Abstract
The autoimmune thyroid diseases (AITDs), comprising Graves disease (GD) and Hashimoto thyroiditis (HT), develop as a result of a complex interaction between predisposing genes and environmental triggers. Previously, we identified six loci that showed evidence for linkage with AITD in a data set of 56 multiplex families. The goals of the present study were to replicate/reject the previously identified loci before fine mapping and sequencing the candidate genes in these regions. We performed a whole-genome linkage study in an expanded data set of 102 multiplex families with AITD (540 individuals), through use of 400 microsatellite markers. Seven loci showed evidence for linkage to AITD. Three loci, on chromosomes 6p, 8q, and 10q, showed evidence for linkage with both GD and HT (maximum multipoint heterogeneity LOD scores [HLOD] 2.0, 3.5, and 4.1, respectively). Three loci showed evidence for linkage with GD: on 7q (HLOD 2.3), 14q (HLOD 2.1), and 20q (LOD 3.3, in a subset of the families). One locus on 12q showed evidence of linkage with HT, giving an HLOD of 3.4. Comparison with the results obtained in the original data set showed that the 20q (GD-2) and 12q (HT-2) loci continued to show evidence for linkage in the expanded data set; the 6p and 14q loci were located within the same region as the previously identified 6p and 14q loci (AITD-1 and GD-1, respectively), but the Xq (GD-3) and 13q (HT-1) loci were not replicated in the expanded data set. These results demonstrated that multiple genes may predispose to GD and HT and that some may be common to both diseases and some are unique. The loci that continue to show evidence for linkage in the expanded data set represent serious candidate regions for gene identification.
Introduction
Two distinct but related autoimmune disorders affect the thyroid gland: Graves disease (GD [MIM #275000]) and Hashimoto thyroiditis (HT [MIM #140300]). In both diseases, thyroid-reactive T cells are formed and infiltrate the thyroid gland. In GD, the majority of the T cells undergo a Th2 differentiation and activate B cells to produce TSH receptor (TSHR) antibodies, which stimulate the thyroid and cause clinical hyperthyroidism (reviewed by Davies 2000). In contrast, HT is characterized by Th1 switching of the thyroid-infiltrating T cells, which induces apoptosis of thyroid follicular cells and clinical hypothyroidism (reviewed by Weetman 1996). Both disorders are common, with a prevalence in the United States of ∼1% (Jacobson et al. 1997; Hollowell et al. 2002).
The etiology of the autoimmune thyroid diseases (AITDs; i.e., GD and HT), and the mechanisms leading to the dichotomy of GD and HT are unknown. However, the AITDs are considered complex diseases, caused by an interaction between susceptibility genes (Tomer et al. 2002a) and nongenetic factors, such as infection (Tomer and Davies 1993). This paradigm is based on epidemiologic evidence demonstrating a genetic predisposition to AITD, including (1) clustering in families (Mather et al. 1980), giving a sibling risk ratio (λs) of >10 (Vyse and Todd 1996; Villanueva et al. 2000); (2) a high concordance rate in MZ twins when compared with DZ twins (Brix et al. 2000, 2001); (3) the presence of thyroid autoantibodies, which are markers of subclinical AITD, in as many as 50% of siblings of patients with AITD (Hall et al. 1972; Burek et al. 1982); and (4) consistent associations with HLA (Stenszky et al. 1985; Tomer et al. 1997).
In the past few years, several groups, including our own, have been trying to identify the AITD susceptibility genes by studies of candidate genes and by whole-genome screening. The candidate-gene studies have identified HLA-DR3 (Farid et al. 1980; Zamani et al. 2000) and CTLA-4 (Yanagawa et al. 1995; Tomer et al. 2001) as putative AITD susceptibility genes, giving relative risks of 2–4. Only two whole-genome screens have been published for AITD, one in 56 multiplex white families (354 individuals) (Tomer et al. 1999) and another in 123 Japanese sib-pair families (236 individuals) (Sakai et al. 2001). In addition, several limited studies of chromosomal regions have also been reported. These studies identified several putative AITD loci (table 1). However, the previous studies have included modest data sets and need to be replicated and confirmed to identify regions likely to harbor AITD susceptibility genes. We have now almost doubled our white data set to include 102 multiplex families and have performed a genome scan on this expanded data set.
Table 1.
Summary of Loci Reported to Show Evidence for Linkage with AITD
| Chromosomeand Locus | Phenotype | Type of Study | Type of Analysisa | Results | Reference |
| 2: | |||||
| 2q33 (CTLA-4) | Thyroid antibodies | Whole-genome screening | Ped-LOD | HLOD 4.2 | Tomer et al. 2001 |
| 2q33 (CTLA-4) | GD | Candidate-locus analysis | ASP-NPL | NPL 3.4 | Vaidya et al. 1999 |
| 5: | |||||
| 5q31 | AITD | Whole-genome screening | ASP-LOD | LOD 3.1 | Sakai et al. 2001 |
| 6: | |||||
| 6p21 (HLA) | GD | Candidate-locus analysis | ASP-NPL | NPL 1.95 | Vaidya et al. 1999 |
| 6p11 (AITD-1) | AITD | Whole-genome screening | Ped-LOD | LOD 2.9 | Tomer et al. 1999 |
| 6p22-6q14 | HT | Whole-genome screening | One large pedigree | LOD 1.52, NPL 7.53 | Alkhateeb et al. 2002 |
| 8: | |||||
| 8q24 (thyroglobulin) | AITD | Whole-genome screening | ASP-LOD | LOD 2.3 | Sakai et al. 2001 |
| 8q24 (thyroglobulin) | AITD | Candidate-locus analysis | Ped-LOD | HLOD 3.5 | Tomer et al. 2002c |
| 12: | |||||
| 12q22 (HT-2) | HT | Whole-genome screening | Ped-LOD | HLOD 2.3 | Tomer et al. 1999 |
| 13: | |||||
| 13q32 (HT-1) | HT | Whole-genome screening | Ped-LOD | LOD 2.1 | Tomer et al. 1999 |
| 14: | |||||
| 14q31 | GD | Whole-genome screening | Ped-LOD | LOD 2.1 | Tomer et al. 1999 |
| 18: | |||||
| 18q21 (IDDM-6) | GD | Candidate-locus analysis | ASP-NPL | NPL 3.1 | Vaidya et al. 2000 |
| 20: | |||||
| 20q11 (GD-2) | GD | Whole-genome screening | Ped-LOD | LOD 3.5 | Tomer et al. 1999 |
| 20q11 (GD-2) | GD | Candidate-locus analysis | ASP-NPL | NPL 2.01 | Pearce et al. 1999 |
| X: | |||||
| Xq21 (GD-3) | GD | Whole-genome screening | Ped-LOD | LOD 2.5 | Tomer et al. 1999 |
| Xp11 (IDDMX) | GD | Candidate-locus analysis | ASP-NPL | NPL 2.01 | Imrie et al. 2001 |
Ped = multiplex multigenerational pedigrees; ASP = affected sib pairs.
Subjects and Methods
Ascertainment of the Study Families
The project was approved by the Mount Sinai School of Medicine institutional review board. One hundred and two families (540 individuals) were analyzed in the current study (54 from North America, 29 from Italy, 10 from Israel, and 9 from the United Kingdom). All families enrolled in the study were multiplex for AITD (i.e., they had more than one affected individual) and/or multigenerational. Families were ascertained through a patient with AITD who confirmed having at least one other first-degree relative with AITD. Although as many relatives as possible were recruited from each family, the minimum requirement for participation in the study was a family consisting of four first-degree relatives (including the proband) from two generations. On the average, our families had 5.3 members.
Clinical assessment
The AITDs include GD and HT. GD was diagnosed by (1) documented clinical and biochemical hyperthyroidism requiring treatment, (2) a diffuse goiter, (3) presence of TSHR antibodies, and/or (4) diffusely increased I-131 uptake in the thyroid gland. HT was diagnosed by (1) documented clinical and biochemical hypothyroidism requiring thyroid hormone replacement and (2) presence of autoantibodies to thyroid peroxidase (TPO). Antithyroglobulin and anti-TPO antibodies were measured by specific radioimmunoassay (Kronus). All family members without AITD, whether thyroid autoantibody–positive or thyroid autoantibody–negative, were defined for this study as “unaffected.” For all subjects, phenotype was determined with the clinician blinded to the individual’s genotype. Each participant was interviewed and examined and gave written informed consent before participating. All the pertinent clinical and laboratory data were recorded and stored in our database. At the time of interview, blood was collected for DNA purification, as well as for thyroid function tests and thyroid antibody testing.
PCR Amplification of Microsatellite Markers
DNA was extracted from whole blood through use of the Puregene kit (Gentra Systems). For the whole-genome screening, we used the Perkin Elmer microsatellite panels (version 2.0, a total of 400 markers). Fluorescently labeled primers were purchased from Applied Biosystems, and microsatellite markers were typed as reported elsewhere (Tomer et al. 1999). In brief, PCRs were performed in 15-μl reaction volumes containing 50 ng of genomic DNA; 5 pmol of each primer (one of which was fluorescently labeled); PCR buffer containing 50 mM KCl, 10 mM Tris HCl (pH 8.3), 1.5 mM MgCl2, and 200 μM each of dATP, dGTP, dTTP, and dCTP; and 1 U of AmpliTaq DNA polymerase (Perkin Elmer, Applied Biosystems). Reaction mixtures were heated to 94°C for 7 min and then cycled 30 times as follows: 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C. The PCR products were diluted 1:20 in ddH2O and pooled. Two microliters of the pooled products were mixed with 0.5 μl of the internal size standard and 10 μl of deionized formamide, denatured, and separated using an ABI 310 genetic analyzer (Applied Biosystems). Allele calling was performed using Genotyper 2.0 software. The marker data were then automatically exported to our database (Ingres Database), where they were integrated with the already existing phenotype information and prepared for linkage analysis.
Linkage Analysis
Linkage analysis was performed using maximum likelihood-based (LOD score) methods of linkage analysis.
Two-point linkage analysis
Two-point LOD scores for the different markers studied were computed using LIPED software (Ott 1976), assuming both dominant and recessive models. Population data and our own previous analyses suggest that a penetrance of 30% is appropriate (Tomer et al. 1999; Brix et al. 2000), but the actual value of the assumed penetrance does not have a major effect on the final LOD score (Greenberg 1989). We considered a LOD score ⩾1.9 in our whole-genome screen as suggestive evidence for linkage (Lander and Kruglyak 1995) and a LOD score ⩾3.3 as evidence for significant linkage, after maximizing the LOD score with respect to dominant and recessive models (Hodge et al. 1997). If a LOD score was suggestive of linkage, we then tested at a higher penetrance (80%), since it was possible that the penetrance of AITD in our families was higher because of our method of ascertainment (multiple affected family members) (table 2). For all loci except two (10q and 12q), this did not result in increased information. Retesting at 80% penetrance resulted in a significant increase in the LOD score only for the 10q and 12q loci (see the “Results” section). All linkage analyses were performed under the assumption of a population prevalence of 1% for both diseases (0.5% for GD and 0.5% for HT), which is based on the disease prevalence data in the literature (Tunbridge et al. 1977; Vanderpump et al. 1995; Hollowell et al. 2002). On the basis of the assumed disease prevalence, the gene frequency was adjusted according to the model used (dominant or recessive) and the penetrance used, under the assumption of Hardy-Weinberg equilibrium (table 2). The phenocopy rate was set at 0; however, previous studies have shown that the actual value of the assumed phenocopy rate has little effect on the maximum LOD score (Durner et al. 1996).
Table 2.
Models Used in Our Analyses
| Mode ofInheritance | AITDDiseaseFrequency | GeneFrequency | Penetrancea | PhenocopyRate |
| Dominant | .01 | .017 and .006 | .3 and .8 | 0 |
| Recessive | .01 | .333 and .125 | .3 and .8 | 0 |
We computed the LOD score at a .8 penetrance only if a marker gave a LOD score suggestive of linkage.
Comparison of the LOD scores obtained in the expanded data set with those obtained in the original data set
We previously performed a whole-genome scan in 56 families (“the first data set”) (Tomer et al. 1999), and the current linkage analysis was performed on an expanded data set that included an additional 46 families (“the second data set”). To examine what statistical information has been gained from the addition of the second data set, we computed the statistic Ztotal−Zorig for all of the markers used in the genome screen. This statistic was computed by calculating the difference between the maximized maximum LOD score (Greenberg et al. 1998) obtained for the expanded data set and the same statistic for the first data set. It should be noted that the Ztotal−Zorig statistic is not identical to the LOD score obtained for the second data set alone, since the maximum LOD scores for the expanded data set and the original data set may not occur under the same inheritance parameters or at exactly the same location.
Heterogeneity testing and multipoint linkage analysis
It was possible that genetic heterogeneity existed in our data set and/or that the simple models used in the analyses (dominant and recessive) were an incomplete description of the complex inheritance of AITD. Thus, we did a multipoint linkage analysis, under the assumption of heterogeneity, for all 23 chromosomes. The marker positions were according to the Genome Database and Généthon chromosomal genetic maps. The order of the markers and recombination fractions in the Genome Database and Généthon maps were verified in our data set. Multipoint LOD scores and multipoint heterogeneity LOD scores (HLODs) were computed by the Genehunter program (Kruglyak et al. 1996), using all the markers on each chromosome. Multipoint linkage analysis yields the maximum marker information for the area of interest. Using Genehunter, we set the inheritance parameters to the values that gave the maximum LOD scores in the two-point analyses. As we did in the two-point linkage analysis, we assumed a population prevalence of 1% for AITD and adjusted the gene frequency accordingly.
Predivided sample test
To try to resolve the heterogeneity in our data set, we applied the predivided sample test (Morton 1956; Ott 1999). In this test, the data set is divided into different groups on the basis of clinical or demographic criteria, and the LOD scores obtained in each subset are compared. If the difference in the LOD scores for the two subsets of families is statistically significant, then it suggests that the two groups are genetically different from one another at the tested locus (Morton 1956; Ott 1999). Since our data set consisted of a large Italian subgroup that could have been different from the rest of our families owing to genetic and/or subtle ascertainment differences, we subdivided our data set into Italian and non-Italian families and applied the predivided sample test.
Affectedness and disease models
The AITDs include GD and HT. It was not clear whether the susceptibility genes for these two disorders were unique or common to both diseases. Indeed, both disorders can occur in the same family. In our data set, 37% of the families included first-degree relatives with GD and HT. Therefore, we analyzed the data using three models:
-
1.
Affectedness status AITD: all patients with AITD were considered as affected (loci identified using this model would confer susceptibility for both GD and HT).
-
2.
Affectedness status GD: only patients with GD were considered as affected (under this model, patients with HT were considered as unaffected even if they had relatives with GD).
-
3.
Affectedness status HT: only patients with HT were considered as affected (under this model, patients with GD were considered as unaffected even if they had relatives with HT).
Analysis for linkage with GD in which HT was considered unknown and analysis for linkage with HT in which GD was considered unknown were also performed, but, in all cases, the changes in the LOD scores were insignificant. Family members with thyroid autoantibodies alone were classified as “unaffected.” In addition, we computed the LOD scores, under the assumption of heterogeneity (HLOD), using Genehunter (Kruglyak et al. 1996).
Results
Characteristics of the Study Sample
Table 3 shows the clinical characteristics of the 102 families studied. Thirty-four (33%) had only GD-affected members, 30 (30%) had only HT-affected members, and 38 (37%) had both GD- and HT-affected first-degree relatives. Of the 250 affected individuals, 208 (83.2%) were female, and the affected female:male (F:M) ratio, 5:1, was in accordance with that reported in the literature (Volpe 1985). Thirty-five percent of the clinically and biochemically unaffected family members had thyroid antibodies, similar to the incidence reported in previous studies (Burek et al. 1982; Aho et al. 1983).
Table 3.
Characteristics of the Study Data Set
| Group | No. | % |
| Families: | 102 | 100 |
| Families with GD | 34 | 33 |
| Families with GD with GOa proband | 11 | 11 |
| Families with HT | 30 | 30 |
| Mixed families (GD+HT) | 38 | 37 |
| Individuals: | 540 | 100 |
| Affected individuals: | 250 | 46b |
| Female | 208 | 83c |
| Male | 42 | 17c |
| Unaffected individuals: | 290 | 54b |
| Female | 138 | 48d |
| Male | 152 | 52d |
GO = Graves ophthalmopathy.
Percentage of total individuals.
Percentage of affected individuals.
Percentage of unaffected individuals.
Whole-Genome Screening for the AITD Genes
Two-point LOD scores
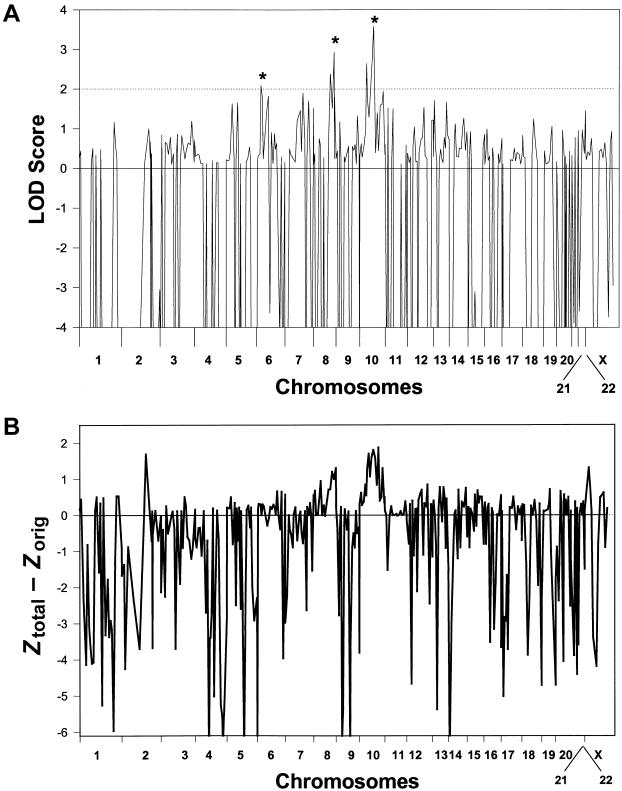
For mapping the AITD genes (i.e., genes causing both GD and HT), individuals with either GD or HT were considered affected. Figure 1A shows the results of whole-genome screening for AITD loci. Three loci, on chromosomes 6p, 8q, and 10q, gave LOD scores >2.0. At the 6p locus, the maximum two-point LOD score was 2.1 for marker D6S422 (34 cM), obtained for the recessive model, at a penetrance of 30% and a recombination fraction (θ) of 0.2 (fig. 1A; table 4). At the 8q locus, the maximum two-point LOD score was 2.9 for marker D8S284 (136.4 cM), obtained for the recessive model, at a penetrance of 30% and a θ of 0.1 (fig. 1A; table 4). The maximum two-point LOD score at the 10q locus was 2.7 for marker D10S537 (93.3 cM), under the assumption of a recessive model, at a penetrance of 30% and a θ of 0.2. At a penetrance of 80%, the LOD score for marker D10S537 increased to 3.6 (fig. 1A; table 4). To determine the increase in information gained from the addition of the second data set, we computed the Ztotal−Zorig for the 400 markers used for whole-genome screening. Figure 1B shows the results of the differences in the two-point LOD scores between the extended and the original data sets (Ztotal−Zorig). An increase in LOD scores was seen at the 8q and 10q loci. The multipoint HLOD scores also increased from 1.8 to 3.5 and 4.1 at 8q and 10q, respectively (table 5). One marker on 2q also showed an increase in the LOD score, but the Ztotal at this locus remained <2.
Figure 1.
A, Whole-genome analysis for loci linked with both GD and HT (AITD). The X-axis shows the relative marker positions on each chromosome, and the Y-axis shows the two-point LOD score obtained for each marker on every chromosome. Three loci, on chromosomes 6p, 8q, and 10q (marked by asterisks [*]), gave two-point LOD scores >2. B, The differences between the two-point LOD scores obtained in the expanded data set and those obtained in the original data set. The X-axis shows the relative marker positions on each chromosome, and the Y-axis shows Ztotal−Zorig for each marker. Two loci, on chromosomes 8q and 10q, showed a notable increase in LOD scores. One marker on 2q also showed an increase in LOD score, but the Ztotal at this locus remained <2.
Table 4.
Two-Point and Multipoint LOD Scores at Loci Showing Evidence for Linkage with AITD (LOD >2)
| Phenotypeand MarkerName | ChromosomalLocation | Two-PointLOD | θ | MultipointHLOD | α |
| AITD: | |||||
| D6S422 | 6p | 2.1 | .2 | 2.0 | .59 |
| D8S284 | 8q | 2.9 | .1 | 3.5 | .6 |
| D10S537 | 10q | 3.6 | .2 | 4.1 | .5 |
| GD: | |||||
| D7S502 | 7q | 2.2 | .1 | 2.3 | .7 |
| D14S258 | 14q | 2.1 | .2 | 2.1 | .32 |
| D20S195 | 20q | 1.2a | .1 | 1.5a | .38 |
| HT: | |||||
| D12S351 | 12q | 2.6 | .2 | 3.4 | .5 |
When we excluded the Italian subset, the two-point LOD score was 2.6 and the multipoint LOD score was 3.3, with no heterogeneity.
Table 5.
Comparison of the Multipoint HLOD Scores Obtained in the First Data Set and in the Expanded Data Set
|
HLOD Score in |
||||
| Locus | Marker | Marker Positiona(cM) | First DataSet (n=56) | Expanded DataSet (n=102) |
| GD-2 | D20S195 | 47.8 | 3.5 | 3.3b |
| GD-3 | DXS8020 | 101.2 | 2.5 | −.5 |
| HT-1 | D13S173 | 103.6 | 2.1 | −.9 |
| HT-2 | D12S346 | 102.0 | 2.3 | 3.4 |
| 7q | D7S502 | 81.2 | 1.4 | 2.3 |
| 8q | D8S284 | 136.4 | 1.8 | 3.5 |
| 10q | D10S537 | 93.3 | 1.8 | 4.1 |
The distance of the marker from the p-terminal end of the chromosome.
This multipoint LOD score was obtained for the white non-Italian families (n=73) without assuming heterogeneity.
Heterogeneity testing and multipoint analysis
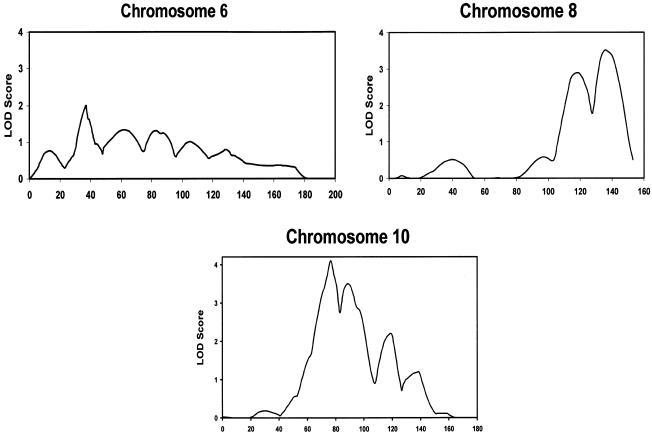
Multipoint analyses using Genehunter showed significant HLOD scores at the same three loci on chromosomes 6p, 8q, and 10q that were identified by the two-point analysis (fig. 2). The maximum multipoint LOD scores at the 6p, 8q, and 10q loci under the assumption of homogeneity were 0.2, 2.2, and −3.1, respectively. However, under the assumption of heterogeneity, the maximum multipoint HLOD scores were 2.0 for the 6p locus, 3.5 for the 8q locus, and 4.1 for the 10q locus (fig. 2; table 4). Thus, our data demonstrated strong evidence for heterogeneity at the 10q locus, whereas it is unclear whether there is genetic heterogeneity at the 6p and 8q loci. However, heterogeneity cannot be quantified by these methods, since the α statistic has been shown to be an inaccurate measure of the percentage of families with linkage (Vieland et al. 2001; Pal and Greenberg 2002).
Figure 2.
Multipoint analysis for chromosomes 6, 8, and 10. The X-axis shows the relative marker position in centimorgans, and the Y-axis shows the multipoint HLOD score. The maximum multipoint HLOD scores were 2.0 for the 6p locus, 3.5 for the 8q locus, and 4.1 for the 10q locus.
Subset analysis by phenotype
We examined subsets of our families (defined by clinical phenotype—i.e., GD or HT) to see whether they contributed most of the positive LOD scores at the three identified AITD loci. We therefore tested these loci separately in our GD, HT, and mixed families (table 6). Subset analysis for the 10q locus showed that all three subsets contributed equally to the total LOD score. However, for the 6p and 8q loci, only the families with HT or GD gave positive LOD scores, whereas the mixed families gave negative LOD scores (table 6). This may imply that the mixed families form a subset of our families that is influenced by different loci than the families with GD or HT. Alternatively, the mixed families may show intrafamilial heterogeneity at the 6p and the 8q loci.
Table 6.
Two Point LOD Scores Obtained for the Families with GD Only, HT Only, and Mixed Families
|
LOD Score in |
||||
| Locus | Familieswith GD(n=34) | Familieswith HT(n=30) | MixedFamilies(n=38) | All Families(N=102) |
| 6p | 1.0 | 1.9 | −.8 | 2.1 |
| 8q | 1.5 | 1.8 | −.4 | 2.9 |
| 10q | 1.0 | 1.5 | 1.1 | 3.6 |
Whole-Genome Screening for GD Genes
For screening the unique GD genes (i.e., genes causing GD but not HT), we defined as “affected” only individuals with GD (therefore, the families with HT were excluded from analysis by definition, only the families with GD and mixed families were screened, and only the GD-affected family members were treated as affected). Two loci gave two-point LOD scores >2.0, on chromosomes 7q and 14q. The maximum two-point LOD score at the 7q locus was 2.1 for marker D7S502 (81.2 cM), obtained for the recessive model, at 0.3 penetrance and a θ of 0.1 (table 4). The maximum two-point LOD score at the 14q locus was 2.1 for marker D14S258 (71.6 cM), obtained for the dominant model, at 0.3 penetrance and a θ of 0.2 (table 4). The previously identified GD-2 (20q) locus (Tomer et al. 1998) showed a maximum two-point LOD score of 1.2 for marker D20S195 (47.8 cM), obtained for the recessive model, at 0.3 penetrance and a θ of 0.1 (table 4). Since our data set consisted of a large subset of Italian families, we reanalyzed this locus with the Italian families excluded (n=29). The GD-2 (20q) locus gave a LOD score of 2.6 without an assumption of heterogeneity when we divided the families into subsets on the basis of the country of origin and excluded Italian families from the analysis (Tomer et al. 2002b). The multipoint LOD score at GD-2 (20q) was 3.3 when we excluded the Italian families (table 4). Application of the predivided-sample test (Morton 1956; Ott 1996) showed that there was statistically significant evidence for linkage heterogeneity at 20q in the Italian families versus the non-Italian white families, with the Italian families showing evidence against linkage at this locus (χ2=9.2; 1 df; P<.01) (Tomer et al. 2002b).
Whole Genome Screening for HT Genes
For screening the unique HT genes (i.e., genes causing HT but not GD), we defined as “affected” only individuals with HT (therefore, the families with GD were excluded from analysis by definition, only the families with HT and mixed families were used, and only the HT-affected family members were classified as affected). One locus showed evidence for linkage only with HT on 12q. The maximum two-point LOD score was 1.9 at marker D12S346 (102 cM), under the assumption of a recessive model, at a penetrance of 0.3 and a θ of 0.2. At a penetrance of 80%, the LOD score increased to 2.6. The maximum multipoint HLOD (recessive model, 80% penetrance) was 3.4 (table 4).
Comparison of the Whole-Genome Screen Results Obtained in the First Data Set with Those Obtained in the Expanded Data Set
Our initial whole-genome screen was performed in 56 families, and the expanded data set included an additional 46 families (a total of 102 families). Comparison of the results obtained for the expanded data set with the results obtained from the original data set showed that, at four of six sites, there is continued evidence for linkage (tables 5 and 7). Two loci (HT-2 and GD-2) continued to show strong evidence of linkage in the expanded data set, with LOD scores >3 (table 5). For two additional loci (AITD-1 and GD-1), we obtained peak LOD scores of >2 ∼20–30 cM away from the original locus (table 7), and, at present, it is unclear whether they represent the same loci or different loci. In contrast, for two additional loci that gave positive LOD scores in the first data set (GD-3 and HT-1), the LOD scores became negative in the expanded data set; thus, these loci could not be confirmed (table 5).
Table 7.
Comparison of the Multipoint HLOD Scores Obtained in the First Data Set and in the Expanded Data Set for the AITD-1 and GD-1 Loci
|
HLOD Score in |
|||
| LocusandMarker | Marker Positiona(cM) | First DataSet (n=56) | Expanded DataSet (n=102) |
| AITD-1: | |||
| D6S257 | 62.8 | 2.9 | 1.5 |
| D6S422 | 34.0 | 2.0 | 2.0 |
| GD-1: | |||
| D14S81 | 91.7 | 2.5 | 1.2 |
| D14S258 | 71.6 | 1.0 | 2.1 |
The distance of the marker from the p-terminal end of the chromosome.
Discussion
The mechanisms underlying the genetic predisposition to AITD are unknown. Although a hereditary component in the pathogenesis of these diseases has long been recognized, the inheritance appears to be complex, involving multiple genes with variable penetrances. Moreover, the genetic relationship between GD and HT is not known, and, even within GD and HT, there exists significant phenotypic heterogeneity. Previous linkage studies by our own group and others have identified several loci as potential genetic regions harboring AITD susceptibility genes (table 1). However, previous studies employed only small numbers of families and/or individuals. We report here the largest whole-genome linkage study performed in AITD to date. Our data demonstrated significant LOD scores (>2) at seven loci: three loci (6p, 8q, and 10q) showed evidence for linkage with both GD and HT, three loci (7q, 14q, and 20q) showed evidence for linkage with GD, and one locus (12q) showed evidence for linkage with HT.
Comparison of the results obtained for this expanded data set with the results obtained for the original data set showed that the addition of the second data set resulted in a notable increase in statistical information (fig. 1B; table 5). Our first genome screen, performed in 56 families, showed evidence for linkage at six sites: 6p (AITD-1) showed evidence for linkage with both GD and HT; 14q (GD-1), 20q (GD-2), and Xq (GD-3) showed evidence for linkage with GD; and 13q (HT-1) and 12q (HT-2) showed evidence for linkage with HT. The results obtained in the expanded data set showed that four of six loci show continued evidence for linkage (tables 5 and 7), whereas two loci, GD-3 and HT-1, gave negative LOD scores in the expanded data set and, thus, could not be confirmed (table 5). The other four loci continued to show evidence for linkage in the expanded data set. Two loci (HT-2 and GD-2) continued to show strong evidence for linkage in the expanded data set (table 5). For HT-2 (12q) the maximum two-point LOD score increased from 2.3 in the original data set to 3.4 in the expanded data set. For GD-2 (20q), the LOD scores remained >2.0 in the expanded data set, but only when we excluded the Italian families from the analysis. Since the original data set had few Italian families and the expanded data set consisted of ∼30% Italian families, this effect was noticeable only in the expanded data set. We have now identified a putative susceptibility gene at the GD-2 locus region, the CD-40 gene (Tomer et al. 2002b). Since the second data set was “enriched” in Italian families, we did similar subset analysis (excluding the Italian families) at the other loci we identified, but in none of them except GD-2 did we resolve the heterogeneity by dividing the families into subsets on the basis of geographic origin (data not shown).
For two additional loci (AITD-1 and GD-1), we obtained peak LOD scores of ⩾2 ∼20–30 cM away from the original locus. Marker D6S257 (AITD-1) gave a maximum multipoint LOD score of 2.9 in the original data set and 1.5 in the expanded data set (table 7). However, in the expanded data set, there was a peak LOD score of 2.0 at marker D6S422 located ∼30 cM telomeric to D6S257 (table 7). Although this is a large distance, it may represent the same locus (AITD-1), and the differences in map positions may be due to inaccuracies of the current genetic maps and/or due to genetic heterogeneity. A similar phenomenon was observed for the GD-1 locus. Marker D14S81 (GD-1) gave a maximum multipoint LOD score of 2.5 in the original data set and 1.2 in the expanded data set (table 7). However, in the expanded data set, marker D14S258, located 30 cM centromeric to D14S81, gave a maximum multipoint LOD score of 2.1 and, again, may represent the same locus. Thus, of the six loci originally identified, two showed increased evidence for linkage (GD-2 and HT-2), two showed significant LOD scores in the same chromosomal regions as the original loci (AITD-1 and GD-1), and two were not replicated (GD-3 and HT-1). In addition, for three loci that gave low positive LOD scores (1.0–2.0) in the original data set (D8S284, D10S537, and D7S502), the LOD scores increased to significance levels (>2.0) in the expanded data set (table 5). As mentioned above, our analyses were performed assuming two modes of inheritance (dominant and recessive) and two penetrances (30% and 80%). Greenberg et al. (1998) have shown that maximizing LOD scores over modes of inheritance, as we did, increases the power of the analysis significantly. However, because of the multiple testing, the recommendation is to subtract 0.3 from the obtained LOD score (Hodge et al. 1997). In our case, that would have resulted in a LOD score of 1.7 for the 6p locus, 3.2 for the 8q locus, 3.8 for the 10q locus, 2.0 for the 7q locus, 1.8 for the 14q locus, 3.0 for the 20q locus, and 3.1 for the 12q locus (see table 4). Thus, at all loci except 6p and 14q, the LOD scores remained above the cutoffs suggested by Lander and Kruglyak (1995). The significance of the 6p and 14q loci will have to be determined by studying additional data sets.
Comparison of our results with those obtained in the genome screen performed by Sakai et al. (2001) demonstrated evidence for linkage, at the 8q locus in their Japanese data set, that was replicated in our data set; however, the 5q locus that gave a LOD score of >3.0 in the Japanese data set did not show evidence for linkage in our data set (two-point LOD 0.54). The AITD-1 (6p) locus is within a broad region that showed evidence for linkage with HT in a genome scan performed in a large family in which vitiligo and HT occurred in numerous individuals (Alkhateeb et al. 2002). Another locus, GD-2 (20q), showed evidence for linkage in a U.K. data set (nonparametric linkage [NPL] score 2.0) (Pearce et al. 1999). Thus, three of the seven loci we identified showed evidence for linkage in independent data sets.
The AITDs are unique; even though their clinical manifestations are different, they share common immunopathogenetic mechanisms. Therefore, two hypotheses have been proposed to explain the etiology of the AITDs. According to one hypothesis, GD and HT share common genetic susceptibility, whereas differing environmental encounters and immunological modulating factors determine the final phenotype (Phillips et al. 1990). The second hypothesis proposes that, even though GD and HT are autoimmune diseases of the same organ (thyroid), they represent separate disease entities caused by distinct genes. Our study shed light on the genetic relationship between GD and HT. Three loci (6p, 8q, and 10q) showed evidence for linkage with both GD and HT, implying that there is significant shared genetic susceptibility to GD and HT. This is also suggested by the fact that more than a third of the families have both diseases (table 3). Moreover, subset analysis showed that these three loci showed positive LOD scores in both the families with GD and the families with HT, again supporting a strong shared genetic susceptibility for GD and HT (table 6). However, the mixed families gave positive LOD scores only for the 10q locus and not for the 6p and 8q loci (table 6). This may imply that, in the mixed families, the development of AITD is influenced by unique loci not contributing to AITD in the families with GD or HT. Alternatively, the mixed families may represent intrafamilial heterogeneity. Therefore, the shared genetic susceptibility to GD and HT may be different in the mixed families than in the families with GD or HT. Preliminary data suggest that the shared genetic susceptibility for GD and HT may involve both immune regulatory genes (e.g., CTLA-4 [Vaidya et al. 1999; Tomer et al. 2001]) and thyroid-specific genes (Tomer et al. 2002c). However, we also found loci that were unique to GD and HT, implying that genetic factors also contribute to the distinct GD and HT phenotypes. Since all loci showed evidence for linkage with heterogeneity, the relative weights of the shared susceptibility loci and the unique loci for GD and HT cannot be assessed until the heterogeneity is resolved.
Some of the loci we have identified harbor interesting positional candidate genes. These include both immune regulatory genes and thyroid-specific genes. The 6p (AITD-1) locus harbors the SOX-4 gene, an immune modulator (Schilham et al. 1997). This locus is also close to a major immune regulatory gene cluster, the human leukocyte antigen (HLA) complex. However, previous studies by our group (Barbesino et al. 1998; Ban et al. 2002) and others (Bode et al. 1973; Hawkins et al. 1985; Shields et al. 1994) have shown strong evidence against linkage of the HLA locus to AITD. These data suggest that the 6p (AITD-1) locus is distinct from the HLA gene cluster. It is interesting that an insulin-dependent diabetes locus (IDDM-15 [MIM #601666]) (Delepine et al. 1997) and a systemic lupus erythematosus (SLE) locus (Gaffney et al. 1998) were identified in the same location as AITD-1, and this may imply that a general autoimmunity susceptibility gene is located in this region. Indeed, AITD is known to be associated with insulin-dependent diabetes (Gray et al. 1983; Kordonouri et al. 2002) and with SLE (Miller et al. 1987; Weetman and Walport 1987) in the same individuals and to cluster together in families. The 8q locus harbors the thyroglobulin gene, and our preliminary data suggest that the thyroglobulin gene may be the AITD susceptibility gene in this region (Tomer et al. 2002c). In addition, the 20q locus (GD-2) harbors the CD40 gene, and we have preliminary data showing that an SNP in the Kozak sequence of CD40 is associated with GD (Tomer et al. 2002b).
Complex diseases are likely to be caused by the interaction of several genes, and their combined effects may differ in different individuals and families (Vyse and Todd 1996). In type I diabetes mellitus, at least 12 susceptibility loci have been identified by several groups (Davies et al. 1994; Hashimoto et al. 1994; Luo et al. 1995, 1996); in AITD, we identified seven loci, and additional loci were identified by others (table 1). It is most likely that these loci interact and that their interactions may influence disease phenotype and severity (Tomer et al. 1999). Indeed, locus interactions were reported in type 2 diabetes (Cox et al. 1999), in murine lupus (Vyse et al. 1997), and in epilepsy (Durner et al. 2001). The molecular basis for the interactions between susceptibility genes in complex diseases is unknown. These interactions could represent the cumulative effect of increased statistical risk, or, alternatively, there may be molecular interactions between the susceptibility genes or their products that ultimately determine disease phenotype. Another unresolved question is that of which loci harbor major genes (i.e., genes that are necessary for developing the disease) and which loci harbor minor modulating genes (i.e., genes that alter the effects of the major genes but are not necessary for disease development). To better understand these effects, we need to identify the susceptibility genes for AITD and the underlying molecular mechanisms by which they allow the induction of thyroid autoimmunity.
Acknowledgments
We thank all the families with AITD, who graciously agreed to participate in the study. Family enrollment was achieved through the collaboration of an International Consortium for the Genetics of Autoimmune Thyroid Disease (ICGA). Members of the ICGA included Drs. Luca Chiovato and Aldo Pinchera (Pisa), Sandra McLachlan (Los Angeles), Bernard Rees Smith (Cardiff, Wales), Fred Clark and Eric Young (Newcastle upon Tyne), Meir Berezin (Tel-Hashomer, Israel), George Carayanniotis (Newfoundland, Canada), and Rhoda Cobin (New York, New York). This work was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases grants DK35764, DK45011, and DK52464 (to T.F.D.), grants DK61659 and DK58072 (to Y.T.), and grants DK31775, NS27941, and MH48858 (to D.A.G.).
Electronic-Database Information
The URLs for data presented herein are as follows:
- Généthon, http://www.genethon.fr/php/index.php
- Genome Database, http://www.gdb.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for GD, HT, and IDDM-15) [DOI] [PubMed]
References
- Aho K, Gordin A, Sievers K, Takala J (1983) Thyroid autoimmunity in siblings: a population study. Acta Endocrinol (Copenh) Suppl 251:11–15 [PubMed] [Google Scholar]
- Alkhateeb A, Stetler GL, Old W, Talbert J, Uhlhorn C, Taylor M, Fox A, Miller C, Dills DG, Ridgway EC, Bennett DC, Fain PR, Spritz RA (2002) Mapping of an autoimmunity susceptibility locus (AIS1) to chromosome 1p31.3-p32.2. Hum Mol Genet 11:661–667 [DOI] [PubMed] [Google Scholar]
- Ban Y, Davies TF, Greenberg DA, Concepcion ES, Tomer Y (2002) The influence of human leucocyte antigen (HLA) genes on autoimmune thyroid disease (AITD): results of studies in HLA-DR3 positive AITD families. Clin Endocrinol (Oxf) 57:81–88 [DOI] [PubMed] [Google Scholar]
- Barbesino G, Tomer Y, Concepcion ES, Davies TF, Greenberg DA (1998) Linkage analysis of candidate genes in autoimmune thyroid disease: I. Selected immunoregulatory genes. J Clin Endocrinol Metab 83:1580–1584 [DOI] [PubMed] [Google Scholar]
- Bode HH, Dorf ME, and Forbes AP (1973) Familial lymphocytic thyroiditis: analysis of linkage with histocompatibility and blood group. J Clin Endocrinol Metab 37:692–697 [DOI] [PubMed] [Google Scholar]
- Brix TH, Kyvik KO, Christensen K, Hegedus L (2001) Evidence for a major role of heredity in Graves' disease: a population-based study of two Danish twin cohorts. J Clin Endocrinol Metab 86:930–934 [DOI] [PubMed] [Google Scholar]
- Brix TH, Kyvik KO, Hegedus L (2000) A population-based study of chronic autoimmune hypothyroidism in Danish twins. J Clin Endocrinol Metab 85:536–539 [DOI] [PubMed] [Google Scholar]
- Burek CL, Hoffman WH, Rose NR (1982) The presence of thyroid autoantibodies in children and adolescents with AITD and in their siblings and parents. Clin Immunol Immunopathol 25:395–404 [DOI] [PubMed] [Google Scholar]
- Cox NJ, Frigge M, Nicolae DL, Concannon P, Hanis CL, Bell GI, Kong A (1999) Loci on chromosomes 2 (NIDDM1) and 15 interact to increase susceptibility to diabetes in Mexican Americans. Nat Genet 21:213–215 [DOI] [PubMed] [Google Scholar]
- Davies JL, Kawauchi Y, Bennet ST, Copeman JB, Cordell HJ, Pritchard LE, Reed PW, et al (1994) A genome-wide search for human type 1 diabetes susceptibility genes. Nature 371:130–136 [DOI] [PubMed] [Google Scholar]
- Davies TF (2000) Graves' diseases: pathogenesis. In: Braverman LE, Utiger RD (eds) Werner and Ingbar's the thyroid: a fundamental and clinical text, 8th ed. Lippincott Williams & Wilkens, Philadelphia, pp 518–530 [Google Scholar]
- Delepine M, Pociot F, Habita C, Hashimoto L, Froguel P, Rotter J, Cambon-Thomsen A, et al (1997) Evidence of a non-MHC susceptibility locus in type I diabetes linked to HLA on chromosome 6. Am J Hum Genet 60:174–187 [PMC free article] [PubMed] [Google Scholar]
- Durner M, Greenberg DA, Hodge SE (1996) Phenocopies versus genetic heterogeneity: can we use phenocopy frequencies in linkage analysis to compensate for heterogeneity? Hum Hered 46:265–273 [DOI] [PubMed] [Google Scholar]
- Durner M, Keddache MA, Tomasini L, Shinnar S, Resor SR, Cohen J, Harden C, et al (2001) Genome scan of idiopathic generalized epilepsy: evidence for major susceptibility gene and modifying genes influencing the seizure type. Ann Neurol 49:328–335 [PubMed] [Google Scholar]
- Farid NR, Stone E, Johnson G (1980) Graves' disease and HLA: clinical and epidemiologic associations. Clin Endocrinol (Oxf) 13:535–544 [DOI] [PubMed] [Google Scholar]
- Gaffney PM, Kearns GM, Shark KB, Ortmann WA, Selby SA, Malmgren ML, Rohlf KE, Ockenden TC, Messner RP, King RA, Rich SS, Behrens TW (1998) A genome-wide search for susceptibility genes in human systemic lupus erythematosus sib-pair families. Proc Natl Acad Sci USA 95:14875–14879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RS, Elton RA, Clarke BF (1983) Familial distribution of thyroid disease and diabetes: further evidence for aetiological heterogeneity of diabetes mellitus. Q J Med 52:244–255 [PubMed] [Google Scholar]
- Greenberg DA (1989) Inferring mode of inheritance by comparison of LOD scores. Am J Med Genet 34:480–486 [DOI] [PubMed] [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet 63:870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R, Dingle PR, Roberts DF (1972) Thyroid antibodies: a study of first degree relatives. Clin Genet 3:319–324 [DOI] [PubMed] [Google Scholar]
- Hashimoto L, Habita C, Beressl JB, Delepine M, Besse C, Cambon-Thomsen A, Deschamps I, et al (1994) Genetic mapping of a susceptibility locus for insulin-dependent diabetes mellitus on chromosome 11q. Nature 371:161–164 [DOI] [PubMed] [Google Scholar]
- Hawkins BR, Ma JT, Lam KS, Wang CC, Yeung RT (1985) Analysis of linkage between HLA haplotype and susceptibility to Graves' disease in multiple-case Chinese families in Hong Kong. Acta Endocrinol (Copenh) 110:66–69 [DOI] [PubMed] [Google Scholar]
- Hodge SE, Abreu PC, Greenberg DA (1997) Magnitude of type I error when single-locus linkage analysis is maximized over models: a simulation study. Am J Hum Genet 60:217–227 [PMC free article] [PubMed] [Google Scholar]
- Hollowell JG, Staehling NW, Flanders WD, Hannon WH, Gunter EW, Spencer CA, Braverman LE (2002) Serum TSH, T(4), and thyroid antibodies in the United States population (1988 to 1994): National Health and Nutrition Examination Survey (NHANES III). J Clin Endocrinol Metab 87:489–499 [DOI] [PubMed] [Google Scholar]
- Imrie H, Vaidya B, Perros P, Kelly WF, Toft AD, Young ET, Kendall-Taylor P, Pearce SHS (2001) Evidence for a Graves' disease susceptibility locus at chromosome Xp11 in a United Kingdom population. J Clin Endocrinol Metab 86:626–630 [DOI] [PubMed] [Google Scholar]
- Jacobson DL, Gange SJ, Rose NR, Graham NM (1997) Epidemiology and estimated population burden of selected autoimmune diseases in the United States. Clin Immunol Immunopathol 84:223–243 [DOI] [PubMed] [Google Scholar]
- Kordonouri O, Klinghammer A, Lang EB, Gruters-Kieslich A, Grabert M, Holl RW (2002) Thyroid autoimmunity in children and adolescents with type 1 diabetes: a multicenter survey. Diabetes Care 25:1346–1350 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Luo DF, Bui MM, Muir A, Maclaren NK, Thomson G, She JX (1995) Affected sibpair mapping of a novel susceptibility gene to insulin-dependent diabetes mellitus (IDDM8) on chromosome 6q25-27. Am J Hum Genet 57:911–919 [PMC free article] [PubMed] [Google Scholar]
- Luo DF, Buzzetti R, Rotter JI, Maclaren NK, Raffel LJ, Nistico L, Giovannini C, Pozzilli P, Thomson G, She JX (1996) Confirmation of three susceptibility genes to insulin-dependent diabetes mellitus: IDDM4, IDDM5 and IDDM8. Hum Mol Genet 5:693–698 [DOI] [PubMed] [Google Scholar]
- Mather BA, Roberts DF, Scanlon MF, Mukhtar ED, Davies TF, Smith BR, Hall R (1980) HLA antigens and thyroid autoantibodies in patients with Graves' disease and their first degree relatives. Clin Endocrinol (Oxf) 12:155–163 [DOI] [PubMed] [Google Scholar]
- Miller FW, Moore GF, Weintraub BD, Steinberg AD (1987) Prevalence of thyroid disease and abnormal thyroid function test results in patients with systemic lupus erythematosus. Arthritis Rheum 30:1124–1131 [DOI] [PubMed] [Google Scholar]
- Morton NE (1956) The detection and estimation of linkage between the genes for elliptocytosis and the Rh blood type. Am J Hum Genet 8:80–96 [PMC free article] [PubMed] [Google Scholar]
- Ott J (1976) A computer program for linkage analysis of general human pedigrees. Am J Hum Genet 28:528–529 [PMC free article] [PubMed] [Google Scholar]
- ——— (1996) Complex traits on the map. Nature 379:772–773 [DOI] [PubMed] [Google Scholar]
- ——— (1999) Analysis of human genetic linkage. 3rd ed. Johns Hopkins University Press, Baltimore [Google Scholar]
- Pal DK, Greenberg DA (2002) Evaluating genetic heterogeneity in complex disorders. Hum Hered 53:216–226 [DOI] [PubMed] [Google Scholar]
- Pearce SH, Vaidya B, Imrie H, Perros P, Kelly WF, Toft AD, McCarthy MI, Young ET, Kendall-Taylor P (1999) Further evidence for a susceptibility locus on chromosome 20q13.11 in families with dominant transmission of Graves disease. Am J Hum Genet 65:1462–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D, McLachlan S, Stephenson A, Roberts D, Moffitt S, McDonald D, Ad'Hiah A, Stratton A, Young E, Clark F (1990) Autosomal dominant transmission of autoantibodies to thyroglobulin and thyroid peroxidase. J Clin Endocrinol Metab 70:742–746 [DOI] [PubMed] [Google Scholar]
- Sakai K, Shirasawa S, Ishikawa N, Ito K, Tamai H, Kuma K, Akamizu T, Tanimura M, Furugaki K, Yamamoto K, Sasazuki T (2001) Identification of susceptibility loci for autoimmune thyroid disease to 5q31-q33 and Hashimoto's thyroiditis to 8q23-q24 by multipoint affected sib-pair linkage analysis in Japanese. Hum Mol Genet 10:1379–1386 [DOI] [PubMed] [Google Scholar]
- Schilham MW, Moerer P, Cumano A, Clevers HC (1997) Sox-4 facilitates thymocyte differentiation. Eur J Immunol 27:1292–1295 [DOI] [PubMed] [Google Scholar]
- Shields DC, Ratanachaiyavong S, McGregor AM, Collins A, Morton NE (1994) Combined segregation and linkage analysis of Graves' disease with a thyroid autoantibody diathesis. Am J Hum Genet 55:540–554 [PMC free article] [PubMed] [Google Scholar]
- Stenszky V, Kozma L, Balazs C, Rochkitz S, Bear JC, Farid NR (1985) The genetics of Graves' disease: HLA and disease susceptibility. J Clin Endocrinol Metab 61:735–740 [DOI] [PubMed] [Google Scholar]
- Tomer Y, Barbesino G, Greenberg DA, Concepcion ES, Davies TF (1998) A new Graves disease-susceptibility locus maps to chromosome 20q11.2. Am J Hum Genet 63:1749–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1999) Mapping the major susceptibility loci for familial Graves' and Hashimoto's diseases: evidence for genetic heterogeneity and gene interactions. J Clin Endocrinol Metab 84:4656–4664 [DOI] [PubMed] [Google Scholar]
- Tomer Y, Barbesino G, Greenberg D, Davies TF (1997) The immunogenetics of autoimmune diabetes and autoimmune thyroid disease. Trends Endocrinol Metab 8:63–70 [DOI] [PubMed] [Google Scholar]
- ——— (2002a) The genetics of the autoimmune thyroid diseases. In: Baxter JD, Melmed S, New MI (eds) Genetics in Endocrinology. Lippincot Williams and Wilkins, Philadelphia, pp 137–152 [Google Scholar]
- Tomer Y, Concepcion E, Greenberg DA (2002b) A C/T single nucleotide polymorphism in the region of the CD40 gene is associated with Graves' disease. Thyroid 12:1129–1135 [DOI] [PubMed] [Google Scholar]
- Tomer Y, Davies TF (1993) Infection, thyroid disease and autoimmunity. Endocr Rev 14:107–120 [DOI] [PubMed] [Google Scholar]
- Tomer Y, Greenberg DA, Barbesino G, Concepcion ES, Davies TF (2001) CTLA-4 and not CD28 is a susceptibility gene for thyroid autoantibody production. J Clin Endocrinol Metab 86:1687–1693 [DOI] [PubMed] [Google Scholar]
- Tomer Y, Greenberg DA, Concepcion E, Ban Y, Davies TF (2002c) Thyroglobulin is a thyroid specific gene for the familial autoimmune thyroid diseases. J Clin Endocrinol Metab 87:404–407 [DOI] [PubMed] [Google Scholar]
- Tunbridge WMG, Evered DC, Hall R, Appleton D, Brewis M, Clark F, Evans JG, Young E, Bird T, Smith PA (1977) The spectrum of thyroid disease in a community: the Whickham survey. Clin Endocrinol (Oxf) 7:481–493 [DOI] [PubMed] [Google Scholar]
- Vaidya B, Imrie H, Perros P, Young ET, Kelly WF, Carr D, Large DM, Toft AD, Kendall-Taylor P, Pearce SH (2000) Evidence for a new Graves disease susceptibility locus at chromosome 18q21. Am J Hum Genet 66:1710–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya B, Imrie H, Perros P, Young ET, Kelly WF, Carr D, Large DM, Toft AD, McCarthy MI, Kendall-Taylor P, Pearce SH (1999) The cytotoxic T lymphocyte antigen-4 is a major Graves' disease locus. Hum Mol Genet 8:1195–1199 [DOI] [PubMed] [Google Scholar]
- Vanderpump MPJ, Tunbridge WMG, French JM, Appleton D, Bates D, Clark F, Grimley Evans J, Hasan DM, Rodgers H, Tunbridge F, Young ET (1995) The incidence of thyroid disorders in the community: a twenty-year follow-up of the Whickham survey. Clin Endocrinol (Oxf) 43:55–68 [DOI] [PubMed] [Google Scholar]
- Vieland VJ, Wang K, Huang J (2001) Power to detect linkage based on multiple sets of data in the presence of locus heterogeneity: comparative evaluation of model-based linkage methods for affected sib pair data. Hum Hered 51:199–208 [DOI] [PubMed] [Google Scholar]
- Villanueva RB, Inzerillo AM, Tomer Y, Barbesino G, Meltzer M, Concepcion ES, Greenberg DA, et al (2000) Limited genetic susceptibility to severe Graves' ophthalmopathy: no role for CTLA-4 and evidence for an environmental etiology. Thyroid 10:791–798 [DOI] [PubMed] [Google Scholar]
- Volpe R (1985) Autoimmune thyroid disease. In: Volpe R (ed) Autoimmunity and endocrine disease. Marcel Dekker, New York, pp 109–285 [Google Scholar]
- Vyse TJ, Rozzo SJ, Drake CG, Izui S, Kotzin BL (1997) Control of multiple autoantibodies linked with a lupus nephritis susceptibility locus in New Zealand black mice. J Immunol 158:5566–5574 [PubMed] [Google Scholar]
- Vyse TJ, Todd JA (1996) Genetic analysis of autoimmune disease. Cell 85:311–318 [DOI] [PubMed] [Google Scholar]
- Weetman AP (1996) Chronic autoimmune thyroiditis. In: Braverman LE, Utiger RD (eds) Werner and Ingbar's the thyroid. Lippincott-Raven, Philadelphia, pp 738–748 [Google Scholar]
- Weetman AP, Walport MJ (1987) The association of autoimmune thyroiditis with systemic lupus erythematosus. Br J Rheumatol 26:359–361 [DOI] [PubMed] [Google Scholar]
- Yanagawa T, Hidaka Y, Guimaraes V, Soliman M, DeGroot LJ (1995) CTLA-4 gene polymorphism associated with Graves' disease in a Caucasian population. J Clin Endocrinol Metab 80:41–45 [DOI] [PubMed] [Google Scholar]
- Zamani M, Spaepen M, Bex M, Bouillon R, Cassiman JJ (2000) Primary role of the HLA class II DRB1*0301 allele in Graves disease. Am J Med Genet 95:432–437 [DOI] [PubMed] [Google Scholar]