Abstract
The multifunctional Nef protein of HIV-1 is important for the progression to AIDS. One action of Nef is to down-regulate surface MHC I molecules, helping infected cells to evade immunity. We found that Nef also down-regulates the macrophage-expressed MHC 1b protein HFE, which regulates iron homeostasis and is mutated in the iron-overloading disorder hemochromatosis. In model cell lines, Nef reroutes HFE to a perinuclear structure that overlaps the trans-Golgi network, causing a 90% reduction of surface HFE. This activity requires a Src-kinase-binding proline-rich domain of Nef and a conserved tyrosine-based motif in the cytoplasmic tail of HFE. HIV-1 infection of ex vivo macrophages similarly down-regulates naturally expressed surface HFE in a Nef-dependent manner. The effect of Nef expression on cellular iron was explored; iron and ferritin accumulation were increased in HIV-1-infected ex vivo macrophages expressing wild-type HFE, but this effect was lost with Nef-deleted HIV-1 or when infecting macrophages from hemochromatosis patients expressing mutated HFE. The iron accumulation in HIV-1-infected HFE-expressing macrophages was paralleled by an increase in cellular HIV-1-gag expression. We conclude that, through Nef and HFE, HIV-1 directly regulates cellular iron metabolism, possibly benefiting viral growth.
Keywords: macrophages, MHC I, AIDS
Individuals infected with a Nef-deficient HIV-1 strain through blood transfusion did not progress to AIDS for >10 years (1). Why Nef is important to HIV-1 pathogenesis is still not clear. Multiple activities have been assigned to Nef (3), including targeting CD4 for degradation (4, 5) and the down-regulation of MHC I molecules (6). This latter function can protect infected cells from attack by HIV-1-specific cytotoxic T lymphocytes (7), although HLA-A and HLA-B down-regulation is only partial, and HLA-C is unaffected by Nef (8).
The MHC 1b protein HFE is structurally homologous to HLA-A2 (9) but does not share the widespread tissue distribution of MHC I. In humans, HFE is strongly expressed by Kupffer cells, circulating monocytes/macrophages, and intestinal crypt cells (10, 11). HFE regulates iron homeostasis; the C282Y mutation of HFE that causes protein misfolding is associated with the iron-overloading disorder hemochromatosis (12, 13). The function of HFE is obscure; in model cell systems, HFE binds the transferrin receptor-1 (TfR) and reduces cellular iron uptake (14, 15), but the significance of this interaction in vivo is uncertain (13). In monocyte and intestinal cell types, HFE increases iron accumulation through the inhibition of iron release, independently of binding TfR (16, 17).
Based on homology between HFE and HLA-A2, we hypothesized that Nef might affect HFE expression. We report that Nef reroutes HFE, interfering with cellular iron-handling in a manner that may have consequences for HIV-1.
Materials and Methods
Cell Culture. HeLa and 293 cells expressing wild-type HFE and HFE mutants were from C. Enns (Oregon Health and Science University, Portland, OR) (15, 18). The 293, THP-1, and U937 cells were from the American Type Culture Collection. Ex vivo macrophages were grown, as described in ref. 19, from peripheral blood taken from healthy volunteers (wild-type HFE-homozygous) and consenting hemochromatosis patients (C282Y HFE-homozygous) with the help of K. Robson [Weatherall Institute of Molecular Medicine (WIMM)].
Antibodies, Plasmids, and Viruses. The following antibodies were used: anti HFE monoclonals 10G4, from Y. Yang (The R. W. Johnson Pharmaceutical Research Institute, San Diego), and 8C10, from R. Ehrlich (Tel Aviv University, Israel) (20), anti-HFE α3-domain polyclonal, from J. Bastin (WIMM), anti-ferritin, anti-Helicobacter pylori polyclonals, MR12 anti-mouse IgG, and anti-TfR (DAKO), anti-EGFR and anti CD29 (Pharmingen), anti-MHC I W6/32, from J. Bastin (WIMM), anti-HIV gag p55/p18 (3D3 and 1D9 mAbs), from R. Ferns (National Institute for Biological Standards and Control, Medical Research Council, U.K.), and anti-trans-Golgi network (TGN)46 (Serotec). We used the second-layer reagents anti-mouse IgG-FITC, anti-mouse IgG-Phycoerythrin (PE), anti-rabbit IgG-PE conjugates (Sigma), anti-mouse IgG-Alexa Fluor 568, and anti-sheep IgG-Alexa Fluor 568 conjugates (Molecular Probes).
pCAGGS plasmids encoding Hck variants and CrkII were from M. Matsuda (Osaka University, Osaka) (21). Plasmids encoding wild-type or mutant Nef-GFP fusion proteins were created by cloning the HIV-1 SF2 Nef gene into pEGFP-N1 or p internal ribosome entry site (IRES)2-eGFP (Clontech) between EcoRI and BamHI. The stop codon in Nef was removed to create the Nef-GFP fusion protein; oligonucleotide-directed site-specific mutagenesis generated the Nef variants. The recombinant lentiviruses encoding either GFP or Nef-IRES-GFP were created, as described in ref. 22, (23), and pelleted viruses were resuspended in 1/10 vol of medium to add to cells. M-tropic HIV-1 and Nef-deleted M-tropic HIV-1 strains with an HIV-1 NL-4-3 backbone and env from HIV-1 Bal, from A. Baur (University of Miami, Coral Gables, FL), were generated as described in ref. 24, and the p24 concentration of viral stocks was assayed by using a p24 ELISA kit (Immunodiagnostics, Woburn, MA).
Transfections, Infections, and Nucleoporations. HeLa-HFE and 293 cells were transfected by using Effectene (Qiagen, Crawley, U.K.). THP-1 and U937 cells were nucleoporated with the Nucleofector device (Amaxa, Cologne, Germany). THP-1 cells at 106 cells per ml or 7-day-old macrophage cultures were cocultured with lentivirus for 4 days. HIV infection of ex vivo macrophages was as described in ref. 19, with 50 ng/ml p24 of HIV or Nef-deleted HIV, for various times.
Flow Cytometry. Transfected HeLa-HFE and 293 cells were stained for surface HFE, TfR, MHC-I, and EGFR. THP-1 and U937 cells were infected with lentiviruses and stained for cell-surface HFE, MHC I, or CD29 or were permeabilized with CytoPerm/CytoFix (Pharmingen) and stained for ferritin. Macrophages were infected with HIV and double-stained for surface p18/55 gag protein and surface HFE or permeabilized and double-stained for p18/55 gag protein and ferritin. Cells were analyzed, in 50,000-cell aliquots, on a Becton Dickinson FACSCalibur flow cytometer running cellquest software.
Immunofluorescence Microscopy. HeLa-HFE cells were permeabilized and stained for HFE or TGN46. Macrophages were permeabilized and stained for HFE. Cells were fixed, slides were mounted in VECTASHIELD containing DAPI (Vector Laboratories), and imaged by using a Zeiss Axiovert microscope equipped with epifluorescence illumination.
Internalization, Recycling, and Transport-to-Cell-Surface Assay. FACS-based assays for internalization, recycling, and transport to the cell surface were performed as described in refs. 6, 25, and 26, with modifications. The recycling and transport-to-cell-surface assays involved a 150-s acid strip of surface β2-microglobulin to destroy reactivity to 8C10. The rate of HFE transport to the cell surface is slower than for MHC I; for this reason, the poststripping culture time was lengthened to 20 h. Cells were cultured in serum-free medium (Pro293a, Cambrex, Wokingham, U.K.), after 20 h of culture non-cycloheximide-treated cells were 95% viable. Percentage of transport to the cell surface was calculated as described in ref. 26.
HFE-Degradation Assay. HeLa-HFE cells were cultured in methionine-free medium at 37°C, labeled with [35S]methionine, and chased with cold methionine for various times. Cytoplasmic proteins were extracted by using Nonidet P-40 lysis buffer (1 M Tris, pH 7.5/0.5 M EDTA/5 M NaCl/0.5% Nonidet P-40). Anti-HFE antibody was added to the lysate at 15 μg/ml and incubated for 1 h at 4°C. 5% wt/vol protein-A-Sepharose beads (Sigma) were added, and the beads were analyzed by using SDS/PAGE. Gels were analyzed by using a Molecular Dynamics Storm 840 PhosphorImager (Amersham Pharmacia).
59Fe-Accumulation Experiments. At 4 h after nucleoporation of THP-1 and U937 cells, [59Fe]transferrin was added at 40 μg/ml; after an additional 14 h of culture, cellular 59Fe was determined as described in ref. 16. Seven-day-old macrophage cultures were inoculated with HIV and, 2 days later, [59Fe]transferrin was added at 33 μg/ml. At 10 days after adding 59Fe, cellular 59Fe was determined.
Results and Discussion
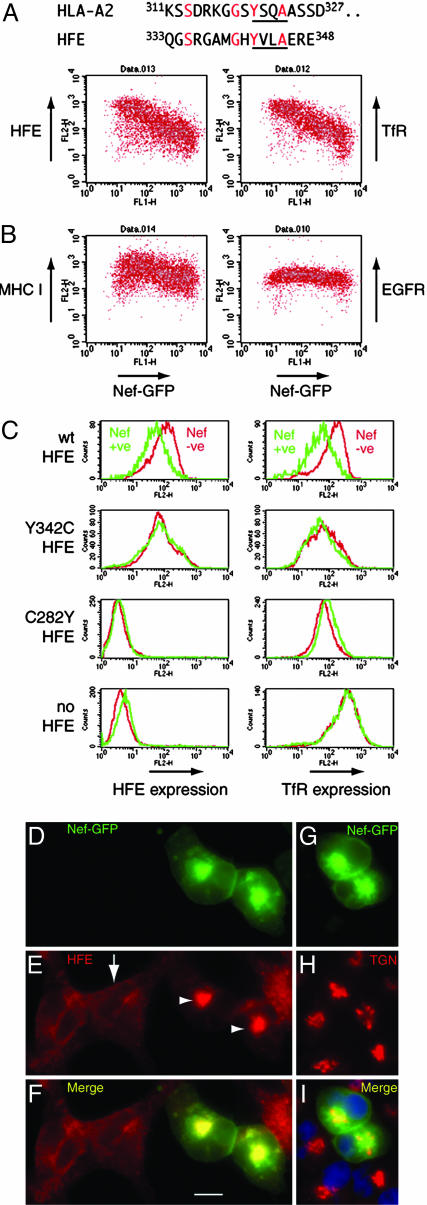
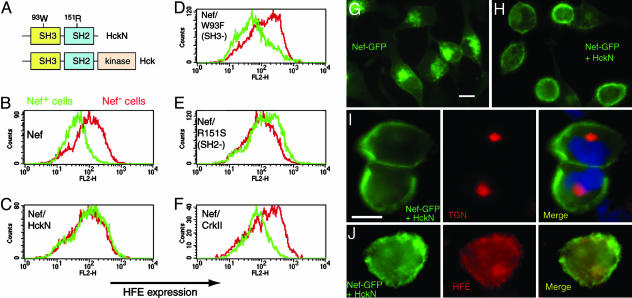
Reduction of Surface HFE by Nef in Model Cell Lines, Monocytic Cell Lines, and ex Vivo Macrophages. Down-regulation of HLA-A and HLA-B proteins by HIV-1 Nef requires a cytoplasmic tyrosine, not present in HLA-C, that forms part of a cryptic sorting motif YxxA (where x is any amino acid) (8). Because this tyrosine-based motif (342YxxA) is conserved in HFE (Fig. 1A), we hypothesized that Nef could down-regulate HFE. To test this idea, we transiently transfected HeLa cells stably expressing HFE with a construct encoding a Nef-GFP fusion protein. Fig. 1B shows that, as Nef expression increases, surface HFE and HFE's ligand TfR (14) are reduced, with maximum down-regulation >10-fold. On the same cells, total surface MHC I (including HLA-C) is reduced 2-fold by Nef; as a specificity-control, epidermal growth factor receptor is unaffected (6). GFP alone did not alter HFE, TfR, or MHC I (data not shown). We then tested how Nef acted in 293 cell lines stably expressing HFE mutants (Fig. 1C). In 293 cells expressing wild-type HFE, Nef reduces surface HFE and TfR. However, in cells expressing Y342C HFE, Nef does not alter surface HFE or TfR. The C282Y variant of HFE does not traffic to the cell surface or bind TfR (14, 27). In cells expressing C282Y HFE, Nef does not alter surface TfR levels, and the same result was found in parental HFE-negative 293 cells (Fig. 1C Bottom Left and Right). These results show that a cytoplasmic tyrosine 342Y is required for Nef to down-regulate surface HFE and indicate that HFE/TfR-binding is required for Nef to down-regulate TfR. Recently, Nef was reported to down-regulate TfR by ≈50% in HeLa and T cells (28), in contrast to previous studies (29, 30). An effect of Nef on TfR may be enhanced by HFE (compare 10-fold reduction of TfR on HeLa-HFE cells with 2-fold reduction on HeLa cells). We investigated the location of HFE in cells expressing Nef-GFP; Fig. 1 D-F shows that HFE is present in a large intracellular structure overlapping Nef-GFP (arrowheads in Fig. 1E), compared with a plasma membrane and endosomal localization in untransfected cells (arrow). Fig. 1 G-I shows that Nef predominantly locates to a perinuclear area stained by antibodies against the TGN.
Fig. 1.
Down-regulation and redistribution of surface HFE and its ligand TfR by HIV-1 Nef in cell lines. (A) Alignment of amino acid sequence of the cytoplasmic portion of HLA-A2 and HFE. Conserved residues are shown in red, and the YxxA motif is underlined. (B) HeLa-HFE cells were transiently transfected with Nef-GFP, stained for surface HFE, TfR, MHC I, or epidermal growth factor receptor (EGFR), and analyzed by flow cytometry. Results show that, as Nef expression increases (along the x axis), surface levels of HFE, TfR, and, to a lesser extent, MHC I decrease (y axis), whereas EGFR expression is unchanged. (C) Aliquots of 293 cells stably expressing HFE or HFE mutants, as indicated, were transiently transfected with Nef-GFP, and Nef-positive and Nef-negative cells were assayed for surface HFE and TfR. The results show that Nef-mediated down-regulation of HFE and TfR depends on a cytoplasmic tyrosine 342Yof HFE and correct folding of HFE (the C282Y mutation misfolds and does not bind TfR). (D-I) HeLa-HFE cells were transiently transfected with Nef-GFP, permeabilized, and stained for HFE (E and F, red) and for TGN46 (reveals the TGN) (H and I, red). Nef (D, F, G, and I, green) locates mostly to a perinuclear structure overlapping the TGN (I, yellow; nuclei stained blue with DAPI). HFE (E and F, red) locates to the cell membrane and endosomes in untransfected cells (E, arrow and F Left) but is redistributed to a perinuclear intracellular region overlapping Nef in transfected cells (E, arrowheads and F, yellow). wt, wild type. (Scale bar, 10 μm.)
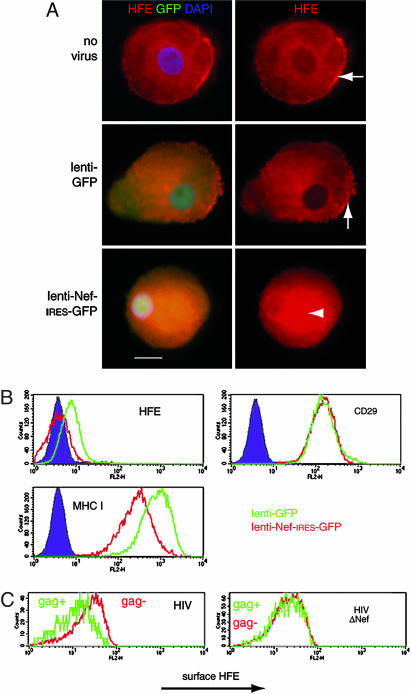
To test whether Nef also down-regulated naturally expressed HFE, we infected ex vivo macrophages and the HFE-expressing monocytic cell line THP-1 (16) with recombinant lentiviruses encoding either GFP (control virus) or Nef, followed by an IRES and GFP. Fig. 2A shows that uninfected macrophages and macrophages infected with the control virus have prominent surface HFE on their membrane ruffles (arrows), but in Nef-virus-infected cells (Fig. 2A Bottom), HFE accumulates intracellularly (arrowhead). Surface expression of HFE on THP-1 cells was unaffected by the control virus; however, surface HFE was undetectable above staining for an isotype control antibody in THP-1 cells infected with the Nef lentivirus (Fig. 2B). Nef also reduced surface MHC I, and, as a negative control, surface CD29 was unaltered by Nef (6). To establish that Nef, in the context of HIV, reduces surface HFE, ex vivo macrophages were infected with HIV-1 containing or lacking nef and double-stained for gag p18/p55 expression and HFE. Cells infected with HIV, but not with Nef-deleted HIV, had reduced surface HFE, compared with uninfected cells in the same culture (Fig. 2C).
Fig. 2.
Down-regulation of endogenous surface HFE in monocytic THP-1 cells and primary macrophages by Nef and HIV. (A) Ex vivo macrophages were infected with recombinant lentivirus-based vectors encoding either GFP (lenti-GFP) or Nef-IRES-GFP (lenti-Nef-IRES-GFP) and stained for HFE (red). Images of representative cells show that surface HFE expression on membrane ruffles in uninfected cells is unaltered in cells infected with the control lenti-GFP virus (Top and Middle Right, arrows) but that HFE is redistributed to a perinuclear region in cells infected with lenti-Nef-IRES-GFP (Bottom Right, arrowhead). (Scale bar, 10 μm.) (B) The cell monocyte/macrophage cell line THP-1 normally expresses low levels of surface HFE that are unchanged by the control lenti-GFP virus, but, in cells infected with lenti-Nef-IRES-GFP, surface HFE is undetectable above the background. Nef does not alter surface CD29 (negative control) but MHC I levels are lowered (positive control). (C) Macrophages were infected for 4 days with M-tropic HIV-1, with (HIV) or without (HIVΔNef) the nef gene, stained for surface HFE (anti-α3 domain) and p18/p55 gag, and analyzed by FACS. HIV-infected gag-expressing cells (Left, green line) express less surface HFE than do uninfected cells from the same culture (Left, red line). HIVΔNef infection (Right) does not alter surface HFE. This finding was repeated on macrophages from multiple donors expressing wild-type HFE.
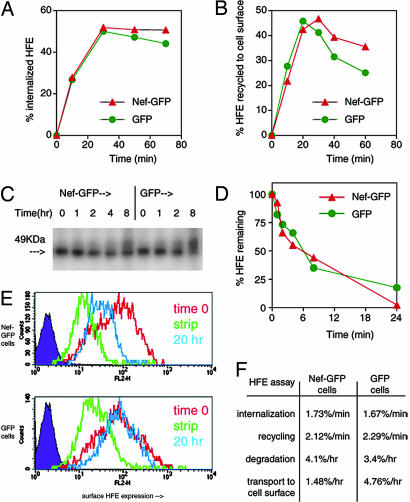
Nef Alters Transport of HFE to the Cell Surface but Does Not Alter HFE Internalization or Recycling. Nef can down-regulate surface MHC I by increasing internalization, inhibiting recycling, and/or inhibiting transport of MHC I to the cell surface (6, 26, 31). We investigated how Nef affected HFE internalization, recycling, stability, and transport. In HeLa cells, HFE internalization proceeds at a 3- to 4-fold-faster rate (1.7%/min, over 30 min) compared with MHC I internalization (0.5%/min, data not shown, and ref. 31). Fig. 3A shows that Nef-GFP-expressing cells internalized HFE from the cell surface at a nearly identical rate to control cells. Using the same assay system, we detected a Nef-induced enhancement of MHC I internalization (data not shown). Recycling of HFE was not significantly altered by Nef (Fig. 3B), in contrast to some results measuring Nef's effects on MHC I recycling (31). The faster constitutive internalization rate of HFE and TfR-binding may be why Nef does not influence HFE internalization or recycling. The pulse-chase experiments in Fig. 3 C and D show that Nef slightly increased HFE degradation over time, as for MHC I in some studies (6, 26). Fig. 3E shows that Nef inhibits the transport of HFE to the cell surface. At 20 h after stripping β2-microglobulin, surface antibody-detectable HFE expression returned to only 30% of the level before stripping in Nef-transfected cells (Fig. 3E Upper, compare blue histogram with red histogram), whereas, in control cells at the same time point, surface HFE was fully restored (Fig. 3E Lower, compare blue histogram with red histogram). As summarized in Fig. 3F, the down-regulation of HFE by Nef is achieved primarily through the inhibition of the transport of HFE to the cell surface, as for some studies investigating MHC I down-regulation (26).
Fig. 3.
Mechanism of Nef-mediated HFE down-regulation. (A) Internalization of HFE. HeLa-HFE cells transfected with Nef-GFP or GFP for 2 days were labeled with anti-HFE antibody at 4°C and then washed and incubated for given times at 37°C to allow internalization. Remaining surface HFE was stained with anti-mouse Ig antibody, and cells were analyzed by FACS. The percentage of internalized HFE was calculated and plotted. (B) Recycling of HFE. Surface β2-microglobulin was stripped from cycloheximide-pretreated Nef-GFP or GFP-expressing HeLa-HFE cells, and the appearance of surface β2-microglobulin-associated HFE from intracellular compartments was assayed, over time, by using FACS. (C and D) Degradation of HFE over time. HeLa-HFE cells transfected with Nef-GFP or GFP were pulsed with [35S]methionine and chased with excess cold methionine for the indicated times. HFE was immunoprecipitated from cell lysates and analyzed by PAGE (C). Numbers above the gel indicate hours of chase after 35S-labeling. Densitometric analysis (D) of gels from several experiments similar to that shown in C allowed quantification of the percentage of remaining HFE. (E) Transport of HFE to the cell surface. Aliquots of HeLa-HFE cells were transfected for 15 h with Nef-GFP (Upper) or GFP (Lower). Cells were stained for surface HFE (red line; at this early time point, Nef has not yet down-regulated HFE), stripped of surface β2-microglobulin and stained again for HFE (green line) or stripped and cultured for 20 h in serum-free medium to allow transport to the cell surface of newly synthesized HFE/β2-microglobulin molecules and stained for HFE (blue line). In Nef-expressing cells, newly synthesized HFE reaching the cell surface is reduced. (F) Summary of A-E: rates of internalization and recycling over 30 min did not differ with expression of Nef. Nef marginally increased HFE degradation, and Nef reduced transport of HFE to the cell surface over 20 h by 3.2-fold.
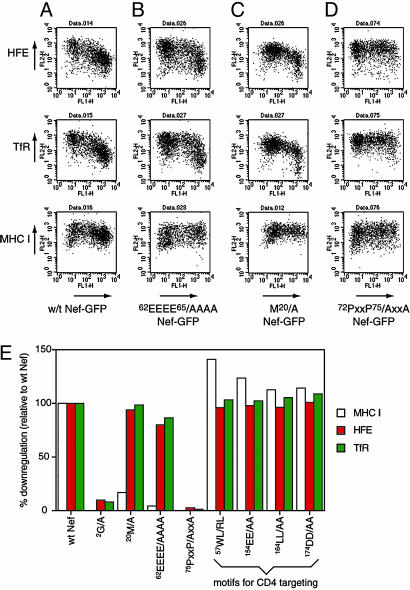
The Src-Kinase-Interacting Proline-Rich Domain of Nef Is Required for HFE Down-Regulation. Three motifs of Nef, 20M, 62EEEE65, and 72PxxP75, are required for MHC I down-regulation (31). We transfected HeLa-HFE cells with wild-type Nef-GFP and Nef-GFP mutants lacking these motifs and analyzed surface HFE (Fig. 4 A-D Top), TfR (Fig. 4 A-D Middle), and MHC I (Fig. 4 A-D Bottom). 62EEEE65/AAAA Nef (Fig. 4B) and 20M/A Nef (Fig. 4C) do not reduce surface MHC I, as found previously (31), but still down-regulated HFE and TfR, whereas 72PxxP75/AxxA Nef failed to down-regulate all three proteins (Fig. 4D). 72PxxP75 is, therefore, a critical region of Nef for HFE down-regulation. That 62EEEE65/AAAA and 20M/A Nef variants still down-regulate HFE is consistent with Nef not altering HFE internalization or recycling (Fig. 3 A and B). Both motifs control these processes in mediating MHC I down-regulation: 62EEEE65, through an interaction with PACS-1 (32), and 20M, by inhibiting MHC I recycling (31). Other regions of Nef were examined for their roles in HFE down-regulation (Fig. 4E). Myristoylation of Nef at position 2 (glycine) is crucial for Nef's association with cell membranes. Mutating the glycine to alanine blocked down-regulation of MHCI, HFE, and TfR. Other Nef motifs, 57WL, 154EE, 164LL, and 174DD, needed for CD4 down-regulation (3), were unnecessary for the reduction of surface MHC I or HFE/TfR.
Fig. 4.
Effects of mutations in Nef on Nef-mediated down-regulation of HFE, TfR, and MHC I. (A-D) HeLa-HFE cells were transfected with Nef-GFP constructs (x axis) encoding wild-type Nef (A), 62EEEE65/AAAA Nef (B), 20M/A Nef (C), and 72PxxP75/AxxA Nef (D) and stained for surface HFE (Top, y axis), TfR (Middle, y axis), and MHC I (Bottom, y axis). The results show that the ability of Nef to down-regulate HFE and TfR is retained by Nef variants lacking 62EEEE65 or 20M, but the 72PxxP75 motif is required for HFE/TfR down-regulation. All three motifs are needed for down-regulation of MHC I. (E) The role of other Nef motifs. HeLa-HFE cells were transfected with the Nef-GFP variants indicated and stained for surface MHC I, HFE, and TfR. The percentage of down-regulation, relative to down-regulation by wild-type Nef-GFP (100%), was calculated. Myristolation and the proline-rich domain are required for MHC I, HFE, and TfR down-regulation; four Nef motifs, needed for CD4 down-regulation, are dispensable.
The 72PxxP75 motif of Nef binds the SH3 domain of Src-family kinases (33). Inhibiting the Nef/Src kinase interaction prevents down-regulation of MHC I (34). To test whether the same was true for HFE down-regulation, we cotransfected HeLa-HFE cells with Nef-GFP and HckN (21), a dominant negative Hck mutant lacking the catalytic region, which blocks potential interactions between Nef and host-cell Src kinases (Fig. 5A). Fig. 5B shows that Nef-GFP down-regulates HFE, but down-regulation is abolished by cotransfection with HckN (Fig. 5C). HckN with a disrupted SH3 domain (W93F, Fig. 5D), had no effect on HFE down-regulation, whereas disrupting the SH2 domain of HckN (R151S, Fig. 5E) still allowed HckN to block Nef activity. CrkII protein that contains SH2 and SH3 domains but does not bind Nef had no effect on HFE down-regulation (Fig. 5F). Fig. 5 G and H shows that, in the presence of HckN, Nef is relocated to the cell periphery and is no longer retained in the TGN (Fig. 5I), and HFE resumes its usual endosomal/cell membrane distribution (Fig. 5J). Figs. 3, 4, 5 show that down-regulation of HFE by Nef is caused by a block in transport to the cell surface through retention in the TGN, in a process requiring Nef 72PxxP75 and, probably, an interaction with host-cell Src kinases. The TGN localization of Nef itself may also require Src kinases.
Fig. 5.
The SH3 domain of dominant negative Hck blocks Nef-mediated down-regulation of HFE. (A) Wild-type Hck has SH3, SH2, and a catalytic domain. Dominant negative HckN lacks the catalytic domain. (B) HeLa-HFE cells transfected with Nef-GFP (green) have reduced surface HFE, compared with Nef-negative cells (red). (C) Cotransfection at 1:1 of Nef-GFP with HckN blocks the effect of Nef on HFE. (D and E) A W93F HckN variant with dysfunctional SH3 domains (D) does not block Nef activity, whereas the blocking effect of HckN is maintained when SH2-disrupted R151S Hck (E) is used. (F) CrkII (which has SH3 domains but does not bind Nef) does not affect HFE down-regulation by Nef. (G) Nef-GFP is normally located perinuclearly, overlapping the TGN, but coexpression of HckN (H) reroutes Nef-GFP to the cell periphery. In the presence of HckN, Nef-GFP no longer colocalizes with the TGN (I, nuclei stained blue with DAPI), and HFE resumes its endosomal localization (J), similar to that in untransfected cells. (Scale bars, 10 μm.)
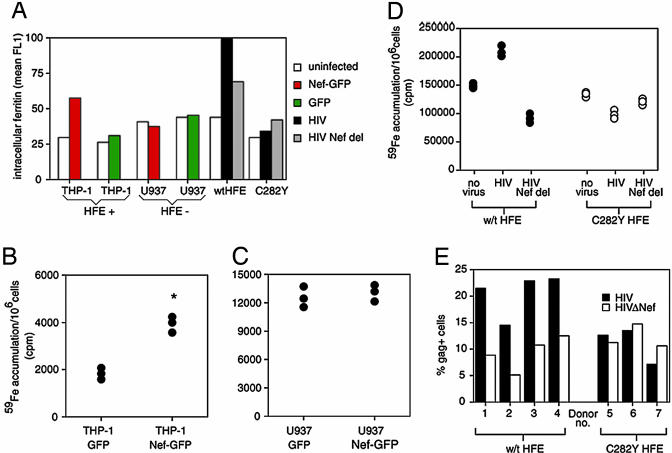
Nef and HIV-1 Induce HFE-Dependent Ferritin Iron Accumulation in Macrophages. In humans, HFE is naturally expressed by macrophages, including Kupffer cells and circulating monocytes/macrophages (10, 35), which are targets for HIV-1 infection. To test the effect of Nef and HFE on cellular iron handling in this setting, we infected THP-1 cells, which model HFE function in macrophages (16), with Nef-GFP or GFP lentiviruses and measured the iron-storage protein ferritin in infected cells, compared with uninfected cells in the same culture. Fig. 6A shows that Nef doubles ferritin in THP-1 cells, indicative of iron accumulation, whereas Nef had no effect on ferritin levels in the monocytic U937 cell line that does not express HFE. This result was confirmed by using [59Fe]transferrin; THP-1 cells expressing Nef accumulated twice as much 59Fe as did control cells (Fig. 6B), whereas Nef had no effect on U937 iron accumulation (Fig. 6C), consistent with a requirement for the Nef/HFE interaction to alter iron accumulation. We also infected macrophages homozygous for either wild-type HFE or C282Y HFE with HIV, with or without Nef, and measured ferritin levels (Fig. 6A) and 59Fe accumulation (Fig. 6D). After 4 days of virus exposure, HIV-infected, wild-type HFE-expressing macrophages contained the highest level of ferritin, which was reduced with Nef-deleted HIV or when infecting macrophages from C282Y hemochromatosis patients. Similarly, Fig. 6D shows that, after 12 days of viral exposure (when most cells are HIV positive, data not shown), HIV increased 59Fe accumulation by macrophages but that either nef deletion or the HFE C282Y mutation abolished this increase. Fig. 6 A-D shows that Nef, alone or in the context of HIV, increases iron accumulation by macrophages in an HFE-dependent manner.
Fig. 6.
The Nef-HFE interaction increases iron accumulation and HIV growth in monocytes/macrophages. (A) THP-1 cells were infected with lentiviral vectors encoding GFP or Nef-GFP and then permeabilized and analyzed for ferritin protein levels. Ex vivo macrophages homozygous for wild-type HFE or C282Y HFE were infected for 4 days with 50 ng/ml p24 HIV or Nef-deleted HIV and stained for p18/p55 gag and ferritin. The mean fluorescence intensity of 30,000-cell aliquots is given. Nef-GFP-expressing THP-1 cells contained double the ferritin of uninfected cells; GFP-infected cells have only a small increase in ferritin. HIV infection increases ferritin only in wild-type-expressing macrophages, and this effect is reduced in the absence of Nef. (B and C) THP-1 (B) and U937 (C), a monocytic cell line that does not express HFE, were transfected by nucleoporation with either Nef-GFP or GFP and incubated with 40 μg/ml [59Fe]transferrin for 14 h, and iron accumulation was measured. Nef-GFP increased intracellular iron in THP-1 but not in U937 cells. The mean accumulation was calculated, and the asterisk indicates significance, compared with GFP control (P < 0.001 by Student's t test). (D) Ex vivo macrophages homozygous for either wild-type or C282Y HFE were infected with wild-type HIV or Nef-deleted HIV and, after 2 days, incubated with 33 μg/ml [59Fe]transferrin for an additional 10 days. The cellular 59Fe was then determined. HIV increased macrophage iron accumulation in a Nef- and wild-type-HFE-dependent manner. Experiments shown in A-D are representative of several giving similar results. (E) Ex vivo macrophages homozygous for wild-type HFE (four donors) or C282Y HFE (three donors) were incubated with 50 ng/ml p24 of M-tropic HIV, with or without nef, for 4 days, stained for p18/p55 gag expression, and analyzed by FACS to calculate the percentage of p18/p55-positive cells in the cultures. The data show that Nef induces a 2- to 3-fold increase in gag-expressing cells in wild-type HFE-expressing macrophages, but this effect is reduced in C282Y-expressing macrophages and is not induced by Nef.
In HIV infection, macrophages are often iron-loaded, and the degree of macrophage iron-loading correlates inversely with survival times (36). Replication of HIV-1 involves the iron-dependent enzyme ribonucleotide reductase and is induced by NF-kappaB (37), which can be activated in macrophages by iron (2). Therefore, we wondered whether the Nef/HFE-mediated iron accumulation would directly benefit HIV-1. Fig. 6E shows that Nef increases the number of HIV-1 gag-positive cells in macrophage cultures expressing wild-type HFE. In macrophages expressing the C282Y variant of HFE, the percentage of gag-positive cells is reduced and not enhanced by Nef. The addition of iron to macrophages expressing C282Y HFE increased the level of gag staining (data not shown). These results raise the possibility that iron accumulation by HIV-1-infected macrophages, influenced by Nef and HFE, benefits the virus.
Summary
We have identified HFE as a target for Nef and shown that HFE down-regulation is more profound than the Nef-mediated down-regulation of total MHC I. We found that HIV-1 interferes in cellular iron handling, sequestering iron in monocytes and macrophages in a process that requires Nef and HFE. More work is needed to investigate whether macrophage iron levels, regulated by Nef and HFE (and altered by HFE mutations), affect the HIV-1 life cycle.
Acknowledgments
We thank C. Enns, R. Ehrlich, Y. Yang, A. Baur, M. Matsuda, and M. Collins for the generous provision of reagents; A. Simmons and A. McMichael for criticizing the manuscript; and A. Mitchison and Q. Sattentau for advice. This work was supported by funding from the Wellcome Trust and the Medical Research Council.
Author contributions: H.D., N.C., H.L., G.S., A.T., and X.-N.X. designed research; H.D., N.C., and H.L. performed research; H.D., N.C., G.S., A.T., and X.-N.X. contributed new reagents/analytic tools; H.D., N.C., H.L., G.S., A.T., and X.-N.X. analyzed data; and H.D., A.T., and X.-N.X. wrote the paper.
Abbreviations: IRES, internal ribosome entry site; TfR, transferrin receptor-1; TGN, trans-Golgi-network.
References
- 1.Deacon, N. J., Tsykin, A., Solomon, A., Smith, K., Ludford-Menting, M., Hooker, D. J., McPhee, D. A., Greenway, A. L., Ellett, A., Chatfield, C., et al. (1995) Science 270, 988-991. [DOI] [PubMed] [Google Scholar]
- 2.She, H., Xiong, S., Lin, M., Zandi, E., Giulivi, C. & Tsukamoto, H. (2002) in Am. J. Physiol. 283, G719-G726. [DOI] [PubMed] [Google Scholar]
- 3.Geyer, M., Fackler, O. T. & Peterlin, B. M. (2001) EMBO Rep. 2, 580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guy, B., Kieny, M. P., Riviere, Y., Le Peuch, C., Dott, K., Girard, M., Montagnier, L. & Lecocq, J. P. (1987) Nature 330, 266-269. [DOI] [PubMed] [Google Scholar]
- 5.Aiken, C., Konner, J., Landau, N. R., Lenburg, M. E. & Trono, D. (1994) Cell 76, 853-864. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz, O., Marechal, V., Le Gall, S., Lemonnier, F. & Heard, J. M. (1996) Nat. Med. 2, 338-342. [DOI] [PubMed] [Google Scholar]
- 7.Collins, K. L., Chen, B. K., Kalams, S. A., Walker, B. D. & Baltimore, D. (1998) Nature 391, 397-401. [DOI] [PubMed] [Google Scholar]
- 8.Le Gall, S., Erdtmann, L., Benichou, S., Berlioz-Torrent, C., Liu, L., Benarous, R., Heard, J. M. & Schwartz, O. (1998) Immunity 8, 483-495. [DOI] [PubMed] [Google Scholar]
- 9.Lebron, J. A., Bennett, M. J., Vaughn, D. E., Chirino, A. J., Snow, P. M., Mintier, G. A., Feder, J. N. & Bjorkman, P. J. (1998) Cell 93, 111-123. [DOI] [PubMed] [Google Scholar]
- 10.Bastin, J. M., Jones, M., O'Callaghan, C. A., Schimanski, L., Mason, D. Y. & Townsend, A. R. (1998) Br. J. Haematol. 103, 931-941. [DOI] [PubMed] [Google Scholar]
- 11.Parkkila, S., Waheed, A., Britton, R. S., Feder, J. N., Tsuchihashi, Z., Schatzman, R. C., Bacon, B. R. & Sly, W. S. (1997) Proc. Natl. Acad. Sci. USA 94, 2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feder, J. N., Gnirke, A., Thomas, W., Tsuchihashi, Z., Ruddy, D. A., Basava, A., Dormishian, F., Domingo, R., Jr., Ellis, M. C., Fullan, A., et al. (1996) Nat. Genet. 13, 399-408. [DOI] [PubMed] [Google Scholar]
- 13.Townsend, A. & Drakesmith, H. (2002) Lancet 359, 786-790. [DOI] [PubMed] [Google Scholar]
- 14.Feder, J. N., Penny, D. M., Irrinki, A., Lee, V. K., Lebron, J. A., Watson, N., Tsuchihashi, Z., Sigal, E., Bjorkman, P. J. & Schatzman, R. C. (1998) Proc. Natl. Acad. Sci. USA 95, 1472-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross, C. N., Irrinki, A., Feder, J. N. & Enns, C. A. (1998) J. Biol. Chem. 273, 22068-22074. [DOI] [PubMed] [Google Scholar]
- 16.Drakesmith, H., Sweetland, E., Schimanski, L., Edwards, J., Cowley, D., Ashraf, M., Bastin, J. & Townsend, A. R. (2002) Proc. Natl. Acad. Sci. USA 99, 15602-15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies, P. S. & Enns, C. A. (2004) J. Biol. Chem. 279, 25085-25092. [DOI] [PubMed] [Google Scholar]
- 18.Roy, C. N., Carlson, E. J., Anderson, E. L., Basava, A., Starnes, S. M., Feder, J. N. & Enns, C. A. (2000) FEBS Lett. 484, 271-274. [DOI] [PubMed] [Google Scholar]
- 19.Collin, M., Montaner, L., Herbein, G. & Gordon, S. (1995) in HIV: A Practical Approach, ed. Karn, J. (Oxford Univ. Press, Oxford), Vol. 1, pp. 63-75. [Google Scholar]
- 20.Ben-Arieh, S. V., Zimerman, B., Smorodinsky, N. I., Yaacubovicz, M., Schechter, C., Bacik, I., Gibbs, J., Bennink, J. R., Yewdell, J. W., Coligan, J. E., et al. (2001) J. Virol. 75, 10557-10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tokunaga, K., Kiyokawa, E., Nakaya, M., Otsuka, N., Kojima, A., Kurata, T. & Matsuda, M. (1998) J. Virol. 72, 6257-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Besnier, C., Takeuchi, Y. & Towers, G. (2002) Proc. Natl. Acad. Sci. USA 99, 11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naldini, L., Blomer, U., Gallay, P., Ory, D., Mulligan, R., Gage, F. H., Verma, I. M. & Trono, D. (1996) Science 272, 263-267. [DOI] [PubMed] [Google Scholar]
- 24.Fackler, O. T., Wolf, D., Weber, H. O., Laffert, B., D'Aloja, P., Schuler-Thurner, B., Geffin, R., Saksela, K., Geyer, M., Peterlin, B. M., et al. (2001) Curr. Biol. 11, 1294-1299. [DOI] [PubMed] [Google Scholar]
- 25.Le Gall, S., Buseyne, F., Trocha, A., Walker, B. D., Heard, J. M. & Schwartz, O. (2000) J. Virol. 74, 9256-9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasper, M. R. & Collins, K. L. (2003) J. Virol. 77, 3041-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feder, J. N., Tsuchihashi, Z., Irrinki, A., Lee, V. K., Mapa, F. A., Morikang, E., Prass, C. E., Starnes, S. M., Wolff, R. K., Parkkila, S., et al. (1997) J. Biol. Chem. 272, 14025-14028. [DOI] [PubMed] [Google Scholar]
- 28.Madrid, R., Janvier, K., Hitchin, D., Day, J., Coleman, S., Noviello, C., Bouchet, J., Benmerah, A., Guatelli, J. & Benichou, S. (2005) J. Biol. Chem. 280, 5032-5044. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz, O., Riviere, Y., Heard, J. M. & Danos, O. (1993) J. Virol. 67, 3274-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenway, A. L., McPhee, D. A., Grgacic, E., Hewish, D., Lucantoni, A., Macreadie, I. & Azad, A. (1994) Virology 198, 245-256. [DOI] [PubMed] [Google Scholar]
- 31.Blagoveshchenskaya, A. D., Thomas, L., Feliciangeli, S. F., Hung, C. H. & Thomas, G. (2002) Cell 111, 853-866. [DOI] [PubMed] [Google Scholar]
- 32.Piguet, V., Wan, L., Borel, C., Mangasarian, A., Demaurex, N., Thomas, G. & Trono, D. (2000) Nat. Cell Biol. 2, 163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saksela, K., Cheng, G. & Baltimore, D. (1995) EMBO J. 14, 484-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang, A. H., O'Shaughnessy, M. V. & Jirik, F. R. (2001) Eur. J. Immunol. 31, 2382-2387. [DOI] [PubMed] [Google Scholar]
- 35.Parkkila, S., Parkkila, A. K., Waheed, A., Britton, R. S., Zhou, X. Y., Fleming, R. E., Tomatsu, S., Bacon, B. R. & Sly, W. S. (2000) Haematologica 85, 340-345. [PubMed] [Google Scholar]
- 36.de Monye, C., Karcher, D. S., Boelaert, J. R. & Gordeuk, V. R. (1999) AIDS 13, 375-380. [DOI] [PubMed] [Google Scholar]
- 37.Nabel, G. & Baltimore, D. (1987) Nature 326, 711-713. [DOI] [PubMed] [Google Scholar]