating genealogy. The age estimate from Risch’s approach will be increased by adding the factor {-(1/r)ln[θer/(er-1)]}, where r is the growth rate of the population and θ is the recombination fraction between APC and the marker used for Risch’s allele-age estimate (Slatkin and Rannala 2000). In accordance with census estimates of the Jewish population between 200 bc and ad 0, we used a growth rate of ∼1.125-fold per generation (ln 1.125 = 0.1178 = r), corresponding to the population growth during the period in which the most recent common ancestor of I1307K is hypothesized to have existed (i.e., approximately the beginning of the Jewish diasporas) (Barnavi 1992).
Test for Selection
Slatkin and Bertorelle (2001) developed a method to test neutrality by examining whether the observed intra-allelic variability is consistent with the allele frequency, given two periods of exponential population growth. The Ashkenazim have experienced at least two periods of massive population growth, separated by ∼1,500 years during which no net population growth occurred. We adapted code from Slatkin and Bertorelle’s double exponential model (kindly provided by M. Slatkin) to allow three periods of exponential growth. From 200 bc to ad 0, the worldwide Jewish population increased from 500,000 to 4.5 million. If an average generation of 25 years is assumed, this increase gives a growth rate over this period of 1.125-fold per generation (r3 = ln 1.125 = 0.1178). Until the mid 1600s, the Jewish population experienced a series of expansions and contractions, but no major phases of population growth occurred, so we assigned r2 to be 0.001 (Slatkin 2000). From the 16th to the 19th centuries, the Ashkenazi population underwent an enormous expansion, with a growth rate of 1.5-fold per generation (r1 = ln 1.5 = 0.4055) (Risch et al. 1995). Input parameters for the triple exponential model were derived from our Ashkenazi data, under the assumption of a 6% carrier frequency of I1307K in the general Ashkenazi population, the dinucleotide microsatellite mutation rate for D5S346 of 0.0005 from Chakraborty et al. (1997), the population growth estimates outlined above, as well as the effective Ashkenazi population size outlined by Slatkin (2000).
Results
The demographic characteristics of cases and controls are shown in table 1. Of the 85 I1307K-positive individuals, 81 (95.3%) are Ashkenazi Jews, including both homozygous positive individuals; 2 (2.4%) are Sephardi Jews; and 2 (2.4%) are Arabs (table 1).
Table 1.
Demographic Data for 83 Matched Pairs
| Individual, APC I1307KStatus, and Ethnicity | n |
| Case: | |
| Positive: | |
| Ashkenazi | 52 |
| Sephardi | 1 |
| Homozygous positive: | |
| Ashkenazi | 1 |
| Negative: | |
| Ashkenazi | 24 |
| Sephardi | 2 |
| Arab | 2 |
| Unknown | 1 |
| Control: | |
| Positive: | |
| Ashkenazi | 27 |
| Sephardi | 1 |
| Arab | 2 |
| Homozygous positive: | |
| Ashkenazi | 1 |
| Negative: | |
| Ashkenazi | 39 |
| Sephardi | 13 |
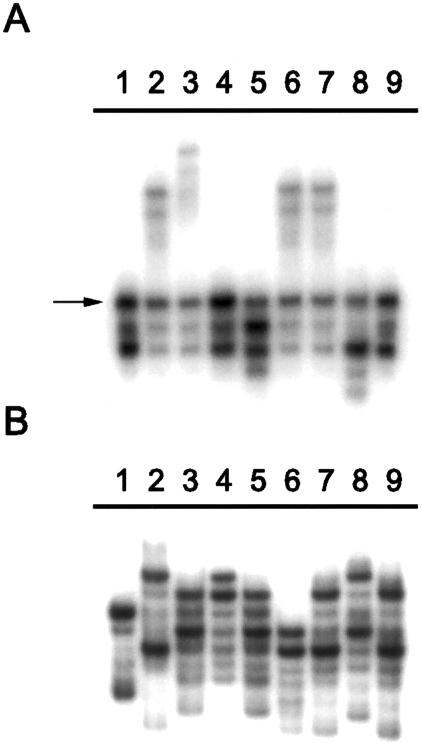
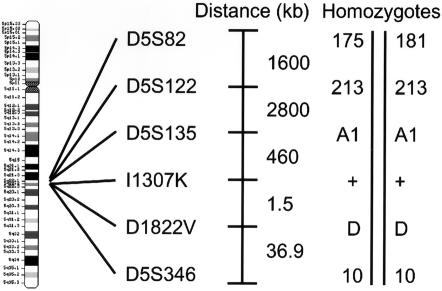
Genotyping data demonstrated conserved alleles at certain marker loci surrounding I1307K. Individuals carrying I1307K share a common allele at the microsatellite marker D5S346 (fig. 1A), but they do not share a common allele at the microsatellite marker D5S82 (fig. 1B), indicating that the progenitor haplotype of the most recent common ancestor of modern-day I1307K alleles incorporates D5S346 but not D5S82. To further identify which markers harbor conserved alleles across I1307K carriers, we evaluated the genotypes of four I1307K-positive homozygotes. All four homozygotes shared the 213-bp allele at D5S122, the A1 allele at D5S135, the D (aspartic acid–encoding) allele at D1822V, and the 10 allele at D5S346 (fig. 2). At D5S82, each homozygote carried one copy of the 175-bp allele and one copy of the 181-bp allele (fig. 2).
Figure 1.
Representative genotypes of APC I1307K carriers. A, Conserved allele at D5S346 among I1307K carriers observed among Ashkenazi Jews (lanes 1–3), Sephardi Jews (lanes 4–6), and Arabs (lanes 7–9) (arrow). B, Absence of conserved genotype at D5S82 among Ashkenazi Jews (lanes 1–3), Sephardi Jews (lanes 4–6), or Arabs (lanes 7–9).
Figure 2.
Conserved alleles at markers linked to I1307K. These alleles, shown for homozygous positive individuals, construct a putative progenitor haplotype of the most recent common ancestor of I1307K.
After constructing haplotypes for each individual within the 83 matched pairs, we analyzed the number of I1307K-positive haplotypes that carried the putative progenitor marker alleles suggested by the genotyping data of the I1307K homozygotes. Of the 87 I1307K-positive haplotypes, 73 (83.9%) at D5S122, 83 (95.4%) at D5S135, 86 (98.9%) at D1822V, and 85 (97.7%) at D5S346 carried the same marker alleles (table 2). At D5S82, 36 (41.4%) carried the 175-bp allele, and 28 (32.2%) carried the 181-bp allele. These putative progenitor alleles are consistent across ethnic heritage (table 2).
Table 2.
Putative Progenitor Marker Alleles on APCI1307K-Positive Haplotypes
| Locus, PutativeProgenitor Allele,and Ethnicity | No. of Haplotypeswith PutativeProgenitorMarker Allele | No. of Haplotypeswith MarkerAlleles Other Than Progenitor |
| D5S82: | ||
| 175 bp: | ||
| Ashkenazi | 34 | 49 |
| Sephardi | 1 | 1 |
| Arab | 1 | 1 |
| D5S82: | ||
| 181 bp: | ||
| Ashkenazi | 26 | 57 |
| Sephardi | 1 | 1 |
| Arab | 1 | 1 |
| D5S122: | ||
| 213 bp: | ||
| Ashkenazi | 72 | 11 |
| Sephardi | 1 | 1 |
| Arab | 0 | 2 |
| D5S135: | ||
| A1: | ||
| Ashkenazi | 79 | 4 |
| Sephardi | 2 | 0 |
| Arab | 2 | 0 |
| D1822V: | ||
| D: | ||
| Ashkenazi | 82 | 1 |
| Sephardi | 2 | 0 |
| Arab | 2 | 0 |
| D5S346: | ||
| 10: | ||
| Ashkenazi | 81 | 2 |
| Sephardi | 2 | 0 |
| Arab | 2 | 0 |
χ2 analyses were performed to formally test whether each marker’s putative progenitor allele was more common on I1307K-positive haplotypes than on I1307K-negative haplotypes (table 3). The A1 allele at D5S135 was significantly more prevalent on I1307K-positive haplotypes (P=.002), as were the D allele and the 10 allele at D1822V and D5S346, respectively (P<.0001). Each of the markers—D5S135, D1822V, and D5S346—demonstrates high LD with I1307K (table 3). At D5S82, the 175-bp allele, but not the 181-bp allele, was significantly more prevalent on I1307K-positive haplotypes (P=.05). However, at D5S122, which is located between I1307K and D5S82, no allele was found more commonly among I1307K carriers (P=.898) (fig. 2). Both D5S82 and D5S122 demonstrate low LDs with I1307K (table 3). These data indicate that the I1307K progenitor haplotype spans across APC I1307K from the centromeric marker D5S135 to the telomeric marker D5S346, constituting a physical distance of ∼500,000 bp.
Table 3.
Analyses of Putative Progenitor Marker Alleles in APC I1307K Carrier and Noncarrier Haplotypes
| Locus (Allele) | No. of Carriers/Noncarriersof Putative ProgenitorAllele in I1307K | No. of Carriers/Noncarriersof All Other Allelesin I1307K | Pa | δ |
| D5S82 (175 bp) | 36/73 | 51/172 | .048 | .165 |
| D5S82 (181 bp) | 28/94 | 59/151 | .304 | −.1 |
| D5S122 (213 bp) | 73/207 | 14/38 | .898 | −.038 |
| D5S135 (A1) | 83/199 | 4/46 | .002 | .755 |
| D1822V (D) | 86/187 | 1/58 | <.0001 | .951 |
| D5S346 (10) | 85/32 | 2/213 | <.0001 | .974 |
By χ2 analysis.
Because a balance between high LD and sufficient recombination is required for age estimation via the intra-allelic variability approach, single-marker age estimates for I1307K could be calculated with data from markers D5S135 and D5S346, using recombination fractions obtained from the Kosambi map function (table 4). On the basis of the data at D5S135 without the Labuda correction, the most recent common ancestor of present-day I1307K alleles existed 60.9 generations ago (ad ∼480). Given the data at D5S346 and without the Labuda correction, the most recent common ancestor of present-day I1307K alleles existed 69.5 generations ago, (ad ∼265). When the data at D5S135 are analyzed using the Labuda correction, the most recent common ancestor of present-day I1307K alleles dates to 87.9 generations ago (∼195 bc); and at D5S346, the Labuda-adjusted age estimate is 118 generations (∼947 bc).
Table 4.
APC I1307K Age Estimates
| Characteristic | D5S135 | D5S346 |
| Physical distance (kb) | 460 | 38.5 |
| Kosambi-calculated θ | .0046 | .000385 |
| Age estimate (in generations) | 60.9 | 69.5 |
| Age estimate (in years)a | 1,523 | 1,738 |
| Date of most recent common ancestor | 480 ad | 265 ad |
| Adjusted age estimate (in generations)b | 87.9 | 118 |
| Adjusted age estimate (in years)a,b | 2,198 | 2,950 |
| Adjusted date of I1307K most recent common ancestor b | 195 bc | 947 bc |
Calculated from the age estimate in generations, assuming a 25-year generation.
Incorporates the Labuda correction factor.
Because recombination rate is one of the major sources of uncertainty inherent in these calculations, table 5 presents the age estimates for the most recent common ancestor of present-day I1307K alleles within our study population at varying recombination rates. At D5S135, a recombination rate of 0.002, half the recombination rate calculated from the Kosambi map function, suggests an age estimate of 140.3 generations ago, which corresponds to 1505 bc. If the Kosambi-calculated recombination fraction is doubled, to 0.009, it provides an age estimate of 31.1 generations ago (∼777 years ago). Varying the recombination fraction at D5S346 yielded similar results (table 5). Use of the recombination fractions previously reported in the literature gave estimates indicating very recent origin of the allele: the recombination fraction of 0.05 at D5S346 indicated that the most recent common ancestor of I1307K existed in ad 1990, and the recombination fraction of 0.02 at D5S135 indicated that the most recent common ancestor existed in ad 1655 (table 5).
Table 5.
Sensitivity Analysis of APCI1307K Age Estimates to Varying Recombination Fractions
| Locus and θ | No. ofGenerationsa | Yearb |
| D5S135: | ||
| θkosambi= .0046 | 60.9 | 481 ad |
| θliteraturec= .02 | 13.9 | 1655 ad |
| θlowd= .002 | 140.3 | 1505 bc |
| θhighe= .04 | 6.9 | 1830 ad |
| D5S346: | ||
| θkosambi= .000385 | 69.5 | 265 ad |
| θliteraturef= .05 | .52 | 1990 ad |
| θlowd= .0002 | 134.0 | 1347 bc |
| θhighe= .0008 | 33.5 | 1165 ad |
Without the Labuda correction factor.
Calculated from the age estimate in generations, assuming a 25-year generation.
Recombination fraction between APC and D5S135 reported by Olschwang et al. (1995).
θlow is half of the lowest estimated recombination fraction, either from the literature or the Kosambi map function calculations.
θhigh is twice the highest estimated recombination fraction, either from the literature or the Kosambi map function calculations.
θliterature is the recombination fraction between APC and D5S346 reported by Nakamura et al. (1988).
Given the low recombination rate between D5S346 and I1307K, mutation might be an important factor underlying the intra-allelic variability observed in our data. In addition, modeling the demographic history of the population, such as population growth and size, might have significant impact on the intra-allelic genealogy of I1307K used to estimate allelic age. When demographic and mutation processes were incorporated, the BATWING algorithm estimated that the most recent common ancestor of I1307K existed 2,490 years ago (∼487 bc).
Age estimates can also be affected by the presence of selection. We tested whether selection was operating at I1307K by examining whether the observed number of I1307K progenitor haplotypes within the Ashkenazi carriers is consistent with the population frequency of the I1307K allele within the Ashkenazi population as a whole. At D5S346 or D5S135, the data do not allow us to reject neutrality (P=.114 and P=.373, respectively). Therefore, we conclude that our data do not suggest selection at I1307K.
Because of the small number of non-Ashkenazi I1307K carriers among our 83 matched pairs, we genotyped 10 additional unmatched I1307K carriers, consisting of 8 cases and 2 controls. All 10 of these I1307K carriers share the I1307K progenitor haplotype, spanning across APC from D5S135 to D5S346. Table 6 shows the ethnic heritage of each individual included in this combination of 10 non-Ashkenazi I1307K carriers with the 2 Sephardi and 2 Arab I1307K carriers from our matched-pair data. These data indicate that the conserved I1307K haplotype exists in individuals from various ethnic backgrounds and geographical locales, including Yemen, Syria, France, Argentina, Morocco, Egypt, Turkey, Palestine, and Israel.
Table 6.
Ethnic and Geographic Origin of 13 Non-Ashkenazi APC I1307K Carriers in the MECC Study[Note]
|
Birth Country of |
|||
| EthnicHeritage | MECCParticipant | Father | Mother |
| Sephardi | Argentina | Syria | Syria |
| Sephardi | Egypt | Syria | Egypt |
| Sephardi | Morocco | Morocco | Morocco |
| Sephardi | Syria | Syria | Syria |
| Sephardi | France | France | France |
| Sephardi | Egypt | Egypt | Egypt |
| Sephardi | Turkey | Turkey | Turkey |
| Sephardi | Israel | Yemen | Yemen |
| Sephardi | Palestine | Palestine | Palestine |
| Sephardi | Israel | Syria | Syria |
| Christian Arab | Israel | Palestine | Palestine |
| Muslim Arab | Palestine | Palestine | Palestine |
| Muslim Arab | Palestine | Palestine | Palestine |
Note.— Information about origin was not available for 1 of the 14 non-Ashkenazi I1307K carriers.
Discussion
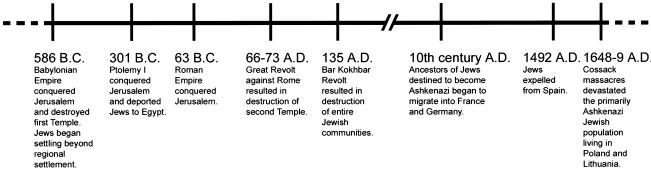
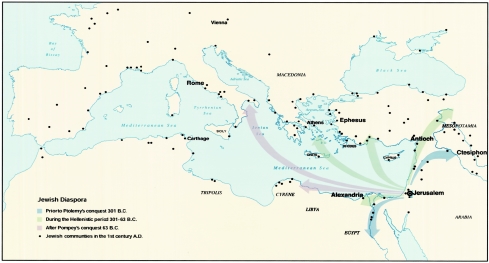
The low prevalence of I1307K in non-Ashkenazi Jews, compared with the high prevalence within the Ashkenazim, suggests one or a combination of three possible explanations: (1) genetic exchange recently occurred, (2) the non-Ashkenazi I1307K carriers have at least one Ashkenazi I1307K-carrying ancestor, or (3) the I1307K mutation preceded or coincided with the Jewish diasporas and was followed by a founder effect specifically in the Ashkenazim. The diasporas refer to the settling of scattered colonies of Jews outside the area of Jerusalem and vicinity. Our research indicates that the most recent common ancestor of present-day I1307K alleles likely existed between 947 bc and 195 bc. Let us consider this range of age estimates within the framework of Jewish history (fig. 3). The fall of Jerusalem in 586 bc to the Babylonian empire spurred the exile of Jews, primarily to Mesopotamia and the banks of the Nile (figs. 3 and 4) (Barnavi 1992). In 301 bc, Ptolemy I conquered Jerusalem in the name of the Greek Empire and deported large numbers of Jews to Egypt. From 301 bc to 63 bc, Jews continued moving into Egypt, as well as into the present-day areas of Syria, Turkey, and Greece (fig. 4). Following the conquest of Jerusalem by the Roman Empire’s General Pompey, in 63 bc, many Jews fled to regions of present-day Italy and northern Africa. The Great Revolt against Rome and the fall of Jerusalem in ad 66–73 caused the destruction of the Second Temple by Titus and the continuation of the diaspora (fig. 4). In ad 135, Emperor Trajan’s forces quelled the Bar Kokhba Revolt in Palestine. This event precipitated numerous persecutions of Jews, which resulted in hundreds of thousands of deaths, as well as the almost total destruction of Jewish communities in Alexandria and northern Africa. Starting in the 1st century ad, small Jewish communities existed in Europe as far north as London; Jewish communities were also present as far west as northern Africa and as far east as western Asia (Barnavi 1992) (fig. 4). However, it was only in the mid-10th century that the Jewish communities destined to become Ashkenazi migrated from southern Europe into the regions of France and Germany (Barnavi 1992). Given this history, the observation that I1307K can be found in non-Ashkenazi individuals is not surprising, because the most recent common ancestor of modern-day I1307K alleles existed sometime between 947 bc and 195 bc, before or near the beginning of the Jewish diaspora. The age estimate for APC I1307K—and its resulting presence in both Ashkenazim and non-Ashkenazim—is consistent with the age estimates and ethnic distributions of other mutations found primarily in Jewish individuals. BRCA1 (MIM 113705) 185delAG dates to 46 generations ago, after the diasporas, and this mutation is found almost exclusively in Ashkenazi individuals (Neuhausen et al. 1996). However, the most recent common ancestor for the F11 (MIM 264900) E117X mutation causing factor XI deficiency type II existed 120 generations ago, before the diasporas, and this mutation is observed in both Ashkenazi and non-Ashkenazi individuals today (Goldstein et al. 1999).
Figure 3.
Timeline of selected events in the history of the Jewish peoples
Figure 4.
The Jewish diasporas. The existence of APC I1307K prior to the Jewish diasporas explains its presence in non-Ashkenazi peoples residing in geographic locales populated by the diasporas. Adapted from Barnavi (1992).
APC I1307K likely represents an example of an allele achieving a high frequency in the Ashkenazim via genetic bottleneck due to the rapid growth of the Ashkenazi population from a small group of founders. The Cossack massacres in 1648–1649 devastated the primarily Ashkenazi Jewish populations living in Poland and Lithuania, with ∼25% of the existing population killed (Weinryb 1972; Barnavi 1992). The census of 1765 estimated that 430,000 Jews lived in Poland and 130,000 lived in Ukraine (Weinryb 1972). By 1900, there were 5 million Jews living in these regions, indicating that the population increased ∼10-fold during these 135 years (Weinryb 1972; Risch et al. 1995). Given massive population growth from a small founder population at least once in the history of the Ashkenazim, the higher prevalence of I1307K in Ashkenazi Jews than in other Jewish populations is consistent with genetic drift alone and does not require the forces of natural selection. CRC primarily affects individuals who are past reproductive age, with an average age at onset of 70.2 years in Ashkenazi I1307K carriers, so selection is unlikely to be responsible for the high allele frequency of APC I1307K (Gryfe et al. 1999; Stern et al. 2001). However, it is arguable that some unknown effect of I1307K could lead to selection for or against the polymorphic allele. To investigate this possibility, we tested the neutrality of I1307K, using the technique developed by Slatkin and Bertorelle (2001), and found no evidence of selection. To investigate genetic drift versus selection as the primary cause of highly prevalent disease mutations in the Ashkenazim, Risch and colleagues (2003) recently compared mutations causing LSDs to mutations causing non–lysosomal-storage diseases (NLSD), including Bloom syndrome, cystic fibrosis, and familial hypercholesterolemia, among others. That study identified no differences between the LSD and the NLSD with respect to the number of mutations, the allele-frequency distributions, or the estimated ages of mutations. Those data indicate that genetic drift due to two founder events (one ∼11 centuries ago and another ∼5 centuries ago), rather than selection, is the most likely explanation for the high frequency of disease mutations in the Ashkenazim (Risch et al. 2003). Our data provide further evidence that genetic drift, not natural selection, is the primary mechanism causing the high prevalence of disease mutations in the Ashkenazim.
The analytical approach used in this research presents a technique to estimate allele age by utilizing intra-allelic variability data. This statistical method has two crucial features. First, the age estimated is that of the most recent common ancestor of all present-day allele copies, not the age(s) of the I1307K mutation event(s). It is possible that many I1307K mutation events have occurred, but we assume that only one event led to all descendant alleles currently in the population. Second, no demographic assumptions, such as the structure of population growth, are required to estimate allele age, although they can be incorporated when using the BATWING algorithm (Wilson and Balding 1998; Slatkin and Rannala 2000).
However, two assumptions are required for the intra-allelic variability aging method to accurately determine the age of the most recent common ancestor of I1307K. First, it is assumed that recombination, not mutation, causes the decay of LD. Second, the method assumes that a progenitor haplotype can be established. The haplotypes in the present study were statistically derived using PHASE; therefore, errors in haplotype determination are possible. However, because our analyses used pairwise comparisons, our results are robust to haplotyping error. When comparing haplotype error in PHASE to the EM algorithm, Stephens et al. (2001) demonstrated that PHASE reduced error rates by 14%–69% when applied to genotypes from pedigree data with unambiguous phase or when applied to molecularly determined haplotypes. While we were establishing the progenitor haplotype, it became evident that the majority of I1307K-positive chromosomes had one of two marker alleles at D5S82 but that no conserved allele was present at D5S122, which is located closer to I1307K. It is likely that, soon after the I1307K polymorphism event, recombination occurred near D5S82, creating I1307K haplotypes with either a 3 allele or a 6 allele at D5S82. Subsequent generations of mutation and recombination have further broken down the LD between D5S82 and I1307K. We concluded that, because no conserved allele exists at D5S122, the present-day I1307K progenitor haplotype spans the distance from the centromeric marker D5S135 to the telomeric marker D5S346. This conclusion is likely to be valid, because a common four-marker haplotype containing the I1307K allele in Ashkenazi and Yemenite Jews has been described elsewhere (Patael et al. 1999). This reported progenitor haplotype includes three APC intragenic markers and D5S346; however, it excludes D5S82, and it was found in 41% of unselected Ashkenazi Jews and 23.4% of unselected Yemenite Jews (Patael et al. 1999).
Because the intra-allelic variability technique used to estimate the age of I1307K relies on the LD statistic and the recombination fraction between I1307K and the marker of interest, errors in either or both of these calculations could significantly alter results. Our LD statistic δ incorporates pD, the proportion of I1307K chromosomes carrying the progenitor allele at the marker locus, and pN, the proportion of noncarrier chromosomes carrying the progenitor allele at the marker locus. Both pD and pN were derived from our sample data, but pN could also be determined from other sources. For D5S346, the CEPH reports an allele 10 frequency of 0.206, compared with .131 within our data. Given a value of 0.206 for pN, the I1307K age estimate, without the Labuda correction, becomes 76.3 generations instead of 69.5 generations. At D5S135, Olschwang et al. (1995) defined an allele frequency of 0.8 for the A1 allele, giving a Labuda-unadjusted age estimate of 56.7 generations, compared with 60.9 generations from our data. These virtually identical age results indicate that our study population provides reasonable estimates of the proportion of noncarrier chromosomes carrying the progenitor allele at the marker locus, and, therefore, our LD calculations are unlikely to adversely affect the allele age estimates. However, age estimates are exquisitely sensitive to recombination fraction estimates. The marker loci used in these analyses are so tightly linked that recombination fractions cannot be accurately estimated from genetic analyses, so recombination fractions were calculated using the Kosambi map function. Because these calculations do not accurately reflect the true recombination fractions either, we reported age estimates from a range of recombination fractions consistent with available data, as was done by Serre et al. (1990), instead of presenting SE estimates. This sensitivity analysis demonstrates that varying the recombination fractions twofold provides drastically different age estimates. If the hypothesis that the most recent common ancestor of I1307K preceded the Jewish diasporas is correct, it would be difficult to distinguish which of the age estimates before the diaspora of the First Temple period (586 bc) through the diaspora of the Second Temple period (ad 70) was most accurate, because it would be difficult to distinguish their outcomes by use of the ethnic distribution of I1307K today. Furthermore, an age estimate more recent than the Cossack massacres in 1648–1649 is unlikely, because a founder effect is likely responsible for the high prevalence of I1307K in the Ashkenazim, especially given that there is no evidence of selection at I1307K.
Because the I1307K ancestor likely existed during a time when the Jewish population size increased dramatically, the I1307K age estimate is biased toward a more recent date (unless corrected) (Labuda et al. 1996). In the 200 years after 200 bc, the Jewish population increased in size, from ∼500,000 to ∼4.5 million (Barnavi 1992). Because this was a rapidly growing population, recombination events involving the I1307K polymorphism that occurred in early generations were less likely than those in later generations, according to Luria-Delbruck theory (Labuda et al. 1996). Therefore, the omission of the contribution of these early generations produces an estimate of the likely number of recombinants, which is the basis for the Labuda correction factor incorporated into our calculations. However, it is likely that the census estimates are inaccurate, and it is possible that the Jewish population size did not increase as drastically as was assumed in our calculations. It is noteworthy that a smaller population growth rate would increase the Labuda correction factor, which would suggest that the most recent common ancestor of I1307K existed earlier than our calculations indicated.
The impact of I1307K on CRC incidence is substantial in the Ashkenazim, with a population-attributable fraction of ∼5% in this group. However, the impact of I1307K is less studied in other Jewish populations. Although further studies are needed to examine the value of presymptomatic testing for I1307K, understanding the evolution of APC I1307K might enable health care providers to better understand which individuals should eventually be offered genetic testing in a clinical setting. Because a conserved I1307K haplotype is shared among Jews and Arabs and because the allele age of I1307K indicates that the most recent ancestor of I1307K existed sometime between 947 bc and 195 bc (prior to the Jewish diasporas), future genetic testing for this polymorphism might be offered to Jewish individuals as well as members of other ethnic populations (Patael et al. 1999). Furthermore, the understanding that APC I1307K did not achieve its high frequency within the Ashkenazim via selective pressure provides additional evidence that genetic drift is the primary cause of the high prevalence of disease mutations, including I1307K, within the Ashkenazim.
Acknowledgments
The authors wish to thank Dr. Montgomery Slatkin for kindly supplying the source code for the double exponential test of neutrality. The authors also wish to thank Diana Duque for sharing her artistic talent. This research has been supported in part by National Institutes of Health grants R01-CA81488 (to S.B.G and G.R.), NIGMS T32 GM07863 (to B.L.N), and T32 HG00040 (to B.L.N.).
Electronic-Database Information
URLs for data presented herein are as follows:
- Centre d’Etude du Polymorphisme Humain (CEPH) database, http://www.cephb.fr
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for APC, BRCA1, and F11)
References
- Barnavi E (ed) (1992) A historical atlas of the Jewish people. Schocken Books, New York [Google Scholar]
- Chakraborty R, Kimmel M, Stivers DN, Davison LJ, Deka R (1997) Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc Natl Acad Sci USA 94:1041–1046 10.1073/pnas.94.3.1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Risch N (1995) A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics 29:311–322 10.1006/geno.1995.9003 [DOI] [PubMed] [Google Scholar]
- Drucker L, Shpilberg O, Neumann A, Shapira J, Stackievicz R, Beyth Y, Yarkoni S (2000) Adenomatous polyposis coli I1307K mutation in Jewish patients with different ethnicity: prevalence and phenotype. Cancer 88:755–760 [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Reich DE, Bradman N, Usher S, Seligsohn U, Peretz H (1999) Age estimates of two common mutations causing factor XI deficiency: recent genetic drift is not necessary for elevated disease incidence among Ashkenazi Jews. Am J Hum Genet 64:1071–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee RT, Hill-Harmon MB, Murray T, Thun M (2000) Cancer statistics, 2000. CA Cancer J Clin 50:7–3310735013 [Google Scholar]
- Gruber SB (2001) Assay for detecting the I1307K susceptibility allele within the adenomatous polyposis coli gene. In: Killeen AA (ed) Methods in molecular medicine. Humana Press, Totowa, NJ [DOI] [PubMed] [Google Scholar]
- Gryfe R, Di Nicola N, Lal G, Gallinger S, Redston M (1999) Inherited colorectal polyposis and cancer risk of the APC I1307K polymorphism. Am J Hum Genet 64:378–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175 [Google Scholar]
- Labuda M, Labuda D, Korab-Laskowska M, Cole DEC, Zietkiewicz E, Weissenbach J, Popowska E, Pronicka E, Root AW, Glorieux FH (1996) Linkage disequilibrium analysis in young populations: pseudo–vitamin D–deficiency rickets and the founder effect in French Canadians. Am J Hum Genet 59:633–643 [PMC free article] [PubMed] [Google Scholar]
- Laken SJ, Petersen GM, Gruber SB, Oddoux C, Ostrer H, Giardiello FM, Hamilton SR, Hampel H, Markowitz A, Klimstra D, Jhanwar S, Winawer S, Offit K, Luce ML, Kinzler KW, Vogelstein B (1997) Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat Genet 17:79–83 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Lathrop M, Leppert M, Dobbs M, Wasmuth J, Wolff E, Carlson M, Fujimoto E, Krapcho K, Sears T, Woodward S, Hughes J, Burt R, Gardner E, Lalouel J, White R (1988) Localization of the genetic defect in familial adenomatous polyposis within a small region of chromosome 5. Am J Hum Genet 43:638–644 [PMC free article] [PubMed] [Google Scholar]
- Neuhausen SL, Mazoyer S, Friedman L, Stratton M, Offit K, Caligo A, Tomlinson G, Cannon-Albright L, Bishop T, Kelsell D, Solomon E, Weber B, Couch F, Struewing J, Tonin P, Durocher F, Narod S, Skolnick MH, Lenoir G, Serova O, Ponder B, Stoppa-Lyonnet D, Easton D, King M, Goldgar D (1996) Haplotype and phenotype analysis of six recurrent BRCA1 mutations in 61 families: results of an international study. Am J Hum Genet 58:271–280 [PMC free article] [PubMed] [Google Scholar]
- Olschwang S, Laurent-Puig P, Melot T, Thuille B, Thomas G (1995) High resolution genetic map of the adenomatous polyposis coli gene (APC) region. Am J Med Genet 56:413–419 [DOI] [PubMed] [Google Scholar]
- Patael Y, Figer A, Gershoni-Baruch R, Papa MZ, Risel S, Schtoyerman-Chen R, Karasik A, Theodor L, Friedman E (1999) Common origin of the I1307K APC polymorphism in Ashkenazi and non-Ashkenazi Jews. Eur J Hum Genet 7:555–559 [DOI] [PubMed] [Google Scholar]
- Prior TW, Chadwick RB, Papp AC, Arcot AN, Isa AM, Pearl DK, Stemmerman G, Percesepe A, Loukola A, Aaltonen LA, de la Chapelle A (1999) The I1307K polymorphism of the APC gene in colorectal cancer. Gastroenterology 116:58–63 [DOI] [PubMed] [Google Scholar]
- Reich DE, Goldstein DB (1999) Estimating the age of mutations using variation at linked markers. In: Goldstein DB, Schotterer C (eds) Microsatellies: evolution and application. Oxford University Press, Oxford [Google Scholar]
- Risch N, de Leon D, Ozelius L, Kramer P, Almasy L, Singer B, Fahn S, Breakefield X, Bressman S (1995) Genetic analysis of idiopathic torsion dystonia in Ashkenazi Jews and their recent descent from a small founder population. Nat Genet 9:152–159 [DOI] [PubMed] [Google Scholar]
- Risch N, Tang H, Katzenstein H, Ekstein J (2003) Geographic distribution of disease mutations in the Ashkenazi Jewish population supports genetic drift over selection. Am J Hum Genet 72:812–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen P, Shomrat R, Strul H, Naiman T, Karminsky N, Legum C, Orr-Urtreger A (1999) Prevalence of the I1307K APC gene variant in Israeli Jews of differing ethnic origin and risk for colorectal cancer. Gastroenterology 116:54–57 [DOI] [PubMed] [Google Scholar]
- Serre JL, Simon-Bouy B, Mornet E, Jaume-Roig B, Balassopoulou A, Schwartz M, Taillandier A, Boue J, Boue A (1990) Studies of RFLP closely linked to the cystic fibrosis locus throughout Europe lead to new considerations in populations genetics. Hum Genet 84:449–454 [DOI] [PubMed] [Google Scholar]
- Slatkin M (2000) Allele age and a test for selection on rare alleles. Philos Trans R Soc Lond B 355:1663–1668 10.1098/rstb.2000.0729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Bertorelle G (2001) The use of intraallelic variability for testing neutrality and estimating population growth rate. Genetics 158:865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Hudson RR (1991) Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129:555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Rannala B (2000) Estimating allele age. Annu Rev Genomics Hum Genet 1:225–249 10.1146/annurev.genom.1.1.225 [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern HS, Viertelhausen S, Hunter AGW, O’Rourke K, Cappelli M, Perras H, Serfas K, Blumenthall A, Dewar D, Baumann E, Lagarde AE (2001) APC I1307K increases risk of transition from polyp to colorectal carcinoma in Ashkenazi Jews. Gastroenterology 120:392–400 [DOI] [PubMed] [Google Scholar]
- Weinryb BD (1972) The Jews of Poland: a social and economic history of the Jewish community of Poland from 1100 to 1880. The Jewish Publication Society of America, Philadelphia [Google Scholar]
- Wilson IJ, Balding DJ (1998) Genealogical inference from microsatellite data. Genetics 150:499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]