Regulation of gene expression by transcriptional repression or activation has long been recognized as an effective means to control a biological process. However, while the initiation of an event can be governed by turning on the gene encoding a key rate-limiting enzyme mediating the respective action, its termination must often be equally tightly controlled by inactivating the responsible factor. Complete destruction is possibly the most effective way of ensuring the irreversible inactivation of a protein; consequently, all organisms employ intracellular proteolytic systems for the selective removal of “unwanted” proteins. This category includes short-lived regulatory factors as well as proteins that have been damaged or incapacitated by heat or other types of stress or toxic agents. In eukaryotes, regulated proteolysis is mediated largely by the 26S proteasome, a multicatalytic protease that consists of a barrel-shaped proteolytic 20S core particle in association with a 19S cap complex (20, 126). In contrast to a large portion of bulk protein turnover, which is mediated by vacuolar or lysosomal proteases, proteolysis by the 26S proteasome is energy dependent, due to the presence of ATPases of the AAA type within the 19S cap, which are responsible for unfolding the target proteins (135). Simpler versions of the 20S core and its associated ATPase subunits are known in archaebacteria and some eubacteria (76, 132). Even those bacteria that lack a conserved 20S particle employ a related strategy for energy-dependent proteolysis, using proteases that resemble the proteasome core particle architecturally and associate with a specific AAA ATPase subunit (6, 93, 132).
While in bacteria the protease itself—possibly in cooperation with accessory factors—is responsible for the recognition of relevant target proteins, eukaryotes have separated substrate selection from the actual proteolytic step, thereby greatly expanding the range and flexibility of possible degradation signals. Here the signal that elicits the degradation of a protein is its modification by ubiquitin, a small, highly conserved polypeptide unique to eukaryotes but ubiquitous among them (41, 89). Ubiquitin is covalently attached to a target protein in the form of polymeric chains, and these multiubiquitin chains generally serve as a recognition signal for the 26S proteasome (13). Thus, the task of correctly identifying and marking a protein for removal falls upon the enzymatic machinery that mediates the ubiquitin conjugation reaction.
The function of the ubiquitin system in protein degradation—target selection—is widely established. It is becoming clear, however, that modification of a protein by ubiquitin may serve other than degradative purposes (91). This review will focus on the actions of the ubiquitin system on eukaryotic chromatin, which include conventional, i.e., proteolytic, as well as possibly nonconventional functions. It will concentrate on studies in the yeast Saccharomyces cerevisiae, which has served as a convenient model system for the investigation of cell autonomous processes such as chromatin metabolism, but parallels to mammalian systems will be pointed out to demonstrate how highly conserved throughout evolution the use of the ubiquitin system appears to be in the context of chromatin. After an overview of the consequences of ubiquitin conjugation and proteolysis on transcription, initiation of replication, and chromosome segregation, I will focus on the RAD6 system and its effects on DNA damage repair, gene silencing, transposition, and meiosis. Finally, I will mention components of the ubiquitin system involved in the pathway of nucleotide excision repair (NER). My account will by no means be complete, as it could be argued that the removal of almost any protein by ubiquitin-dependent proteolysis has some influence on chromatin structure (if only its resynthesis by turning on transcription of the relevant gene). Rather than attempting full coverage of all possible areas, this review will therefore concentrate on a few examples that highlight the diversity of ubiquitin-associated processes in the context of chromatin.
CONSEQUENCES OF UBIQUITINATION
Ubiquitin is conjugated to target proteins in a multistep reaction that involves a cascade of enzymes (41, 89). In an ATP-dependent reaction, the ubiquitin-activating enzyme, E1, undergoes a thioester linkage between a cysteine residue within its active site and the carboxy terminus of ubiquitin. The activated ubiquitin moiety is then transferred to the active-site cysteine of a ubiquitin-conjugating enzyme (UBC, also known as E2), which mediates the formation of an isopeptide bond between ubiquitin and the ɛ-amino group of a lysine residue within the target protein. This conjugation reaction normally involves additional factors, ubiquitin protein ligases (also known as E3s), which are responsible for target recognition and in some cases also take part in the thioester transfer reaction. Repeated rounds of conjugation of ubiquitin to an internal lysine of the preceding ubiquitin moiety may result in the formation of multiubiquitin chains. While in most organisms a single E1 is responsible for activation of the entire cellular ubiquitin pool, all eukaryotic genomes analyzed encode several E2s and a large number of E3s with differing substrate specificities and subcellular localizations. Thus, target selectivity results from the cooperation of E2s and E3s in a combinatorial fashion.
The fate of ubiquitinated proteins depends on the nature of the modification itself: in contrast to multiubiquitination, attachment of a single molecule or a few molecules of ubiquitin to a number of plasma membrane transporters and receptors does not result in proteasomal degradation but rather in endocytosis and subsequent proteolysis in the lysosome or vacuole (42). Multiubiquitin chains, in turn, may potentially convey different signals depending on their topology (46, 90). Of the seven lysine residues in ubiquitin, at least three—K29, K48, and K63—are known to be used in vivo for further ubiquitination, and since they each reside on opposing or orthogonal faces of the molecule, the resulting chains most likely assume different structures (1, 55, 112). Chains linked via K48 are the principal signal for recognition by the 26S proteasome (13). The importance of this lysine is demonstrated by the lethality of the K48R mutation in S. cerevisiae (32). The K29 linkage has also been shown to be involved in proteasomal targeting in a number of contexts; however, efficient formation of long ubiquitin chains required the presence of K48 in at least some of the conjugates analyzed (55, 65). K63, on the other hand, has been shown to be involved in a puzzling variety of cellular functions not obviously related to proteasomal degradation: endocytosis and transport to the yeast vacuole (36, 114), mitochondrial inheritance (33), ribosome function (111), IκB kinase activation (23), and—last but not least—DNA damage repair (112). Although the biological function of K63-linked multiubiquitin chains is by no means understood, a unique and novel role in signaling unrelated to proteolysis is an attractive hypothesis.
REGULATION OF TRANSCRIPTION BY THE UBIQUITIN/PROTEASOME SYSTEM
Regulation of transcription initiation is one of the key control mechanisms for biological processes within a cell. Consequently, the levels and activities of the transcriptional activators and repressors themselves should be tightly controlled. It is therefore no surprise that many transcription factors have been found to be extremely unstable proteins; their short half-lives allow for rapid modulation of their concentrations by adjustment of the rate of either their synthesis or their degradation (108). This strategy is used by gram-negative bacteria, where the activities of several RNA polymerase-associated sigma factors are regulated on the basis of protein stability (109, 116, 121), but even more widely by yeast and higher eukaryotes. Here, the ubiquitin/proteasome system is mainly responsible for the degradation of short-lived transcription factors (45). In S. cerevisiae, these include the bZIP proteins GCN4 (69) and MET4 (100), regulators of amino acid biosynthesis and sulfur assimilation, respectively, as well as the homeodomain proteins CUP9 (12), a mediator of peptide import and copper homeostasis, and MATα2, which in combination with a number of different dimerization partners acts as a repressor of a- and haploid-specific genes, thereby controlling mating type identity in yeast (44). With half-lives as short as 3 to 5 min under destabilizing conditions, these proteins exemplify the flexibility of the transcriptional machinery, which can quickly adapt to environmental changes. In mammals, the number of transcription factors whose abundance is controlled by the ubiquitin/proteasome system is even larger, including such key regulators as p53, c-Jun, c-Fos, MyoD, and many others (45).
Despite a common degradation mechanism, the signals conveying ubiquitination as well as the enzymes involved in their recognition are quite diverse, and in many cases more than a single UBC contributes to the ubiquitination of a particular transcription factor (Table 1). For example, ubiquitination of CUP9 is mediated by the ubiquitin ligase UBR1, which has been identified as the cognate E3 of the N-end rule pathway, a degradation system based on the identity of a protein's amino-terminal residue that directly contacts the ligase (125). UBR1 generally acts in conjunction with the UBC RAD6/UBC2 (4, 25), and in fact, degradation of CUP9 has been found to depend on this E2; however, ubiquitination of the protein is independent of its amino terminus, indicating that an internal site within CUP9 serves as a ubiquitination signal for UBR1 in this case (12). Moreover, UBC4 in addition to RAD6 contributes to CUP9 turnover.
TABLE 1.
Targets of the ubiquitin/proteasome system involved in yeast chromatin metabolism
| Target proteina | E2b | E3b | Reference(s) |
|---|---|---|---|
| Transcription factors | |||
| GCN4 | CDC34 | SCFCDC4 | 69, 79 |
| RAD6 | ? | ||
| MET4 | CDC34 | SCFMET30 | 60, 100 |
| CUP9 | RAD6 | UBR1 | 12 |
| UBC4 | ? | ||
| MATα2 | UBC6, UBC7 | DOA10 | 16 |
| UBC4, UBC5 | ? | ||
| Replication initiation factors | |||
| CDC6 | CDC34 | SCFCDC4 | 27, 28, 105 |
| DBF4 | ? | APC | 17, 85, 130 |
| Kinetochore components | |||
| CTF13 | CDC34 | SCFCDC4 | 61 |
| CBF2 | CDC34 | ? | 133 |
| Anaphase regulators | |||
| PDS1 | ? | APC | 18, 48, 53 |
| SCC* | RAD6 ? | UBR1 | 96 |
| REC8* | RAD6 ? | UBR1 | 11 |
| Other proteins | |||
| RPB1 | ? | RSP5 | 5, 51 |
| HO | CDC34 | SCFUFO1 | 62 |
| RAD6 | RAD18 ? |
*, proteolytic fragments resulting from cleavage by ESP1.
?, identity of enzyme unknown.
No fewer than four E2s participate in the ubiquitination of MATα2: UBC4, UBC5, UBC6, and UBC7 (16). These UBCs respond to distinct ubiquitination signals within the α2 protein. While UBC4 and UBC5, which appear to be largely redundant, depend on a poorly defined internal signal, UBC6 and UBC7 respond to a defined, transplantable signal termed Deg1, which resides within the first 67 amino acids of α2 and overlaps with the hydrophobic face of the amphipathic α-helix responsible for dimerization (16).
GCN4 ubiquitination apparently involves two UBCs, based on the fact that the protein is partially stabilized in the respective mutants (69). One of them is RAD6; however, in contrast to CUP9 ubiquitination, the presence of UBR1 is not required here. The other is CDC34/UBC3, the E2 responsible for cell cycle progression at the G1/S transition. CDC34 is known to function in conjunction with a modular type of ubiquitin ligase, the SCF complex, which employs exchangeable subunits, the F-box proteins, for substrate recognition (24, 87). Ubiquitination of GCN4 is mediated by the F-box protein CDC4 (79). Other prominent targets of SCFCDC4 are the cyclin-dependent kinase inhibitors SIC1 and FAR1 (30, 40, 110).
Like GCN4, MET4 is ubiquitinated by CDC34 in conjunction with the SCF complex. The cognate F-box subunit in this case is MET30 (100). Interestingly, it is an unresolved question whether MET4 ubiquitination causes its degradation by the proteasome or conveys an alternative signal: while Rouillon et al. (100) determined a half-life of less than 10 min under destabilizing conditions and inhibition of degradation in proteasome mutants, Kaiser et al. (60) report that ubiquitination of MET4 by CDC34 and SCFMET30 did not lead to immediate proteolysis but instead inhibited its activity as a transcriptional activator of methionine biosynthesis genes. Thus, a nonproteolytic function of MET4 ubiquitination remains a possibility.
Intriguingly, in a recent report, ubiquitination via MET30 was demonstrated to contribute not to the inactivation, but instead to the activation, of a transcription factor: Salghetti et al. found that the function of the short-lived VP16 transcriptional activation domain (TAD) in yeast was dependent on the SCFMET30 ubiquitin ligase, as deletion of MET30 resulted in stabilization of the TAD in an inactive form (104). Linear fusion of ubiquitin to the TAD restored transcriptional activation function in the absence of MET30 without destabilizing the protein, indicating that activation and destruction are two separable consequences of ubiquitination. This dual role of ubiquitin conjugation may provide a “licensing” of the transcriptional activator by linking its activation to its rapid destruction (104).
Finally, the ubiquitin/proteasome system appears to be involved in the transcriptional elongation step in a nonconventional, i.e., nonproteolytic way. Ferdous et al. observed an inhibition of transcription elongation in vivo and in vitro by inactivation of two 19S cap subunits and the restoration of elongation by addition of purified 19S complex (31). This effect was independent of the proteolytic activity of the 20S core, and coimmunoprecipitation of 19S components with the elongation factor CDC68 suggested that the regulatory particle of the proteasome exerts its influence by means of physical interactions.
While these examples demonstrate the importance of the ubiquitin system for the regulation of transcription, they also raise the question how closely the components of the ubiquitination machinery really interact with the chromatin itself. Apart from its influence on transcription elongation (31), the 26S proteasome has not been observed in direct association with chromatin, arguing against proteolysis “in situ.” However, its exact subcellular localization remains a matter of debate (29, 103). It seems likely, however, that many short-lived transcription factors are ubiquitinated while they are bound to their recognition elements. At least in the case of MATα2, ubiquitination appears to occur preferentially in the nucleus, as inhibition of nuclear import results in the abolishment of ubiquitin conjugation and a significant stabilization of the protein (73). A nuclear localization was also demonstrated for the F-box protein MET30 (100). Similarly, mammalian p53 is ubiquitinated in the nucleus; however, its degradation apparently requires its export from the nucleus to the cytoplasm (120). Thus, it remains to be seen how close to their sites of action DNA-binding proteins are being recognized and modified by the ubiquitin system.
INFLUENCE OF THE UBIQUITIN SYSTEM ON CELL CYCLE-REGULATED CHROMOSOME DYNAMICS
Ubiquitination and subsequent degradation of cyclins and cyclin-dependent kinase inhibitors ensure orderly progression through the eukaryotic cell cycle (66, 82). However, the ubiquitin system has an even more direct influence on chromosome dynamics throughout the cell cycle, ranging from initiation of replication to chromosome segregation. Replication initiation is controlled by the association of a set of proteins, the prereplication complex, with the origins of replication, a process called licensing. Two essential DNA-associated factors involved in this function, CDC6 and DBF4, have been shown to be substrates of the ubiquitin/proteasome system (27, 130). CDC6 mediates the assembly of the prereplication complex by increasing its specificity for the replication origins (115). Transcription of the CDC6 gene peaks at the M/G1 transition, and the instability of the protein necessitates its resynthesis in each round of DNA replication. Its degradation once the prereplication complex is assembled is mediated by CDC34 and SCFCDC4 after phosphorylation by the cyclin-dependent kinase CDC28 (27, 28, 105). The observation that deletion of its nuclear localization signal stabilizes the protein again argues for recognition and modification by the ubiquitin system within the nucleus, possibly still in association with the DNA (28).
DBF4 is recruited to the chromatin as a positive regulator of the CDC7 kinase, which is responsible for release of the prereplication complex during S phase by phosphorylation of several of its subunits, preceding the assembly of the replication machinery (115). Gene expression peaks in S phase and persists until after mitosis (17). The protein is rapidly degraded prior to START. The E3 responsible for ubiquitination of DBF4 is the anaphase-promoting complex (APC) (17, 130), a multisubunit ubiquitin ligase that mediates the destruction of mitotic cyclins at the metaphase-to-anaphase transition (88). The fact that overproduction of DBF4 is lethal in combination with a mutation in a gene encoding one of the APC subunits, apc1-1, indicates the importance of timely removal of DBF4 (85).
Several observations point to an influence of the ubiquitin system on kinetochore assembly, yet the nature of this effect remains poorly understood. Connelly and Hieter identified SKP1, the F-box binding subunit of the SCF complex, as a component of the CBF3 complex, a protein assembly that mediates the connection of centromere DNA to the mitotic spindle (19). SKP1 was found to promote the DNA association of CTF13, another CBF3 subunit (61). This activation coincided with the phosphorylation of CTF13, and Kaplan et al. propose that a SKP1-associated kinase may be responsible for this activity. CTF13 apparently contacts SKP1 directly by means of its F-box domain. Moreover, CTF13 is a short-lived protein whose degradation depends on CDC34 and CDC4, suggesting that SKP1, as part of an SCF-type ubiquitin ligase, promotes the ubiquitin-dependent removal of CTF13 following phosphorylation (61). Kaplan et al. hypothesize that CTF13 proteolysis may contribute to the destruction of incorrectly assembled kinetochores. How exactly SKP1 activates CTF13 is not fully understood; however, an additional component required for this process has been shown to be SGT1, which apparently acts as another SKP1-interacting subunit of the SCFCDC4 complex and is also required for ubiquitination of other SCF substrates (64). Consequently, sgt1 mutants are defective in CBF3 assembly. Yet another kinetochore component, CBF2, was found to be ubiquitinated in vivo by CDC34 (133); however, the significance of this modification remains unknown, as the protein does not appear to be degraded (61).
Arguably the most important contribution of the ubiquitin system to progression of the cell cycle at the level of chromosome dynamics is the control over sister chromatid separation at the onset of anaphase, a function that is highly conserved among eukaryotes (83). Cohesion between sister chromatids after replication of the genome is essential for the symmetric distribution of the chromatids to opposite poles of the mitotic spindle. During metaphase, this cohesion between the sisters results in a tension that is created by the pulling force of the spindle fibers at the kinetochores and is relieved in anaphase when sister chromatid separation allows the movement of the chromatids to the opposing spindle poles. The observation that a ubiquitin ligase, the APC (see above), is required for the onset of anaphase first led to the hypothesis that a proteinaceous bridge mediates cohesion between sisters (48, 53). This bridge was identified as a multiprotein complex now called cohesin (122). However, the APC does not, as was initially believed (53), directly trigger the destruction of cohesin by ubiquitination. In fact, it turned out that its substrate is a protein called PDS1, an inhibitor of the cysteine protease ESP1, also called separin, which in turn cleaves one of the subunits of cohesin, SCC1, and thereby mediates sister chromatid separation (18). As separin is also responsible for cleaving REC8, a meiotic counterpart of SCC1, the ubiquitin system exerts its effect on the dissociation of homologous chromosomes during meiosis I as well (11). Despite the absence of sequence homology, the interplay between vertebrate securin and separin has been found to be controlled by the APC in the same manner (134).
Interestingly, ubiquitin-dependent proteolysis plays a role in chromosome segregation even after SCC1 and REC8 are cleaved: Rao et al. were able to show that the carboxy-terminal fragment of SCC1 resulting from cleavage by separin is short-lived and is degraded in a UBR1-dependent manner (96). In fact, this fragment bears a destabilizing amino terminus and thus represents the first known physiological substrate of the N-end rule pathway in yeast that is truly recognized by its amino terminus (see above). Proteolysis appears to be important for mitosis, as overexpression of a stable analog of the respective SCC1 fragment is lethal to mitotic cells. Even when expressed from the native promoter, the stable fragment leads to a greatly increased frequency of missegregation and chromosome loss, similar to that observed with the ubr1 deletion itself (96). Besides REC8, whose proteolytic fragment has been confirmed as another N-end rule substrate (11), there are several other proteins in S. cerevisiae bearing potential ESP1 consensus recognition sites where cleavage would generate destabilizing amino termini, suggesting a more general relevance of the N-end rule in vivo (96).
ACTIONS OF RAD6 ON CHROMATIN
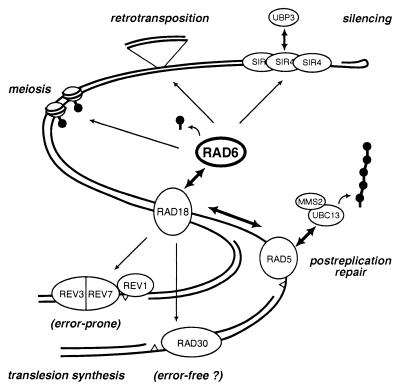
RAD6/UBC2 is the E2 that cooperates with UBR1 in N-end rule-dependent ubiquitin conjugation in yeast (25). Independently of UBR1, RAD6 is intimately involved in many aspects of chromatin metabolism, judging by the highly pleiotropic phenotype of rad6 mutants. First isolated in a screen for mutants sensitive to UV irradiation (21), rad6 cells in fact display an extremely high sensitivity to a broad spectrum of DNA-damaging agents, including UV light, X rays, and virtually all chemical mutagens tested (70). They are incapable of DNA damage-induced mutagenesis but have an elevated spontaneous-mutation rate (71, 72, 95). Homozygous diploids fail to sporulate but are hyperactive in mitotic recombination (63, 81). Moreover, rad6 mutants are deficient in silent information regulator (SIR)-dependent silencing (50) and exhibit elevated levels of Ty1 transposition, along with an altered site preference for transposon integration (74, 92). These properties identify RAD6 as a key player in the maintenance of DNA integrity (Fig. 1), and genetic analysis has since provided a lot of information about the pathways controlled by this UBC (10, 70). Yet its mechanism of action on a biochemical level still remains an enigma. As a matter of fact, its connection to the ubiquitin system was established only when RAD6 was identified as a UBC by means of its ability to covalently attach to a ubiquitin affinity column (54). The observation that mutation of its conserved active-site cysteine, responsible for ubiquitin thioester formation, results in the same phenotype as the rad6 null mutant confirmed that its catalytic activity as an E2 is essential for in vivo function (50, 117, 118). However, most of the relevant RAD6 substrates have yet to be identified.
FIG. 1.
Actions of RAD6 on yeast chromatin. DNA-associated processes and the factors involved are represented schematically. Thick, double-headed arrows, physical interactions; thin arrows, indirect or genetically inferred influences. Lollipops, ubiquitin; triangles, DNA lesions.
In higher eukaryotes ubiquitination is one of the prominent histone modifications that has been correlated with an alteration of nucleosome structure and activation of transcription (7, 113). Purified yeast RAD6 is in fact able to efficiently ubiquitinate histones H2A and H2B in vitro (54, 119). This observation prompted Robzyk et al. to examine histone ubiquitination in yeast (98). They observed that a significant fraction of H2B is modified by a single ubiquitin at one of the conserved lysines in its carboxy-terminal tail, K123. This situation is different in mammals, where H2A is the most prevalent ubiquitinated histone (113). H2B modification is absent in the rad6 mutant and correlates with the ability to undergo meiosis, as the H2B(K123R) mutant is, like the rad6 mutant, defective in sporulation. In addition, the mutation confers a slight growth defect that is shared by the rad6 mutant. Thus, ubiquitination of H2B by RAD6 is likely a prerequisite for meiosis and optimal mitotic growth in yeast (98). No E3 has been found to cooperate with RAD6 in this process in vivo or in vitro, suggesting that the highly acidic carboxy-terminal tail of RAD6 may be sufficient to promote an interaction with the basic histone substrate (119). Deletion of the terminal 23 amino acids indeed confers a sporulation and growth defect similar to that of the rad6 null mutant, but without affecting other RAD6 functions. The acidic tail is apparently unique to yeast; as a consequence, homologs of other species, including Schizosaccharomyces pombe, mice, and humans, which lack this extension, fully or partially complement the UV repair deficiency, but not the sporulation defect, of the yeast rad6 mutant (68).
Interestingly, mammalian RAD6 nevertheless seems to play an important role in meiosis. Two RAD6 homologs have been identified in both mice and humans: the X-linked HR6A and the autosomal HR6B (119). Expression of HR6A is strongly repressed during spermatogenesis, but the HR6B transcript and protein are highly enriched in the testis, and transcription peaks at the time of histone-to-protamine transition (67). Deletion of HR6B in mice results in male sterility, accompanied by severe aberrations in spermatogenesis (99). Extensive ubiquitination of histone H2A is indeed observed during the pachytene stage of prophase I and again in elongating spermatids, and Baarends et al. have proposed that ubiquitination by HR6B may trigger the removal of histones, which is required for chromatin condensation and packaging (2). Intriguingly, this attractive model is contradicted by the troubling finding that the pattern of modification appears unchanged in HR6B knockout mice, arguing that HR6B alone may not be responsible for histone ubiquitination after all. Thus, Baarends et al. conclude that dysregulation of other, yet unidentified HR6B substrates is likely to cause the impairment of spermatogenesis in HR6B knockout mice (2). A possible candidate for this alternative HR6B substrate is histone H3, which has been observed in a ubiquitinated form in elongating spermatids as well (14). However, the extent of H3 modification has not yet been reported in the HR6B knockout.
Additional substrates of RAD6 must exist in yeast as well, since lack of histone H2B ubiquitination is not responsible for the UV sensitivity of rad6 mutants (98). This defect is primarily due to the inability of rad6 mutants to restore high-molecular-weight DNA by replicative synthesis on templates that have been damaged by irradiation in the absence of NER (94). Due to this phenotype, the RAD6 pathway has also been called postreplication repair, and it is believed to confer damage tolerance in a situation where the DNA replication machinery is blocked by lesions in the template strand that have not been removed by other repair systems (70). Genetic analysis has revealed two apparently distinct activities of RAD6 that contribute to this function: one of these is responsible for damage-induced mutagenesis and is therefore termed error-prone, whereas the other is an apparently error-free mode of repair (70, 81). Numerous other repair genes have been found by classical epistasis analysis to be dependent on RAD6, and based on the ability of the respective mutants to undergo damage-induced mutagenesis, they have been assigned to either the error-free or the error-prone branch.
Biochemical characterization of the factors involved has revealed that error-prone repair entails translesion synthesis very similar to the SOS response of Escherichia coli. A damage-tolerant DNA polymerase, Polζ, encoded by the two RAD6-dependent repair genes REV3 and REV7, is able to bypass a broad range of lesions, including photoadducts and abasic sites (84). Polζ cooperates with the REV1 gene product, a deoxycytidyl transferase with homology to the E. coli UmuC protein. REV1 can incorporate dCMP opposite a lesion such as an abasic site, producing a 3" terminus that is efficiently extended by Polζ. A second damage-tolerant polymerase, Polη, encoded by RAD30, has recently been identified as another member of the RAD6 pathway (58). Polη, which shares homology with the E. coli DinB protein, was found to correctly insert adenine opposite a thymine dimer; consequently, Johnson et al. termed Polη-dependent repair error-free translesion synthesis (58). However, the overall fidelity of Polη on nondamaged templates is much lower than that of replicative polymerases (128). Moreover, other types of lesions were found to cause a mutagenic bypass in cooperation with Polζ (56). Accordingly, there is disagreement about the extent of damage-induced mutagenesis in rad30 mutants (78, 101). Homologs of both Polζ and Polη exist in mammals, indicating that translesion synthesis is a highly conserved process in eukaryotes (127); nevertheless, the role of RAD6 in this process is not at all understood.
In error-free repair, RAD6 cooperates with a number of factors that include additional components of the ubiquitin system. An early contribution came from the discovery that mutant yeast cells in which lysine 63 of ubiquitin was replaced by arginine (K63R) displayed a UV-sensitive phenotype that falls into the RAD6 epistasis group, arguing that K63-linked multiubiquitin chains are important for postreplication DNA repair (112). The E2 responsible for the assembly of those chains was later identified as a heterodimer of UBC13 and the E2 homolog MMS2 (46). MMS2 had been cloned independently by complementation of a mutant sensitive to the DNA-damaging agent methyl methane sulfonate (MMS) and was found to be a member of the error-free RAD6-dependent repair system (9). Both mms2 and ubc13 cells in fact exhibit UV sensitivities identical to that of the ubiquitin K63R mutant (47). A close cooperation between RAD6 and UBC13/MMS2 can be inferred from the identification of physical interactions within the error-free system (124). RAD6 is known to form a stable complex with the DNA-binding protein RAD18, and although its DNA-binding properties are not fully characterized, it has been suggested that RAD18 serves to recruit RAD6 to sites of damage for both error-free and error-prone repair (3). Recently, another member of the error-free repair system, RAD5, was found to act as a mediator between the RAD6-RAD18 complex and UBC13/MMS2: RAD5, a DNA-dependent ATPase with homology to the SNF2/SWI2 family of helicases and chromatin remodelling factors (57, 59), is able to promote the association of UBC13 with the chromatin in a manner similar to the recruitment of RAD6 by RAD18 (124). In addition, RAD5 physically associates with RAD18 in a way that is permissive for the RAD5-UBC13 interaction, resulting in a heteromeric complex in which the two E2s, RAD6, and UBC13/MMS2 may directly cooperate in the ubiquitination of a common target protein (124).
In contrast to the concept of translesion synthesis, however, the molecular mechanism of error-free damage tolerance remains largely obscure. Current models of the events at a stalled replication fork mostly invoke a transient template switch to the nondamaged sister chromatid, possibly in a recombination-mediated fashion (10, 70, 123). Identification of the target proteins ubiquitinated by RAD6 and UBC13/MMS2 is expected to shed some light on the function of ubiquitin in this process. In particular the role of the nonconventional K63-linked multiubiquitin chains is of interest, as analyses of proteasome mutants have indicated that proteolysis is not required for postreplication DNA repair (26, 46). Instead, ubiquitination may serve to recruit additional repair factors to the site of damage or promote the disassembly of a multimeric complex by inducing changes in the conformation of the target protein.
Intriguingly, both RAD18 and RAD5 harbor a RING domain, a specialized type of zinc finger that has been identified as part of a growing number of ubiquitin ligases, including UBR1, the SCF complex, and the APC (34, 75). In many, but not all, cases, the RING domain mediates interaction with the E2; similarly, RAD5 contacts UBC13 by means of this domain (124). It is therefore attractive to speculate that RAD18 and RAD5 may actually not only function as recruiting factors but directly take part in ubiquitin conjugation as E3s. Consistent with this hypothesis is the notion that an intact RING domain is essential for the function of both factors in DNA repair (unpublished data), even though the RAD18 RING finger is not required for binding to either RAD6 or RAD5. A possible E3 function of RAD18 is also suggested by its recent implication in the proteasome-dependent degradation of HO (62), the endonuclease that initiates mating type switching in yeast (39). Two E2s, CDC34 and RAD6, as well as components of the SCF complex and a previously uncharacterized F-box protein, UFO1, have also been found to contribute to the protein's short half-life (62). Hence, if the ubiquitin ligase activity of RAD18 can indeed be demonstrated with HO as a substrate, this would indicate that the RAD6-RAD18 complex can function in a conventional fashion to mark a short-lived protein for proteasomal degradation.
A more general effect on chromatin structure is suggested by the effects of the rad6 mutation on silencing and the rate and site bias of Ty1 transposition. Ty1 is a yeast retrotransposon that integrates preferentially into AT-rich sequences and tRNA genes, with a strong bias toward promoters and 5" ends of coding regions (106). In rad6 null mutants, this target site bias is abolished for all sites analyzed and overall transposition frequency is elevated without a concomitant increase in Ty1 message, arguing that deletion of RAD6 leads to an alteration of chromatin structure that facilitates transposon integration at random sites (49, 74, 92). A derangement of chromatin structure is similarly suggested by the loss of silencing at the telomeres, the rDNA cluster, and the silent mating type loci observed in rad6 mutants (50). In a phenotypic analysis of a broad spectrum of rad6 alleles, Freiberg et al. found a correlation between defects in silencing and elevated retrotransposition in most mutants, whereas effects on DNA damage repair, sporulation, growth rate, and N-end rule activity were genetically separable (35). Since neither UBR1 nor RAD18 is required for silencing and transposition (50), Freiberg et al. propose that these activities may be manifestations of the same aspect of RAD6 function, possibly mediated by a third, yet unidentified interaction partner (35). Interestingly, not only ubiquitination but also deubiquitination affects silencing. In contrast to the rad6 mutation, deletion of the ubiquitin hydrolase gene UBP3 results not in a defect, but in an enhancement, of silencing (80). Since UBP3 was found to interact directly with the DNA-binding silencing protein SIR4, it is very likely that its target proteins are chromatin components associated with the silenced mating type loci and telomeres (80). Thus, ubiquitin deconjugation apparently counteracts the effects of ubiquitin conjugation at these sites. It is unknown whether RAD6 and UBP3 act on the same set of target proteins, but it is attractive to speculate that they may function in opposite ways to regulate the extent of chromatin silencing by means of ubiquitination.
FUNCTIONS OF THE UBIQUITIN SYTEM IN NER
In contrast to postreplication repair, NER is mechanistically well understood and has been reconstituted in vitro from purified components (22). Yet ubiquitin and the proteasome appear to play a regulatory role in this process that is still an issue of controversy. The first hint that the ubiquitin system may be involved in NER came from the identification of a ubiquitin-like domain at the amino terminus of the repair factor RAD23 (129). This protein, like its two human homologs (77), acts in complex with another NER protein, RAD4, which is necessary for the incision step in both transcription-coupled and global NER (38); however, its catalytic function is unknown. The ubiquitin-like domain is important for efficient repair, and its removal results in a UV sensitivity intermediate between the sensitivity with the wild-type gene (wt) and that with complete deletion of the gene. Interestingly, this domain can be replaced by ubiquitin itself without affecting repair efficiency, although the protein is quite stable and is not subject to proteasomal degradation (129). Schauber et al. (107) were able to show that the ubiquitin-like domain in fact mediates an interaction of RAD23 with the 26S proteasome. In a recent study from the same laboratory, RAD23 was found to interfere with the formation of long multiubiquitin chains in vitro and in vivo, its overexpression leading to a stabilization of several otherwise short-lived proteins (86). Apparently, RAD23 exerts this inhibitory role by directly binding to substrates bearing short chains and inhibiting chain elongation (15, 86, 131). Surprisingly, however, its ubiquitin-like domain is dispensable for this activity (86). Suppression of the UV sensitivity of rad23Δ cells by deletion of the E2 UBC4 or the chain elongation factor UFD2 suggested antagonistic roles of RAD23 and the ubiquitin conjugation machinery, leading to the conclusion that RAD23 may have a novel antiproteolytic function for DNA repair, possibly the regulation of RAD4 stability (86).
These findings stand in contrast to those of Russell et al. who confirmed the interaction of RAD23 with the 26S proteasome but found a stimulatory role of the 19S particle for NER in cell extracts, independent of the 20S proteolytic activity, thus suggesting a nonproteolytic, possibly chaperone-like function of the proteasome in repair (102) reminiscent of its effect on transcription elongation (31). However, the same group recently reported that in vivo the 19S particle actually had an inhibitory rather than a stimulatory effect on the removal of lesions from the DNA by NER that was independent of RAD23, despite an increased UV sensitivity of mutants with impaired 19S function (37). These conflicting results could be partially reconciled if ubiquitination and its inhibition by RAD23 served a nonproteolytic function in the context of NER, while the effect of RAD23 overexpression on the degradation of unrelated proteins could be a nonphysiological side effect. Nevertheless, how this activity relates to the recruitment of the proteasome by RAD23's ubiquitin-like domain or even a RAD23-independent action of the 19S cap on the chromatin remains an open question.
A second contribution of the ubiquitin system to NER appears to be limited to transcription-coupled repair, a special form of NER that promotes the preferential repair of lesions within the transcribed strand of transcriptionally active DNA. The process is dependent on RNA polymerase II (PolII), and the large subunit of this polymerase has been identified as a substrate for ubiquitination in response to DNA damage in both mammals and yeast (8, 51). In yeast, ubiquitination is dependent on the ligase RSP5, which is capable of ubiquitinating the human polymerase in vitro as well (5, 51). Both mammalian and yeast PolII's are indeed subject to proteasomal degradation following damage-induced ubiquitin conjugation (5, 97). However, the relevance for transcription-coupled repair is not entirely clear yet. On the one hand, ubiquitination of PolII was found to be absent in two different cell lines derived from Cockayne syndrome patients that were defective in transcription-coupled repair; on the other hand, it was not determined whether this lack of ubiquitination was the cause or a consequence of the repair defect (8). Moreover, RAD26, the yeast homolog of CS-B, the factor missing in one of the Cockayne syndrome cell lines, was found to have no influence on the ubiquitination of PolII, and mutation of RSP5 did not cause a defect in transcription-coupled repair (5).
CONCLUDING REMARKS
Proteolytic functions of the ubiquitin/proteasome system obviously play an essential role in the regulation of chromatin-associated processes: in particular, the degradation of short-lived regulatory factors has direct consequences for transcription, initiation of replication, and chromosome segregation. The conjugation factors involved in the modification of the target proteins reflect the full range and the diversity of the ubiquitination machinery that is active even in one of the least complex eukaryotes (Table 1).
However, the maintenance of chromatin integrity appears to invoke a number of additional, less conventional aspects of the ubiquitin system, best exemplified by the influence of RAD6 on DNA repair, transposition, silencing, and meiosis (Fig. 1), but also by the activity of RAD23. Unfortunately, as most of the relevant ubiquitination targets are still unknown, any predictions about the consequences of ubiquitin conjugation remain speculative, and we are not even close to understanding the nature of this potential nonproteolytic regulatory signal. Where proteasome involvement can be excluded, alternative models would invoke functions such as the recruitment of other factors by means of an affinity for ubiquitin chains or ubiquitinated proteins, the modulation of a protein's activity, and the disassembly or structural alteration of a multiprotein complex due to a conformational change induced by ubiquitination of a subunit. An even more speculative, but not entirely impossible, scenario would be the conjugation of ubiquitin not to a proteinaceous substrate but to a different biomolecule, potentially even DNA. One precedent for this option was found in the case of a remote cousin of ubiquitin, APG8, which is conjugated by its carboxy terminus not to a protein but to the amino group of a lipid (52). On the whole, ubiquitination is emerging more and more as a general and multipurpose protein modification system much like phosphorylation or acetylation (42, 91).
Components of the ubiquitin system in different organisms are often highly conserved, and in many cases the human homologs are able to complement the phenotypes of the corresponding yeast mutants. This conservation is particularly striking for the chromatin-related aspects of ubiquitin, raising the question how and in what order the ubiquitin system could have acquired its degradative as well as nonproteolytic functions. Possible hints come from ubiquitin's relatives, a growing set of ubiquitin-like modifiers that cover an extremely wide range of cellular activities (43). Obvious mechanistic parallels in their conjugation machineries suggest an evolutionary origin entirely unrelated to proteolysis, arguing that the specialization in degradation may have been a later acquisition. An intriguing question is how, then, ubiquitin was able to assume control in eukaryotes over processes that in prokaryotes are organized in a very similar manner, but without the need for ubiquitin—for example, mutagenic translesion DNA synthesis. The fact that the basic principle of this process still appears to be conserved from prokaryotes to eukaryotes (127) supports the notion that the ubiquitin system mainly acts in a regulatory manner to fine-tune the events at the chromatin. Considering that the set of conjugation factors and substrates described here reflects only a fraction of the known ubiquitin-dependent aspects of metabolism, we might expect to find an even broader scope of nonconventional ubiquitin functions in other areas of cell biology, illustrating once more the pervasiveness of the ubiquitin system throughout the eukaryotic cell.
Acknowledgments
I thank Regine Kahmann for generous support. Work in this laboratory is supported by grants from the Deutsche Forschungsgemeinschaft (DFG), the German Ministry for Education and Research (BMBF), and the German-Israeli Foundation for Scientific Research and Development (GIF).
REFERENCES
- 1.Arnason, T., and M. J. Ellison. 1994. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol. Cell. Biol. 14:7876-7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baarends, W. M., J. W. Hoogerbrugge, H. P. Roest, M. Ooms, J. Vreeburg, J. H. Hoeijmakers, and J. A. Grootegoed. 1999. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 207:322-333. [DOI] [PubMed] [Google Scholar]
- 3.Bailly, V., S. Lauder, S. Prakash, and L. Prakash. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272:23360-23365. [DOI] [PubMed] [Google Scholar]
- 4.Bartel, B., I. Wünning, and A. Varshavsky. 1990. The recognition component of the N-end rule pathway. EMBO J. 9:3179-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaudenon, S. L., M. R. Huacani, G. Wang, D. P. McDonnell, and J. M. Huibregtse. 1999. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6972-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bochtler, M., C. Hartmann, H. K. Song, G. P. Bourenkov, H. D. Bartunik, and R. Huber. 2000. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature 403:800-805. [DOI] [PubMed] [Google Scholar]
- 7.Bradbury, E. M. 1992. Reversible histone modifications and the chromosome cell cycle. Bioessays 14:9-16. [DOI] [PubMed] [Google Scholar]
- 8.Bregman, D. B., R. Halaban, A. J. van Gool, K. A. Henning, E. C. Friedberg, and S. L. Warren. 1996. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc. Natl. Acad. Sci. USA 93:11586-11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broomfield, S., B. L. Chow, and W. Xiao. 1998. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl. Acad. Sci. USA 95:5678-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broomfield, S., T. Hryciw, and W. Xiao. 2001. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 486:167-184. [DOI] [PubMed] [Google Scholar]
- 11.Buonomo, S. B., R. K. Clyne, J. Fuchs, J. Loidl, F. Uhlmann, and K. Nasmyth. 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103:387-398. [DOI] [PubMed] [Google Scholar]
- 12.Byrd, C., G. C. Turner, and A. Varshavsky. 1998. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 17:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chau, V., J. W. Tobias, A. Bachmair, D. Marriott, D. J. Ecker, D. K. Gonda, and A. Varshavsky. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576-1583. [DOI] [PubMed] [Google Scholar]
- 14.Chen, H. Y., J. M. Sun, Y. Zhang, J. R. Davie, and M. L. Meistrich. 1998. Ubiquitination of histone H3 in elongating spermatids of rat testes. J. Biol. Chem. 273:13165-13169. [DOI] [PubMed] [Google Scholar]
- 15.Chen, L., U. Shinde, T. G. Ortolan, and K. Madura. 2001. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, P., P. Johnson, T. Sommer, S. Jentsch, and M. Hochstrasser. 1993. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell 74:357-369. [DOI] [PubMed] [Google Scholar]
- 17.Cheng, L., T. Collyer, and C. F. Hardy. 1999. Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol. Cell. Biol. 19:4270-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen-Fix, O., J. M. Peters, M. W. Kirschner, and D. Koshland. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10:3081-3093. [DOI] [PubMed] [Google Scholar]
- 19.Connelly, C., and P. Hieter. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coux, O., K. Tanaka, and A. L. Goldberg. 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65:801-847. [DOI] [PubMed] [Google Scholar]
- 21.Cox, B. S., and J. M. Parry. 1968. The isolation, genetics and survival characteristics of ultraviolet light-sensitive mutants in yeast. Mutat. Res. 6:37-55. [DOI] [PubMed] [Google Scholar]
- 22.de Laat, W. L., N. G. Jaspers, and J. H. Hoeijmakers. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13:768-785. [DOI] [PubMed] [Google Scholar]
- 23.Deng, L., C. Wang, E. Spencer, L. Yang, A. Braun, J. You, C. Slaughter, C. Pickart, and Z. J. Chen. 2000. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351-361. [DOI] [PubMed] [Google Scholar]
- 24.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell. Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 25.Dohmen, R. J., K. Madura, B. Bartel, and A. Varshavsky. 1991. The N-end rule is mediated by the UBC2 (RAD6) ubiquitin-conjugating enzyme. Proc. Natl. Acad. Sci. USA 88:7351-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dor, Y., B. Raboy, and R. G. Kulka. 1996. Role of the conserved carboxy-terminal alpha-helix of Rad6p in ubiquitination and DNA repair. Mol. Microbiol. 21:1197-1206. [DOI] [PubMed] [Google Scholar]
- 27.Drury, L. S., G. Perkins, and J. F. Diffley. 1997. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16:5966-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elsasser, S., Y. Chi, P. Yang, and J. L. Campbell. 1999. Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell 10:3263-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enenkel, C., A. Lehmann, and P. M. Kloetzel. 1998. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 17:6144-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldman, R. M., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221-230. [DOI] [PubMed] [Google Scholar]
- 31.Ferdous, A., F. Gonzalez, L. Sun, T. Kodadek, and S. A. Johnston. 2001. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol. Cell 7:981-991. [DOI] [PubMed] [Google Scholar]
- 32.Finley, D., S. Sadis, B. P. Monia, P. Boucher, D. J. Ecker, S. T. Crooke, and V. Chau. 1994. Inhibition of proteolysis and cell cycle progression in a multiubiquitin-deficient yeast mutant. Mol. Cell. Biol. 14:5501-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fisk, H. A., and M. P. Yaffe. 1999. A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 145:1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freemont, P. S. 2000. Ubiquitination: RING for destruction? Curr. Biol. 10:R84-R87. [DOI] [PubMed]
- 35.Freiberg, G., A. D. Mesecar, H. Huang, J. Y. Hong, and S. W. Liebman. 2000. Characterization of novel rad6/ubc2 ubiquitin-conjugating enzyme mutants in yeast. Curr. Genet. 37:221-233. [DOI] [PubMed] [Google Scholar]
- 36.Galan, J., and R. Haguenauer-Tsapis. 1997. Ubiquitin Lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 16:5847-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillette, T. G., W. Huang, S. J. Russell, S. H. Reed, S. A. Johnston, and E. C. Friedberg. 2001. The 19S complex of the proteasome regulates nucleotide excision repair in yeast. Genes Dev. 15:1528-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guzder, S. N., P. Sung, L. Prakash, and S. Prakash. 1999. Synergistic interaction between yeast nucleotide excision repair factors NEF2 and NEF4 in the binding of ultraviolet-damaged DNA. J. Biol. Chem. 274:24257-24262. [DOI] [PubMed] [Google Scholar]
- 39.Haber, J. E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561-599. [DOI] [PubMed] [Google Scholar]
- 40.Henchoz, S., Y. Chi, B. Catarin, I. Herskowitz, R. J. Deshaies, and M. Peter. 1997. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 11:3046-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 42.Hicke, L. 1999. Gettin' down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 9:107-112. [DOI] [PubMed] [Google Scholar]
- 43.Hochstrasser, M. 2000. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2:E153-E157. [DOI] [PubMed]
- 44.Hochstrasser, M., M. J. Ellison, V. Chau, and A. Varshavsky. 1991. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc. Natl. Acad. Sci. USA 88:4606-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hochstrasser, M., and D. Kornitzer. 1998. Ubiquitin-dependent degradation of transcription regulators, p. 279-302. In J.-M. Peters, J. R. Harris, and D. Finley (ed.), Ubiquitin and the biology of the cell. Plenum, New York, N.Y.
- 46.Hofmann, R. M., and C. M. Pickart. 2001. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem. 276:27936-27943. [DOI] [PubMed] [Google Scholar]
- 47.Hofmann, R. M., and C. M. Pickart. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96:645-653. [DOI] [PubMed] [Google Scholar]
- 48.Holloway, S. L., M. Glotzer, R. W. King, and A. W. Murray. 1993. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell 73:1393-1402. [DOI] [PubMed] [Google Scholar]
- 49.Huang, H., J. Y. Hong, C. L. Burck, and S. W. Liebman. 1999. Host genes that affect the target-site distribution of the yeast retrotransposon Ty1. Genetics 151:1393-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang, H., A. Kahana, D. E. Gottschling, L. Prakash, and S. W. Liebman. 1997. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6693-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huibregtse, J. M., J. C. Yang, and S. L. Beaudenon. 1997. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 94:3656-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ichimura, Y., T. Kirisako, T. Takao, Y. Satomi, Y. Shimonishi, N. Ishihara, N. Mizushima, I. Tanida, E. Kominami, M. Ohsumi, T. Noda, and Y. Ohsumi. 2000. A ubiquitin-like system mediates protein lipidation. Nature 408:488-492. [DOI] [PubMed] [Google Scholar]
- 53.Irniger, S., S. Piatti, C. Michaelis, and K. Nasmyth. 1995. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 81:269-278. [DOI] [PubMed] [Google Scholar]
- 54.Jentsch, S., J. P. McGrath, and A. Varshavsky. 1987. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329:131-134. [DOI] [PubMed] [Google Scholar]
- 55.Johnson, E. S., P. C. Ma, I. M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442-17456. [DOI] [PubMed] [Google Scholar]
- 56.Johnson, R. E., L. Haracska, S. Prakash, and L. Prakash. 2001. Role of DNA polymerase ζ in the bypass of a (6-4) TT photoproduct. Mol. Cell. Biol. 21:3558-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson, R. E., S. T. Henderson, T. D. Petes, S. Prakash, M. Bankmann, and L. Prakash. 1992. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol. 12:3807-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson, R. E., S. Prakash, and L. Prakash. 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol η. Science 283:1001-1004. [DOI] [PubMed] [Google Scholar]
- 59.Johnson, R. E., S. Prakash, and L. Prakash. 1994. Yeast DNA repair protein RAD5 that promotes instability of simple repetitive sequences is a DNA-dependent ATPase. J. Biol. Chem. 269:28259-28262. [PubMed] [Google Scholar]
- 60.Kaiser, P., K. Flick, C. Wittenberg, and S. I. Reed. 2000. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 102:303-314. [DOI] [PubMed] [Google Scholar]
- 61.Kaplan, K. B., A. A. Hyman, and P. K. Sorger. 1997. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell 91:491-500. [DOI] [PubMed] [Google Scholar]
- 62.Kaplun, L., Y. Ivantsiv, D. Kornitzer, and D. Raveh. 2000. Functions of the DNA damage response pathway target Ho endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc. Natl. Acad. Sci. USA 97:10077-10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kern, R., and F. K. Zimmermann. 1978. The influence of defects in excision and error prone repair on spontaneous and induced mitotic recombination and mutation in Saccharomyces cerevisiae. Mol. Gen. Genet. 161:81-88. [DOI] [PubMed] [Google Scholar]
- 64.Kitagawa, K., D. Skowyra, S. J. Elledge, J. W. Harper, and P. Hieter. 1999. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4:21-33. [DOI] [PubMed] [Google Scholar]
- 65.Koegl, M., T. Hoppe, S. Schlenker, H. D. Ulrich, T. U. Mayer, and S. Jentsch. 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96:635-644. [DOI] [PubMed] [Google Scholar]
- 66.Koepp, D. M., J. W. Harper, and S. J. Elledge. 1999. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97:431-434. [DOI] [PubMed] [Google Scholar]
- 67.Koken, M. H., J. W. Hoogerbrugge, I. Jasper-Dekker, J. de Wit, R. Willemsen, H. P. Roest, J. A. Grootegoed, and J. H. Hoeijmakers. 1996. Expression of the ubiquitin-conjugating DNA repair enzymes HHR6A and -B suggests a role in spermatogenesis and chromatin modification. Dev. Biol. 173:119-132. [DOI] [PubMed] [Google Scholar]
- 68.Koken, M. H., P. Reynolds, I. Jaspers-Dekker, L. Prakash, S. Prakash, D. Bootsma, and J. H. Hoeijmakers. 1991. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc. Natl. Acad. Sci. USA 88:8865-8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kornitzer, D., B. Raboy, R. G. Kulka, and G. R. Fink. 1994. Regulated degradation of the transcription factor Gcn4. EMBO J. 13:6021-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lawrence, C. 1994. The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? Bioessays 16:253-258. [DOI] [PubMed] [Google Scholar]
- 71.Lawrence, C. W., and R. Christensen. 1976. UV mutagenesis in radiation-sensitive strains of yeast. Genetics 82:207-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lawrence, C. W., J. W. Stewart, F. Sherman, and R. Christensen. 1975. Influence of repair on the specificity of ultraviolet-induced reversion of an ochre allele of the structural gene for iso-1-cytochrome c. Basic Life Sci. 5A:397-398. [DOI] [PubMed]
- 73.Lenk, U., and T. Sommer. 2000. Ubiquitin-mediated proteolysis of a short-lived regulatory protein depends on its cellular localization. J. Biol. Chem. 275:39403-39410. [DOI] [PubMed] [Google Scholar]
- 74.Liebman, S. W., and G. Newnam. 1993. A ubiquitin-conjugating enzyme, RAD6, affects the distribution of Ty1 retrotransposon integration positions. Genetics 133:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama, and A. M. Weissman. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96:11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lupas, A., and W. Baumeister. 1998. The 20S proteasome, p. 127-146. In J.-M. Peters, J. R. Harris, and D. Finley (ed.), Ubiquitin and the biology of the cell. Plenum, New York, N.Y.
- 77.Masutani, C., K. Sugasawa, J. Yanagisawa, T. Sonoyama, M. Ui, T. Enomoto, K. Takio, K. Tanaka, P. J. van der Spek, D. Bootsma, et al. 1994. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 13:1831-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McDonald, J. P., A. S. Levine, and R. Woodgate. 1997. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147:1557-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meimoun, A., T. Holtzman, Z. Weissman, H. J. McBride, D. J. Stillman, G. R. Fink, and D. Kornitzer. 2000. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol. Biol. Cell 11:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moazed, D., and A. D. Johnson. 1996. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell 86:667-677. [DOI] [PubMed] [Google Scholar]
- 81.Montelone, B. A., S. Prakash, and L. Prakash. 1981. Recombination and mutagenesis in rad6 mutants of Saccharomyces cerevisiae: evidence for multiple functions of the RAD6 gene. Mol. Gen. Genet. 184:410-415. [DOI] [PubMed] [Google Scholar]
- 82.Nasmyth, K. 1996. At the heart of the budding yeast cell cycle. Trends Genet. 12:405-412. [DOI] [PubMed] [Google Scholar]
- 83.Nasmyth, K., J. M. Peters, and F. Uhlmann. 2000. Splitting the chromosome: cutting the ties that bind sister chromatids. Science 288:1379-1385. [DOI] [PubMed] [Google Scholar]
- 84.Nelson, J. R., C. W. Lawrence, and D. C. Hinkle. 1996. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272:1646-1649. [DOI] [PubMed] [Google Scholar]
- 85.Nougarede, R., F. Della Seta, P. Zarzov, and E. Schwob. 2000. Hierarchy of S-phase-promoting factors: yeast Dbf4-Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol. Cell. Biol. 20:3795-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ortolan, T. G., P. Tongaonkar, D. Lambertson, L. Chen, C. Schauber, and K. Madura. 2000. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat. Cell Biol. 2:601-608. [DOI] [PubMed] [Google Scholar]
- 87.Patton, E. E., A. R. Willems, and M. Tyers. 1998. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 14:236-243. [DOI] [PubMed] [Google Scholar]
- 88.Peters, J.-M. 1999. Subunits and substrates of the anaphase-promoting complex. Exp. Cell Res. 248:339-349. [DOI] [PubMed] [Google Scholar]
- 89.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 90.Pickart, C. M. 1997. Targeting of substrates to the 26S proteasome. FASEB J. 11:1055-1066. [DOI] [PubMed] [Google Scholar]
- 91.Pickart, C. M. 2001. Ubiquitin enters the new millennium. Mol. Cell 8:499-504. [DOI] [PubMed] [Google Scholar]
- 92.Picologlou, S., N. Brown, and S. W. Liebman. 1990. Mutations in RAD6, a yeast gene encoding a ubiquitin-conjugating enzyme, stimulate retrotransposition. Mol. Cell. Biol. 10:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Porankiewicz, J., J. Wang, and A. K. Clarke. 1999. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32:449-458. [DOI] [PubMed] [Google Scholar]
- 94.Prakash, L. 1981. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 184:471-478. [DOI] [PubMed] [Google Scholar]
- 95.Prakash, L. 1974. Lack of chemically induced mutation in repair-deficient mutant of yeast. Genetics 78:1101-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rao, H., F. Uhlmann, K. Nasmyth, and A. Varshavsky. 2001. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410:955-959. [DOI] [PubMed] [Google Scholar]
- 97.Ratner, J. N., B. Balasubramanian, J. Corden, S. L. Warren, and D. B. Bregman. 1998. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J. Biol. Chem. 273:5184-5189. [DOI] [PubMed] [Google Scholar]
- 98.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 99.Roest, H. P., J. van Klaveren, J. de Wit, C. G. van Gurp, M. H. Koken, M. Vermey, J. H. van Roijen, J. W. Hoogerbrugge, J. T. Vreeburg, W. M. Baarends, D. Bootsma, J. A. Grootegoed, and J. H. Hoeijmakers. 1996. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 86:799-810. [DOI] [PubMed] [Google Scholar]
- 100.Rouillon, A., R. Barbey, E. E. Patton, M. Tyers, and D. Thomas. 2000. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30) complex. EMBO J. 19:282-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roush, A. A., M. Suarez, E. C. Friedberg, M. Radman, and W. Siede. 1998. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol. Gen. Genet. 257:686-692. [DOI] [PubMed] [Google Scholar]
- 102.Russell, S. J., S. H. Reed, W. Huang, E. C. Friedberg, and S. A. Johnston. 1999. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol. Cell 3:687-695. [DOI] [PubMed] [Google Scholar]
- 103.Russell, S. J., K. A. Steger, and S. A. Johnston. 1999. Subcellular localization, stoichiometry, and protein levels of 26S proteasome subunits in yeast. J. Biol. Chem. 274:21943-21952. [DOI] [PubMed] [Google Scholar]
- 104.Salghetti, S. E., A. A. Caudy, J. G. Chenoweth, and W. P. Tansey. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science 293:1651-1653. [DOI] [PubMed] [Google Scholar]
- 105.Sanchez, M., A. Calzada, and A. Bueno. 1999. The Cdc6 protein is ubiquitinated in vivo for proteolysis in Saccharomyces cerevisiae. J. Biol. Chem. 274:9092-9097. [DOI] [PubMed] [Google Scholar]
- 106.Sandmeyer, S. B., L. J. Hansen, and D. L. Chalker. 1990. Integration specificity of retrotransposons and retroviruses. Annu. Rev. Genet. 24:491-518. [DOI] [PubMed] [Google Scholar]
- 107.Schauber, C., L. Chen, P. Tongaonkar, I. Vega, D. Lambertson, W. Potts, and K. Madura. 1998. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391:715-718. [DOI] [PubMed] [Google Scholar]
- 108.Schimke, R. T. 1973. Control of enzyme levels in mammalian tissues. Adv. Enzymol. 37:135-187. [DOI] [PubMed] [Google Scholar]
- 109.Schweder, T., K. H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (sigma s) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 111.Spence, J., R. R. Gali, G. Dittmar, F. Sherman, M. Karin, and D. Finley. 2000. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102:67-76. [DOI] [PubMed] [Google Scholar]
- 112.Spence, J., S. Sadis, A. L. Haas, and D. Finley. 1995. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Spencer, V. A., and J. R. Davie. 1999. Role of covalent modifications of histones in regulating gene expression. Gene 240:1-12. [DOI] [PubMed] [Google Scholar]
- 114.Springael, J. Y., J. M. Galan, R. Haguenauer-Tsapis, and B. Andre. 1999. NH4+-induced down-regulation of the Saccharomyces cerevisiae Gap1p permease involves its ubiquitination with lysine-63-linked chains. J. Cell Sci. 112:1375-1383. [DOI] [PubMed] [Google Scholar]
- 115.Stillman, B. 1996. Cell cycle control of DNA replication. Science 274:1659-1664. [DOI] [PubMed] [Google Scholar]
- 116.Straus, D. B., W. A. Walter, and C. A. Gross. 1987. The heat shock response of E. coli is regulated by changes in the concentration of sigma 32. Nature 329:348-351. [DOI] [PubMed] [Google Scholar]
- 117.Sung, P., E. Berleth, C. Pickart, S. Prakash, and L. Prakash. 1991. Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J. 10:2187-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sung, P., S. Prakash, and L. Prakash. 1990. Mutation of cysteine-88 in the Saccharomyces cerevisiae RAD6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc. Natl. Acad. Sci. USA 87:2695-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sung, P., S. Prakash, and L. Prakash. 1988. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 2:1476-1485. [DOI] [PubMed] [Google Scholar]
- 120.Tao, W., and A. J. Levine. 1999. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc. Natl. Acad. Sci. USA 96:3077-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tomoyasu, T., J. Gamer, B. Bukau, M. Kanemori, H. Mori, A. J. Rutman, A. B. Oppenheim, T. Yura, K. Yamanaka, H. Niki, et al. 1995. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor sigma 32. EMBO J. 14:2551-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Toth, A., R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, and K. Nasmyth. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ulrich, H. D. 2001. The srs2 suppressor of UV sensitivity acts specifically on the RAD5- and MMS2-dependent branch of the RAD6 pathway. Nucleic Acids Res. 29:3487-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ulrich, H. D., and S. Jentsch. 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19:3388-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Varshavsky, A. 1996. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 93:12142-12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015-1068. [DOI] [PubMed] [Google Scholar]
- 127.Wang, Z. 2001. Translesion synthesis by the UmuC family of DNA polymerases. Mutat. Res. 486:59-70. [DOI] [PubMed] [Google Scholar]
- 128.Washington, M. T., R. E. Johnson, S. Prakash, and L. Prakash. 1999. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J. Biol. Chem. 274:36835-36838. [DOI] [PubMed] [Google Scholar]
- 129.Watkins, J. F., P. Sung, L. Prakash, and S. Prakash. 1993. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol. Cell. Biol. 13:7757-7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Weinreich, M., and B. Stillman. 1999. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 18:5334-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wilkinson, C. R., M. Seeger, R. Hartmann-Petersen, M. Stone, M. Wallace, C. Semple, and C. Gordon. 2001. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 3:939-943. [DOI] [PubMed] [Google Scholar]
- 132.Wollenberg, K., and J. C. Swaffield. 2001. Evolution of proteasomal ATPases. Mol. Biol. Evol. 18:962-974. [DOI] [PubMed] [Google Scholar]
- 133.Yoon, H. J., and J. Carbon. 1995. Genetic and biochemical interactions between an essential kinetochore protein, Cbf2p/Ndc10p, and the CDC34 ubiquitin-conjugating enzyme. Mol. Cell. Biol. 15:4835-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zou, H., T. J. McGarry, T. Bernal, and M. W. Kirschner. 1999. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285:418-422. [DOI] [PubMed] [Google Scholar]
- 135.Zwickl, P., W. Baumeister, and A. Steven. 2000. Dis-assembly lines: the proteasome and related ATPase-assisted proteases. Curr. Opin. Struct. Biol. 10:242-250. [DOI] [PubMed] [Google Scholar]