Abstract
Plasma proteins represent an important part of the human proteome. Although recent proteomics research efforts focus largely on determining the overall number of proteins circulating in plasma, it is equally important to delineate protein variations among individuals, because they can signal the onset of diseases and be used as biological markers in diagnostics. To date, there has been no systematic proteomics effort to characterize the breadth of structural modifications in individual proteins in the general population. In this work, we have undertaken a population proteomics study to define gene- and protein-level diversity that is encountered in the general population. Twenty-five plasma proteins from a cohort of 96 healthy individuals were investigated through affinity-based mass spectrometric assays. A total of 76 structural forms/variants were observed for the 25 proteins within the samples cohort. Posttranslational modifications were detected in 18 proteins, and point mutations were observed in 4 proteins. The frequency of occurrence of these variations was wide-ranged, with some modifications being observed in only one sample, and others detected in all 96 samples. Even though a relatively small cohort of individuals was investigated, the results from this study illustrate the extent of protein diversity in the human population and can be of immediate aid in clinical proteomics/biomarker studies by laying a basal-level statistical foundation from which protein diversity relating to disease can be evaluated.
Keywords: mass spectrometry, population proteomics, protein modifications
It is now becoming clear that the complexity of gene products within humans extends dramatically beyond the 20,000-25,000 protein-coding genes in the human genome (1). Foremost, transcriptional variations predict the number of encoded proteins to be well in excess of 100,000. Additionally, ample empirical data are now indicating that a large number of single-nucleotide polymorphisms have minor allele frequencies of >20% in the general population (2), further increasing the number of non-wild-type proteins. These gene-level variations are subsequently exacerbated by a myriad of endogenous posttranslational modifications (3). Finally, environmental changes can influence both the structure and expression level of proteins. In all, it is accurate from first principles to view variations in the human protein complement as structurally, quantitatively, and temporally dynamic.
This statement is not to say that the study of protein diversity in humans is intractable, just more complicated from an analytical point of view than is often acknowledged. Importantly, proteomics investigations have now been driven to the point of being applied clinically in the detection of several diseases (4, 5), although not without major debate and controversies (6, 7). As these studies have proceeded to the doorstep of application in diagnostics, fundamental differences in proteins occurring naturally between individuals, and stemming from the aforementioned causes, have been largely overlooked. Given that numerous previously characterized diseases have been linked to minor structural variants and quantitative differences, it stands to reason that proteomics investigations need to account for variations between individuals, whether for a single or a group of protein(s), before application in a diagnostic setting. The fundamental points of comparison for such application are the basal-level differences found in the general, healthy population. In this study, we have undertaken a concerted research effort to delineate such differences in a small cohort of healthy individuals. Twenty-five plasma proteins from 96 plasma samples were assayed through affinity-based mass spectrometry approaches, and protein variants were identified and characterized in each sample.
Materials and Methods
Commercially available antibodies to all 25 proteins were obtained from several sources. Rabbit anti-human polyclonal antibodies to α-1-antitrypsin (catalog no. A0012), β2-microglobulin (O9100), C-reactive protein (A0073), cerruloplasmin (A0031), cystatin C (A0451), GC Globulin (A0021), lysozyme (A0099), orosomucoid (A0011), plasminogen (A0081), retinol binding protein (A0040), serum amyloid P component (A0302), transferrin (A0061), transthyretin (A0002), and urine protein 1 (A0257) were purchased from DakoCytomation (Carpinteria, CA); rabbit anti-human polyclonal serum albumin antibody (A1327-45) was purchased from US Biologicals (Swampscott, MA). Rabbit anti-human polyclonal antibodies to insulin-like growth factor I (PA0362) and insulin-like growth factor II (PA0382) were purchased from Cell Sciences (Norwood, MA). Mouse anti-human monoclonal antibody to serum amyloid A (MO-C40028A) was purchased from Anogen (Mississauga, ON, Canada). Goat anti-human polyclonal antibody to antithrombin III (ACL20016AP) was purchased from Accurate Chemical (Westbury, NY). Goat anti-human polyclonal antibodies to apolipoprotein A-I (11A-2b), apolipoprotein A-II (12A-G1b), apolipoprotein C-I (31A-G1b), apolipoprotein C-II (32A-G2b), apolipoprotein C-III (33A-G2b), and apolipoprotein E (50A-G1b) were purchased from Academy Bio-Medical (Houston). 1,1′-Carbonyldiimidazole-activated affinity pipettes (Intrinsic Bioprobes, Tempe, AZ) were prepared and derivatized with the antibodies as described in ref. 8. Proteolytic enzyme-derivatized MALDI targets (Intrinsic Bioprobes) were manufactured and used as described in ref. 9.
Ninety-six samples of human plasma (in 5-ml volumes) were obtained through ProMedDX (Norton, MA). The samples were collected at certified blood donor and medical centers and designated as normal based on their nonreactivity for common blood infectious agents and donor information. Samples were provided labeled only with a barcode and an accompanying specification sheet containing information about the gender, age, and ethnicity of each donor, thus ensuring proper privacy protection. Of the 96 sample donors, 49 declared themselves as Caucasians (26 males and 23 females), and 47 as African Americans (25 males and 22 females), with ages ranging from 18 to 65 (median age of 36). The plasma samples were divided into 100-μl aliquots and kept at -75°C until use. For the analyses of most of the proteins, a 100-μl aliquot of each of the 96 samples was placed into a particular 2-ml well on a 96-well Masterblock plate (Greiner, Longwood, FL, catalog no. 780271) and diluted with 900 μl of Hepes-buffered saline (HBS) physiological buffer (HBS/10 mM Hepes, pH 7.4/150 mM NaCl) to a 10-fold working sample dilution. Two sample trays were prepared in this way for the analysis of 18 proteins; three more trays were prepared differently for the analysis of the other 7 proteins. Specifically, for the analysis of serum amyloid A, ceruloplasmin, retinol binding protein, and urine protein 1, a sample tray was prepared where the 100-μl plasma aliquots were diluted with 400 μl of HBS buffer to a 5-fold working sample dilution to obtain a better signal/quality mass spectra. For the analysis of apolipoprotein E, a separate sample tray was prepared consisting of 10-fold diluted plasma samples to which Tween 20 (Sigma, to a final concentration of 0.1% vol/vol) was added to help disassociate apolipoprotein E from the HDLs and LDLs (8). And for the final sample tray used in the analysis of IGF-I and IGF-II, 40 μl of each human plasma sample was mixed with 60 μl HBS and 60 μl of 0.5% SDS, incubated at room temperature for 10 min, and brought up to a 1 ml working volume with an 840-μl aliquot of HBS (10) to disrupt their in vivo protein complexes. In all, five 96-well trays containing diluted plasma samples were prepared and used for assaying the 25 proteins. Accordingly, multiple protein assays were performed from a single sample tray by sequentially addressing the tray with successive extractions, each targeting a different protein. As each assay depletes the samples of a specific protein, there is essentially no limitation as to how many times each sample tray can be used in consecutive analyses. The rationale behind the use of two sample trays for analysis of the 18 proteins that followed the general sample preparation protocol was to avoid prolonged storage of the sample trays at 4°C, which we limited to 1 week.
Each protein assay involved parallel processing of the 96 plasma aliquots. Beckman Multimek Automated 96-Channel Pipettor (Beckman Coulter) was used for the parallel processing of the assays from start (affinity capture of the target protein in the affinity pipettes) to finish (elution of the captured proteins onto a MALDI target). The protein extraction/affinity capture process followed protocols in refs. 9 and 11. Brief ly, antibody-derivatized affinity pipettes were mounted onto the head of the Multimek pipettor and initially rinsed with 100 μl of HBS buffer (10 cycles, each cycle consisting of a single aspiration and dispensing through the affinity pipette). Next, the pipettes were immersed into the sample tray, and 150 aspirations and dispense cycles (100 μl volumes each) were performed, allowing for affinity capture of the targeted protein. After affinity capture, the pipettes were rinsed with HBS (10 cycles), water (10 cycles), 2 M ammonium acetate/acetonitrile (3:1 vol/vol) mixture (10 cycles), and two final water rinses (10 cycles each). The affinity pipettes containing the retrieved protein were then rinsed with 1 mM N-octyl glucoside (single cycle with a 150-μl aliquot) to homogenize the subsequent matrix draw and elution by completely wetting the porous affinity supports inside the pipettes (9). For elution of the captured proteins, an adequate MALDI matrix was prepared [either α-cyano-4-hydroxycinnamic acid (6 g/liter) or sinapic acid (10 g/liter), in aqueous solution containing 33% (vol/vol) acetonitrile, 0.4% (vol/vol) trifluoroacetic acid], and 6-μl aliquots were aspired into each affinity pipette. After a 10-second delay (to allow for the dissociation of the protein from the capturing antibody, which is triggered by the low pH and chaotropic effects of the matrix), the eluates from all 96 affinity pipettes containing the targeted protein were dispensed directly onto a 96-well formatted MALDI target. After air drying and visual inspection of the sample spots, linear mass spectra were acquired on Bruker Biflex III and Autoflex MALDI-TOF mass spectrometers (Bruker Daltonics, Billerica, MA). The mass spectra were evaluated by using the root Bruker software (flexanalysis) and proteome analyzer (Intrinsic Bioprobes, Tempe, AZ, codeveloped with Beavis Informatics, Winnipeg, MB, Canada). The proteome analyzer is a mass spectrometry data visualization MS tool that, when used in conjunction with paws (sequence display and manipulation software from Proteo-Metrics, New York), allows for rapid identification of protein sequence modifications through display of mass values and differences between peaks. A peak was assigned to a specific modification if the observed m/z value differed by <0.02% from that empirically predicted. For higher mass glycosylated proteins, a mass value within 0.1% from that empirically calculated was deemed acceptable.
For those proteins exhibiting modifications, further validation experiments were performed to delineate the modification. Trypsin-derivatized mass spectrometer probes were prepared as described in refs. 9 and 12 and used for peptide mapping experiments. Mass spectra in reflectron mode were acquired on the Biflex III Bruker instrument. For validation of the apolipoprotein E phenotypes, SNPs analyses were performed by the Molecular Diagnostics Laboratory at the University of Massachusetts Medical School (Worcester, MA). For verification of apolipoprotein E cysteinylation, in situ reduction of the plasma samples (before affinity retrieval) was performed. Two microliters of 20 mM DTT were added to each 20-μl human plasma aliquot, incubated at room temperature for 30 min, and diluted with 150 μl of HBS/20 μl of 1% Tween 20/10 μl of 6.6 M guanidine hydrochloride. After affinity capture and MS analysis, the disappearance of the cysteinylated peak in the mass spectra was monitored and signaled the suggested cysteinylation.
Results and Discussion
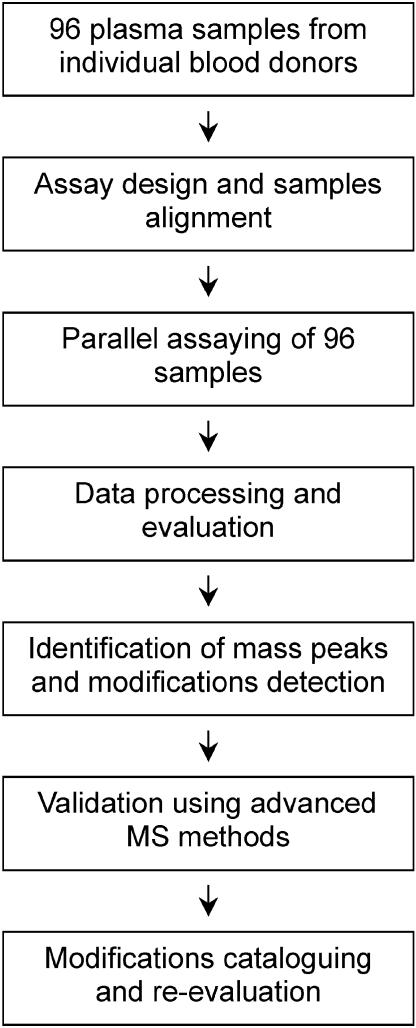
The steps that were followed for the analysis of the 25 proteins from the 96-samples cohort are outlined in Fig. 1. The core of the method is a top-down proteomics approach comprising protein affinity-extraction, followed by rigorous characterization with MALDI-TOF mass spectrometry. We have developed this method over the past few years and have recently demonstrated its robustness, reproducibility, and limits of detection when applied in a high-throughput format (9-11). The general approach centers on initial interrogation of intact protein masses, followed (if necessary) by subsequent characterization with proteolytic enzymes and peptide mapping. Specifically, when looking for possible protein modifications, it is imperative to evaluate the mass of the intact protein first. The major signal in the mass spectrum that corresponds to the targeted protein should be within a reasonable range (e.g., error of measurement of <0.05%) from the value of the empirically calculated mass obtained from the sequence of the protein deposited in the swiss-prot data bank. Once this mass value is confirmed (or observed to be shifted), the presence of protein modifications can be delineated by the appearance of other signals in the mass spectra (usually in the vicinity of the native protein peaks) or by the pure fact that the major signal has a different mass from that predicted. It is also important to note that any protein under investigation has the potential of registering as both wild-type and variant species in the same analysis. Modifications are tentatively identified by accurate measurement of the observed mass shifts (from the wild-type protein signals and/or in silico calculated mass) and knowledge of the protein sequence and possible modifications (e.g., a removal of an N- or C-terminal residue would result in a signal shifted down by a value that matches the mass of the removed amino acid). The tentative identity of the modifications is then verified by using mass mapping approaches in combination with high-performance mass spectrometry.
Fig. 1.
Diagram of the study design.
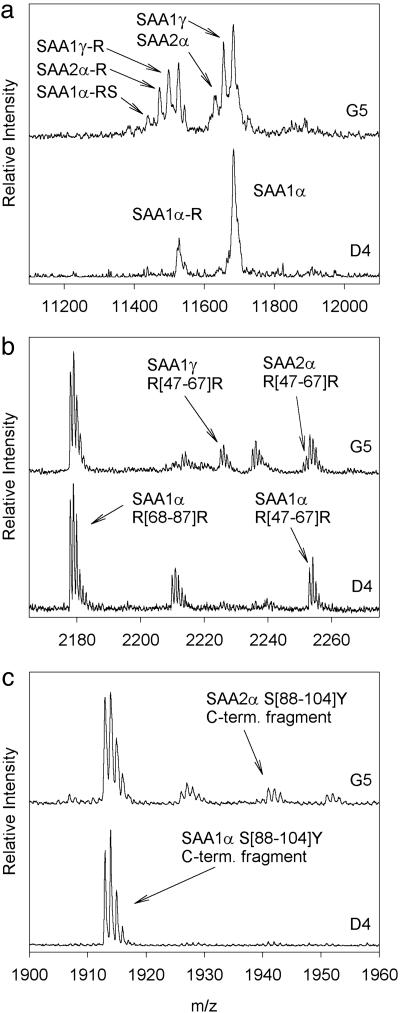
To illustrate the overall protocol and data interpretation, shown in Fig. 2 are two mass spectra resulting from the analysis of serum amyloid A (SAA) from samples D4 and G5 (the letter and number designate the position of the sample on the 96-well tray). In the analysis of the native SAA protein (Fig. 2a), the D4 sample yielded two major signals in the mass spectrum, corresponding to SAA1α and a truncated form missing the N-terminal arginine (SAA1α-R). The G5 sample, in addition to these two signals, yielded other signals corresponding to SAA1γ, SAA2α, their N-terminal arginine truncated forms (-R), and SAA1α missing an arginine and serine from the N terminus (-RS). These assignments were made based on the observed m/z values of the signals and the known sequences of the protein products of all four SAA genes and the existing polymorphisms within each gene (13, 14). To confirm and validate the observed modifications, on-target tryptic digests/reflectron MALDI TOF MS analyses were performed on the eluted SAA proteins. Two regions of the peptide maps are shown in Fig. 2 b and c, showing the peptides that are specific to the SAA1γ and SAA2α allelic variants.
Fig. 2.
Mass spectra resulting from the serum amyloid A (SAA) assays of samples D4 and G5. (a) Linear mass spectra showing intact protein signals. (b and c) Reflectron mass spectra of the digested samples. The numbers in the brackets represent the sequence of the amino acid residues. SAA1γ differs from SAA1α at residue 52 (Val52Ala), whereas SAA2α differs from SAA1α in several positions, two of which are depicted in the peptide digests maps (Asp60Asn and Lys90Arg).
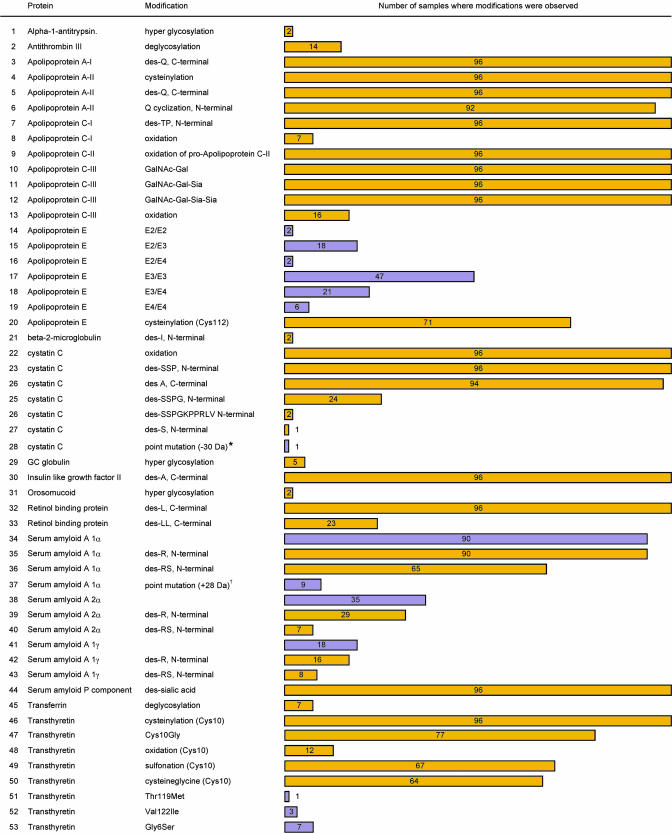
Following the approaches and protocols described above, we analyzed 25 proteins from a cohort of 96 plasma samples obtained from healthy individuals. Among others, the proteins included transport proteins (retinol binding protein, transthyretin, and transferrin), growth factors (insulin-like growth factors), enzyme inhibitors (cystatin C and α-1-antitrypsin), proteins involved in cellular recognition (apolipoproteins), and inflammatory response markers (serum amyloid A and C-reactive protein). Although categorically grouped by function, all of the proteins under investigation have been studied clinically for either diagnostic or prognostic monitoring of a number of diseases (15, 16). Fig. 3 summarizes the results of the study, listing the modifications observed for 18 of the 25 proteins studied and showing the frequency of each modification found in the 96-sample cohort (also see Fig. 5 and Table 1, which are published as supporting information on the PNAS web site, for examples of mass spectra showing all of the modifications and their predicted and observed masses). A total of 53 protein variants were observed for these 18 proteins, stemming from point mutations or posttranslational modifications.† Seven proteins did not exhibit any readily observable modifications.‡ Wild-type protein signals (as determined by the main protein sequence listed in swiss-prot) were observed for all 25 proteins (Note that several apolipoprotein E and serum amyloid A forms can be considered wild type). Considering both the wild-type and the protein variants, a total of 76 protein species were detected within the 96 samples cohort with the 25 protein assays. It is important to note that, other than for albumin, transthyretin, and some of the high-level apolipoproteins, none of the other 25 proteins assayed through the affinity-MS approach could be detected by direct MS analysis of human plasma. The fractionation through immobilized antibodies is critical in obtaining enough of the protein in a pure form for the subsequent MS detection,§ the signals of which are otherwise masked in the mass spectra of whole human plasma by the presence of the high-level proteins such as albumin.
Fig. 3.
Modifications observed in 18 of the 25 proteins analyzed from 96 human plasma samples. Yellow, posttranslational modifications; purple, point mutations. The wild-type signals were observed in all 96 samples for all of the 25 proteins [Note that several forms of apolipoprotein E (E2, E3, and E4) and serum amyloid A (1α, 2α, and 1γ) can be considered wild types and are listed in this figure]. Modifications were not detected for albumin, cerruloplasmin, C-reactive protein, insulin-like growth factor I, lysozyme, plasminogen, and urine protein 1. *, The cystatin C point mutation could not be assigned because of the <50% sequence coverage resulting from the peptide mapping. †, The SAA1α point mutation was tentatively assigned as Lys90Arg substitution.
Point mutations were observed in 4 of the 25 proteins (apolipoprotein E, cystatin C, serum amyloid A, and transthyretin). Prominently, a high incidence of point mutations was noted for apolipoprotein E and for transthyretin, consistent with genomic studies that have found these proteins to be highly polymorphic (21, 22) (Note that the apolipoprotein E phenotypes depicted through the MS assays were additionally confirmed through SNPs analyses). Posttranslational modifications were observed in 18 proteins. The largest number of variants was found to be C- or N-terminal truncated versions of the targeted protein. Deglycosylation, oxidation, and cysteinylation were also observed among several of the proteins. All four of the proteins found to contain point mutations also exhibited numerous posttranslational modifications with these “compound” (mix of gene- and protein-level) modifications accounting for more than half of the catalogued modifications. It is these proteins that need to be studied in greater detail and from a larger cohort of samples to get a more representative view of the distribution of these modifications in the general population.
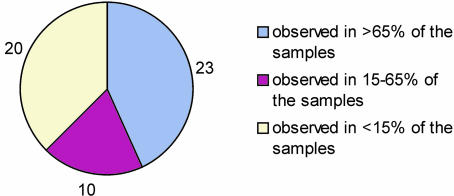
The data presented in Fig. 3 clearly show that some modifications were detected at high frequency among the 96 samples. Fourteen modifications (within the 25-protein panel) were observed in all 96 samples, which suggest that they must be regarded as wild-type protein forms. Most notably, insulin-like growth factor II was detected in all 96 individuals as two species, the wild-type and a des-Ala form. Based on a very small population study, we recently reported this truncated form as a naturally occurring variant present in human plasma (10, 23), but the striking result from this larger study is that the truncation appears in 100% of the samples. Likewise, various distinct glycoforms of apolipoprotein C-III, and oxidation and N-terminal truncations of cystatin C, were observed in all 96 samples. Several other protein modifications also occurred with high frequency such as the serum amyloid A and apolipoproteins truncations. Overall, 23 of the modifications were observed in >65% of the samples, and 20 in <15% of the 96 samples analyzed (Fig. 4). It is those low frequency protein modifications that are most likely to be of great significance as potential biomarkers of disease.
Fig. 4.
Occurrence of specific modifications in the 96 samples cohort.
Because the 96 samples could be divided into subgroups based on sex, age, and ethnicity, we searched for any correlations in the frequency and character of the observed modifications within these subgroups. The only significant correlation observed was in regards to the Gly6Ser mutation in transthyretin. This modification was detected only in individuals of Caucasian origin (two males and five females), which is consistent with existing knowledge about this common nonamyloidogenic population polymorphism in Caucasians (24). The correlation and agreement with existing literature data validates the approach used in this study and demonstrates its efficacy in delineating modifications stemming from genetic events. However, an alternate future use of studies such as these may be that of assisting in economizing gene sequencing efforts focusing on large populations by denoting individuals specifically in need of follow-up investigation.
An additional result of the study was that of viewing similar interprotein variations between the individuals, which is exemplified by an apparent correlation between deglycosylated transferrin and deglycosylated antithrombin III. Of the seven individuals for which transferrin deglycosylation was noted, five were Caucasian males and two African American females. For antithrombin III, deglycosylation was seen in four African American males, seven Caucasian males, and in two African American females. However, all seven individuals observed to have deglycosylated transferrin had deglycosylated antithrombin III as well. Thus, it is quite possible that these individuals are unknowingly suffering from carbohydrate-deficient glycoprotein syndrome (25), which is recognized by carbohydrate-deficient isoforms of glycoproteins, including both transferrin and antithrombin III (26). Chronic alcohol consumption can also lead to carbohydrate-deficient transferrin (27). Importantly, these modifications occurred at the posttranslational level and are recognized (for these two proteins) as relatively large (≈2 kDa) mass shift that are easily observable during the mass interrogation of the intact proteins, thus underscoring the utility of affinity extraction/direct intact mass measurement in a clinical environment.
Although it is true that many of the modifications described here have been reported previously, it is important to note that the aim of this study was to identify variants in multiple plasma proteins (whether previously known) and to delineate their incidence in a subset of the general population, a task which to date has not been undertaken. Recently, Anderson et al. (28) published a nonredundant list of 1,175 distinct human plasma proteins compiled from four separate sources. However, 195 of those proteins were included in more than one of the four data sets, and only 46 proteins appeared in all four sources. Of the 25 proteins investigated in our study, 14 can be found among the 46 Anderson et al. discovered to be overlapping in the four data sets. Anderson et al. projects that (compilation of) “a large list of proteins actually observed in plasma paves the way for top-down, targeted proteomics approaches to the discovery of disease markers.” We view the data and approach given here as fitting with this goal by (sensibly) starting with the most frequently identified plasma proteins and, with time, addressing more proteins from a larger population cohort.
In summary, we believe this study to be a systematic investigation of structural diversity found in multiple plasma proteins in the general population. Importantly, the study involved the mass spectrometric immunoassay (29) characterization of proteins from numerous individuals, which is crucial given that any protein observed in a significantly large population will register predominantly as the wild-type protein and only a minor number of times as a recognizable variant. True to form, although only 25 proteins were under investigation, 76 forms were found to be present when viewing the 96-individual cohort collectively. Such findings are considered significant from both a purely biological point of view as well as in application. Foremost, they yield an accurate definition of the “wild-type” protein as present in the general human population. In comparison to the wild-type protein, basal-level differences in proteins (i.e., non-wild-type forms) and their frequency of occurrence can be established, which is of clear importance in the long-term study of protein diversity in populations. However, knowledge of such protein diversity stands to have an immediate impact in the field of clinical proteomics. In that clinical proteomics studies are now reporting diagnostic sensitivities and specificities approaching 100%, it is important to be aware of proteins that fluctuate naturally in the general population. Given knowledge from studies such as the one presented here, proteins known to vary naturally with high incidence (e.g., the 23 modifications observed in >65% of the cohort) can be either eliminated as diagnostic markers or more rigorously scrutinized for unique variants specific to disease. In contrast, proteins found to vary at a low frequency become of interest in biomarker discovery efforts that target diseases occurring naturally at the same frequency. In these regards, we believe the approach and findings given in this report will find increased use in the further understanding of protein diversity in humans with subsequent applications in the clinical proteomics and diagnostic arenas.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants Nos. 5 R44 CA99117-03 and 5 R44 HL072671-03.
Author contributions: D.N. designed research; D.N., U.A.K., E.E.N., K.A.T., and R.W.N. performed research; K.A.T. contributed new reagents/analytic tools; D.N., U.A.K., E.E.N., and R.W.N. analyzed data; and D.N. and R.W.N. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HBS, Hepes-buffered saline; SAA, serum amyloid A.
Footnotes
It is highly unlikely that the observed protein modifications are a byproduct of the extraction process, because the purification conditions employed in the assay were essentially those traditionally used in protein purification by using immunoprecipitation. It is possible that some of the oxidized protein forms were enhanced by prolonged sample storage at -75°C, but most of the modifications observed here have also been detected in fresh nonfrozen plasma samples (8, 11, 17-20).
Seven proteins, albumin, cerruloplasmin, C-reactive protein, insulin-like growth factor I, lysozyme, plasminogen, and urine protein 1, were not observed to exhibit any modifications, as indicated by the absence of any other peaks in the mass spectra except for the wild-type protein signal. That is not to say that there were not, in fact, any modifications present in these proteins, especially in albumin, plasminogen, and ceruloplasmin. It should be noted that the method we utilized detailed peptide mapping experiments only in those samples where visible deviations from the wild-type molecular mass were observed in the mass spectra. Because all three of the above-mentioned proteins are high molecular mass proteins, glycosylated (plasminogen and ceruloplasmin), and highly heterogeneous (albumin), the resulting wild-type signals in the mass spectra are relatively wide and of reduced resolution, making the observation of small mass shifts resulting from, e.g., single amino acid substitution, extremely difficult to delineate from the native mass signals.
The mass spectra resulting from these plasma assays were dominated by signals from the targeted protein. In few instances, signals from nonspecifically bound (to the support) apolipoprotein Cs appeared in the mass spectra. These few additional peaks do not interfere with the overall analysis, as long as their existence is acknowledged and their character (m/z value) is known. Moreover, they can be used advantageously to internally calibrate the mass spectra for a more accurate m/z peak assignment.
References
- 1.International Human Genome Sequencing Consortium (2004) Nature 431, 931-945. [DOI] [PubMed] [Google Scholar]
- 2.Marth, G., Yeh, R., Minton, M., Donaldson, R., Li, Q., Duan, S., Davenport, R., Miller, R. D. & Kwok, P. Y. (2001) Nat. Genet. 27, 371-372. [DOI] [PubMed] [Google Scholar]
- 3.Mann, M. & Jensen, O. N. (2003) Nat. Biotechnol. 21, 255-261. [DOI] [PubMed] [Google Scholar]
- 4.Johann, D. J., Jr., McGuigan, M. D., Patel, A. R., Tomov, S., Ross, S., Conrads, T. P., Veenstra, T. D., Fishman, D. A., Whiteley, G. R., Petricoin, E. F., 3rd, & Liotta, L. A. (2004) Ann. N. Y. Acad. Sci. 1022, 295-305. [DOI] [PubMed] [Google Scholar]
- 5.Petricoin, E. F., Ardekani, A. M., Hitt, B. A., Levine, P. J., Fusaro, V. A., Steinberg, S. M., Mills, G. B., Simone, C., Fishman, D. A., Kohn, E. C. & Liotta, L. A. (2002) Lancet 359, 572-577. [DOI] [PubMed] [Google Scholar]
- 6.Diamandis, E. P. (2004) Mol. Cell Proteomics 3, 367-378. [DOI] [PubMed] [Google Scholar]
- 7.Sorace, J. M. & Zhan, M. (2003) BMC Bioinformatics 4, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niederkofler, E. E., Tubbs, K. A., Kiernan, U. A., Nedelkov, D. & Nelson, R. W. (2003) J. Lipid Res. 44, 630-639. [DOI] [PubMed] [Google Scholar]
- 9.Nedelkov, D., Tubbs, K. A., Niederkofler, E. E., Kiernan, U. A. & Nelson, R. W. (2004) Anal. Chem. 76, 1733-1737. [DOI] [PubMed] [Google Scholar]
- 10.Nelson, R. W., Nedelkov, D., Tubbs, K. A. & Kiernan, U. A. (2004) J. Proteome Res. 3, 851-855. [DOI] [PubMed] [Google Scholar]
- 11.Kiernan, U. A., Nedelkov, D., Tubbs, K. A., Niederkofler, E. E. & Nelson, R. W. (2004) Clin. Proteomics J. 1, 7-16. [DOI] [PubMed] [Google Scholar]
- 12.Kiernan, U. A., Black, J. A., Williams, P. & Nelson, R. W. (2002) Clin. Chem. 48, 947-949. [PubMed] [Google Scholar]
- 13.Yamada, T. (1999) Clin. Chem. Lab. Med. 37, 381-388. [DOI] [PubMed] [Google Scholar]
- 14.Uhlar, C. M. & Whitehead, A. S. (1999) Eur. J. Biochem. 265, 501-523. [DOI] [PubMed] [Google Scholar]
- 15.Ritchie, R. F. (1999) (Found. for Blood Res., Scarborough, ME).
- 16.Craig, W. Y., Ledue, T. B. & Ritchie, R. F. (2000) Plasma Proteins: Clinical Utility and Interpretation (Found. for Blood Res., Scarborough, ME).
- 17.Kiernan, U. A., Tubbs, K. A., Gruber, K., Nedelkov, D., Niederkofler, E. E., Williams, P. & Nelson, R. W. (2002) Anal. Biochem. 301, 49-56. [DOI] [PubMed] [Google Scholar]
- 18.Kiernan, U. A., Tubbs, K. A., Nedelkov, D., Niederkofler, E. E. & Nelson, R. W. (2002) Biochem. Biophys. Res. Commun. 297, 401-405. [DOI] [PubMed] [Google Scholar]
- 19.Kiernan, U. A., Tubbs, K. A., Nedelkov, D., Niederkofler, E. E. & Nelson, R. W. (2003) FEBS Lett. 537, 166-170. [DOI] [PubMed] [Google Scholar]
- 20.Kiernan, U. A., Nedelkov, D., Tubbs, K. A., Niederkofler, E. E. & Nelson, R. W. (2004) Proteomics 4, 1825-1829. [DOI] [PubMed] [Google Scholar]
- 21.Connors, L. H., Lim, A., Prokaeva, T., Roskens, V. A. & Costello, C. E. (2003) Amyloid 10, 160-184. [DOI] [PubMed] [Google Scholar]
- 22.Mahley, R. W. & Huang, Y. (1999) Curr. Opin. Lipidol. 10, 207-217. [DOI] [PubMed] [Google Scholar]
- 23.Nedelkov, D., Nelson, R. W., Kiernan, U. A., Niederkofler, E. E. & Tubbs, K. A. (2003) FEBS Lett. 536, 130-134. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson, D. R., Alves, I. L., Saraiva, M. J., Thibodeau, S. N. & Buxbaum, J. N. (1995) Hum. Genet. 95, 308-312. [DOI] [PubMed] [Google Scholar]
- 25.Carchon, H., Van Schaftingen, E., Matthijs, G. & Jaeken, J. (1999) Biochim. Biophys. Acta 1455, 155-165. [DOI] [PubMed] [Google Scholar]
- 26.Stibler, H., Holzbach, U. & Kristiansson, B. (1998) Scand. J. Clin. Lab. Invest. 58, 55-61. [DOI] [PubMed] [Google Scholar]
- 27.Fleming, M. F., Anton, R. F. & Spies, C. D. (2004) Alcohol Clin. Exp. Res. 28, 1347-1355. [DOI] [PubMed] [Google Scholar]
- 28.Anderson, N. L., Polanski, M., Pieper, R., Gatlin, T., Tirumalai, R. S., Conrads, T. P., Veenstra, T. D., Adkins, J. N., Pounds, J. G., Fagan, R. & Lobley, A. (2004) Mol. Cell Proteomics 3, 311-326. [DOI] [PubMed] [Google Scholar]
- 29.Nelson, R. W., Krone, J. R., Bieber, A. L. & Williams, P. (1995) Anal. Chem. 67, 1153-1158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.