Abstract
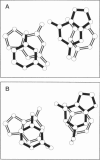
Base-stacking interactions in canonical and crystal B-DNA and in Z-DNA steps are studied using the ab initio quantum-chemical method with inclusion of electron correlation. The stacking energies in canonical B-DNA base-pair steps vary from -9.5 kcal/mol (GG) to -13.2 kcal/mol (GC). The many-body nonadditivity term, although rather small in absolute value, influences the sequence dependence of stacking energy. The base-stacking energies calculated for CGC and a hypothetical TAT sequence in Z-configuration are similar to those in B-DNA. Comparison with older quantum-chemical studies shows that they do not provide even a qualitatively correct description of base stacking. We also evaluate the base-(deoxy)ribose stacking geometry that occurs in Z-DNA and in nucleotides linked by 2',5'-phosphodiester bonds. Although the molecular orbital analysis does not rule out the charge-transfer n-pi* interaction of the sugar 04' with the aromatic base, the base-sugar contact is stabilized by dispersion energy similar to that of stacked bases. The stabilization amounts to almost 4 kcal/mol and is thus comparable to that afforded by normal base-base stacking. This enhancement of the total stacking interaction could contribute to the propensity of short d(CG)n sequences to adopt the Z-conformation.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aida M. An ab initio molecular orbital study on the sequence-dependency of DNA conformation: an evaluation of intra- and inter-strand stacking interaction energy. J Theor Biol. 1988 Feb 7;130(3):327–335. doi: 10.1016/s0022-5193(88)80032-8. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Birdsall D. L., Leslie A. G., Ratliff R. L. Left-handed DNA helices. Nature. 1980 Feb 21;283(5749):743–745. doi: 10.1038/283743a0. [DOI] [PubMed] [Google Scholar]
- Berger I., Egli M., Rich A. Inter-strand C-H...O hydrogen bonds stabilizing four-stranded intercalated molecules: stereoelectronic effects of O4' in cytosine-rich DNA. Proc Natl Acad Sci U S A. 1996 Oct 29;93(22):12116–12121. doi: 10.1073/pnas.93.22.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger I., Kang C., Fredian A., Ratliff R., Moyzis R., Rich A. Extension of the four-stranded intercalated cytosine motif by adenine.adenine base pairing in the crystal structure of d(CCCAAT). Nat Struct Biol. 1995 May;2(5):416–425. doi: 10.1038/nsb0595-416. [DOI] [PubMed] [Google Scholar]
- Berman H. M., Olson W. K., Beveridge D. L., Westbrook J., Gelbin A., Demeny T., Hsieh S. H., Srinivasan A. R., Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992 Sep;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R. Principles of sequence-dependent flexure of DNA. J Mol Biol. 1986 Dec 20;192(4):907–918. doi: 10.1016/0022-2836(86)90036-7. [DOI] [PubMed] [Google Scholar]
- DEVOE H., TINOCO I., Jr The stability of helical polynucleotides: base contributions. J Mol Biol. 1962 Jun;4:500–517. doi: 10.1016/s0022-2836(62)80105-3. [DOI] [PubMed] [Google Scholar]
- Dang L. X., Pearlman D. A., Kollman P. A. Why do A.T base pairs inhibit Z-DNA formation? Proc Natl Acad Sci U S A. 1990 Jun;87(12):4630–4634. doi: 10.1073/pnas.87.12.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli M., Gessner R. V. Stereoelectronic effects of deoxyribose O4' on DNA conformation. Proc Natl Acad Sci U S A. 1995 Jan 3;92(1):180–184. doi: 10.1073/pnas.92.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florián J., Leszczyński J. Theoretical investigation of the molecular structure of the pi kappa DNA base pair. J Biomol Struct Dyn. 1995 Apr;12(5):1055–1062. doi: 10.1080/07391102.1995.10508797. [DOI] [PubMed] [Google Scholar]
- Friedman R. A., Honig B. The electrostatic contribution to DNA base-stacking interactions. Biopolymers. 1992 Feb;32(2):145–159. doi: 10.1002/bip.360320205. [DOI] [PubMed] [Google Scholar]
- Gabb H. A., Sanghani S. R., Robert C. H., Prévost C. Finding and visualizing nucleic acid base stacking. J Mol Graph. 1996 Feb;14(1):6-11, 23-4. doi: 10.1016/0263-7855(95)00086-0. [DOI] [PubMed] [Google Scholar]
- Gessner R. V., Frederick C. A., Quigley G. J., Rich A., Wang A. H. The molecular structure of the left-handed Z-DNA double helix at 1.0-A atomic resolution. Geometry, conformation, and ionic interactions of d(CGCGCG). J Biol Chem. 1989 May 15;264(14):7921–7935. doi: 10.2210/pdb1dcg/pdb. [DOI] [PubMed] [Google Scholar]
- Gorin A. A., Zhurkin V. B., Olson W. K. B-DNA twisting correlates with base-pair morphology. J Mol Biol. 1995 Mar 17;247(1):34–48. doi: 10.1006/jmbi.1994.0120. [DOI] [PubMed] [Google Scholar]
- Halgren T. A. Potential energy functions. Curr Opin Struct Biol. 1995 Apr;5(2):205–210. doi: 10.1016/0959-440x(95)80077-8. [DOI] [PubMed] [Google Scholar]
- Harvey S. C. DNA structural dynamics: longitudinal breathing as a possible mechanism for the B in equilibrium Z transition. Nucleic Acids Res. 1983 Jul 25;11(14):4867–4878. doi: 10.1093/nar/11.14.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U., Alings C. Crystallographic study of one turn of G/C-rich B-DNA. J Mol Biol. 1989 Nov 20;210(2):369–381. doi: 10.1016/0022-2836(89)90337-9. [DOI] [PubMed] [Google Scholar]
- Herbert A. G., Spitzner J. R., Lowenhaupt K., Rich A. Z-DNA binding protein from chicken blood nuclei. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3339–3342. doi: 10.1073/pnas.90.8.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C. A. Sequence-dependent DNA structure. The role of base stacking interactions. J Mol Biol. 1993 Apr 5;230(3):1025–1054. doi: 10.1006/jmbi.1993.1217. [DOI] [PubMed] [Google Scholar]
- Jaworski A., Hsieh W. T., Blaho J. A., Larson J. E., Wells R. D. Left-handed DNA in vivo. Science. 1987 Nov 6;238(4828):773–777. doi: 10.1126/science.3313728. [DOI] [PubMed] [Google Scholar]
- Kagawa T. F., Stoddard D., Zhou G. W., Ho P. S. Quantitative analysis of DNA secondary structure from solvent-accessible surfaces: the B- to Z-DNA transition as a model. Biochemistry. 1989 Aug 8;28(16):6642–6651. doi: 10.1021/bi00442a017. [DOI] [PubMed] [Google Scholar]
- Kang C. H., Berger I., Lockshin C., Ratliff R., Moyzis R., Rich A. Crystal structure of intercalated four-stranded d(C3T) at 1.4 A resolution. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11636–11640. doi: 10.1073/pnas.91.24.11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr I. M., Brown R. E. pppA2'p5'A2'p5'A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc Natl Acad Sci U S A. 1978 Jan;75(1):256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R., Seshadri T. P. Stereochemistry of 2'-5' linked nucleic acids: crystal and molecular structure of ammonium adenylyl-2',5'-adenosine tetrahydrate: a core fragment of 2'-5' oligo A's produced by interferon induced adenylate synthetase. J Biomol Struct Dyn. 1993 Feb;10(4):727–745. doi: 10.1080/07391102.1993.10508003. [DOI] [PubMed] [Google Scholar]
- Kudritskaya Z. G., Danilov V. I. Quantum mechanical study of bases interactions in various associates in atomic dipole approximation. J Theor Biol. 1976 Jul 7;59(2):303–318. doi: 10.1016/0022-5193(76)90172-7. [DOI] [PubMed] [Google Scholar]
- Lavery R., Hartmann B. Modelling DNA conformational mechanics. Biophys Chem. 1994 May;50(1-2):33–45. doi: 10.1016/0301-4622(94)85018-6. [DOI] [PubMed] [Google Scholar]
- Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J Biomol Struct Dyn. 1988 Aug;6(1):63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- Lavery R., Sklenar H., Zakrzewska K., Pullman B. The flexibility of the nucleic acids: (II). The calculation of internal energy and applications to mononucleotide repeat DNA. J Biomol Struct Dyn. 1986 Apr;3(5):989–1014. doi: 10.1080/07391102.1986.10508478. [DOI] [PubMed] [Google Scholar]
- Lukomski S., Wells R. D. Left-handed Z-DNA and in vivo supercoil density in the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):9980–9984. doi: 10.1073/pnas.91.21.9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooers B. H., Schroth G. P., Baxter W. W., Ho P. S. Alternating and non-alternating dG-dC hexanucleotides crystallize as canonical A-DNA. J Mol Biol. 1995 Jun 16;249(4):772–784. doi: 10.1006/jmbi.1995.0336. [DOI] [PubMed] [Google Scholar]
- Müller V., Takeya M., Brendel S., Wittig B., Rich A. Z-DNA-forming sites within the human beta-globin gene cluster. Proc Natl Acad Sci U S A. 1996 Jan 23;93(2):780–784. doi: 10.1073/pnas.93.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson W. K., Srinivasan A. R., Marky N. L., Balaji V. N. Theoretical probes of DNA conformation examining the B leads to Z conformational transition. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):229–241. doi: 10.1101/sqb.1983.047.01.028. [DOI] [PubMed] [Google Scholar]
- Pearlman D. A., Kollman P. A. The calculated free energy effects of 5-methyl cytosine on the B to Z transition in DNA. 1990 Jul-Aug 5Biopolymers. 29(8-9):1193–1209. doi: 10.1002/bip.360290810. [DOI] [PubMed] [Google Scholar]
- Privé G. G., Yanagi K., Dickerson R. E. Structure of the B-DNA decamer C-C-A-A-C-G-T-T-G-G and comparison with isomorphous decamers C-C-A-A-G-A-T-T-G-G and C-C-A-G-G-C-C-T-G-G. J Mol Biol. 1991 Jan 5;217(1):177–199. doi: 10.1016/0022-2836(91)90619-h. [DOI] [PubMed] [Google Scholar]
- Quadrifoglio F., Manzini G., Yathindra N. Short oligodeoxynucleotides with d(G-C)n sequence do not assume left-handed conformation in high salt conditions. J Mol Biol. 1984 May 25;175(3):419–423. doi: 10.1016/0022-2836(84)90358-9. [DOI] [PubMed] [Google Scholar]
- Rahmouni A. R., Wells R. D. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989 Oct 20;246(4928):358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- Rich A. Speculation on the biological roles of left-handed Z-DNA. Ann N Y Acad Sci. 1994 Jul 29;726:1–17. doi: 10.1111/j.1749-6632.1994.tb52792.x. [DOI] [PubMed] [Google Scholar]
- Saenger W., Heinemann U. Raison d'être and structural model for the B-Z transition of poly d(G-C).poly d(G-C). FEBS Lett. 1989 Nov 6;257(2):223–227. doi: 10.1016/0014-5793(89)81539-x. [DOI] [PubMed] [Google Scholar]
- Sponer J., Hobza P. G.C base pair in parallel-stranded DNA--a novel type of base pairing: an ab initio quantum chemical study. J Biomol Struct Dyn. 1994 Dec;12(3):671–680. doi: 10.1080/07391102.1994.10508766. [DOI] [PubMed] [Google Scholar]
- Sponer J., Kypr J. Base pair buckling can eliminate the interstrand purine clash at the CpG steps in B-DNA caused by the base pair propeller twisting. J Biomol Struct Dyn. 1990 Jun;7(6):1211–1220. doi: 10.1080/07391102.1990.10508560. [DOI] [PubMed] [Google Scholar]
- Sponer J., Kypr J. Different intrastrand and interstrand contributions to stacking account for roll variations at the alternating purine-pyrimidine sequences in A-DNA and A-RNA. J Mol Biol. 1991 Oct 5;221(3):761–764. doi: 10.1016/0022-2836(91)80172-q. [DOI] [PubMed] [Google Scholar]
- Sponer J., Kypr J. Relationships among rise, cup, roll and stagger in DNA suggested by empirical potential studies of base stacking. J Biomol Struct Dyn. 1993 Aug;11(1):27–41. doi: 10.1080/07391102.1993.10508707. [DOI] [PubMed] [Google Scholar]
- Sponer J., Kypr J. Theoretical analysis of the base stacking in DNA: choice of the force field and a comparison with the oligonucleotide crystal structures. J Biomol Struct Dyn. 1993 Oct;11(2):277–292. doi: 10.1080/07391102.1993.10508726. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wang S., Kool E. T. Origins of the large differences in stability of DNA and RNA helices: C-5 methyl and 2'-hydroxyl effects. Biochemistry. 1995 Mar 28;34(12):4125–4132. doi: 10.1021/bi00012a031. [DOI] [PubMed] [Google Scholar]
- Wölfl S., Martinez C., Rich A., Majzoub J. A. Transcription of the human corticotropin-releasing hormone gene in NPLC cells is correlated with Z-DNA formation. Proc Natl Acad Sci U S A. 1996 Apr 16;93(8):3664–3668. doi: 10.1073/pnas.93.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]