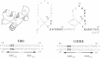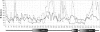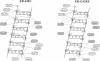Abstract
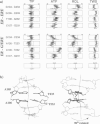
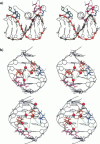
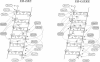
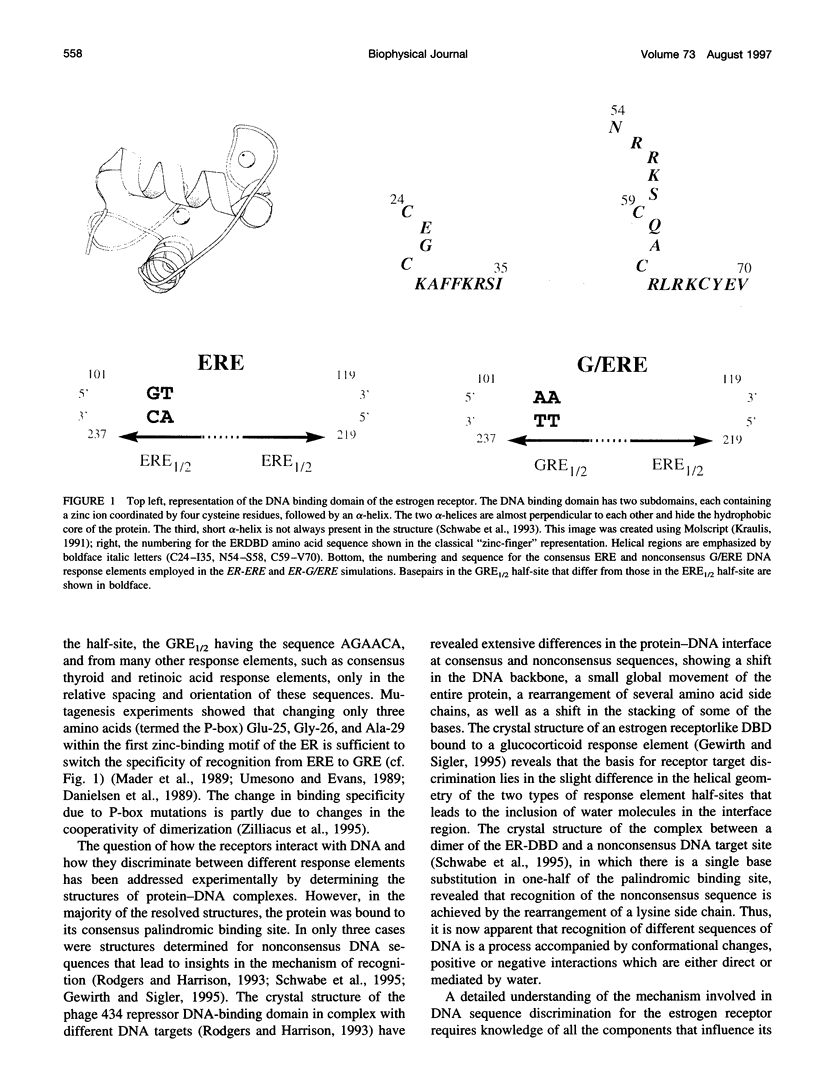
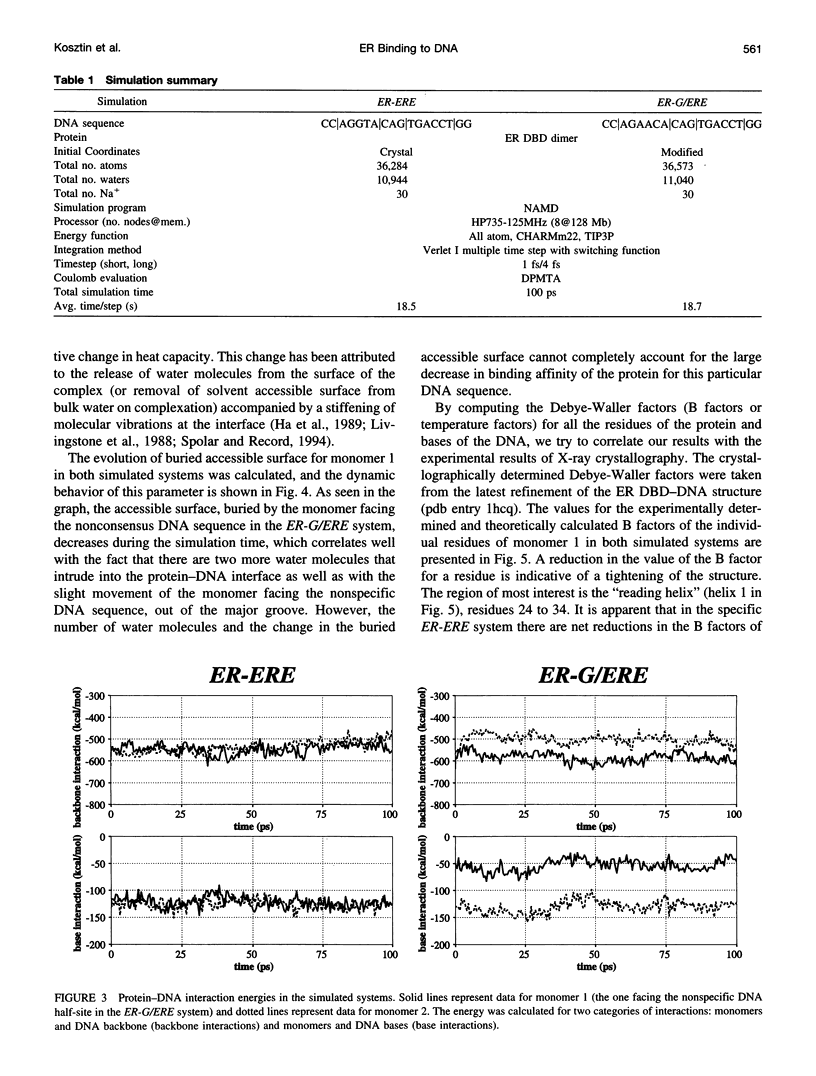
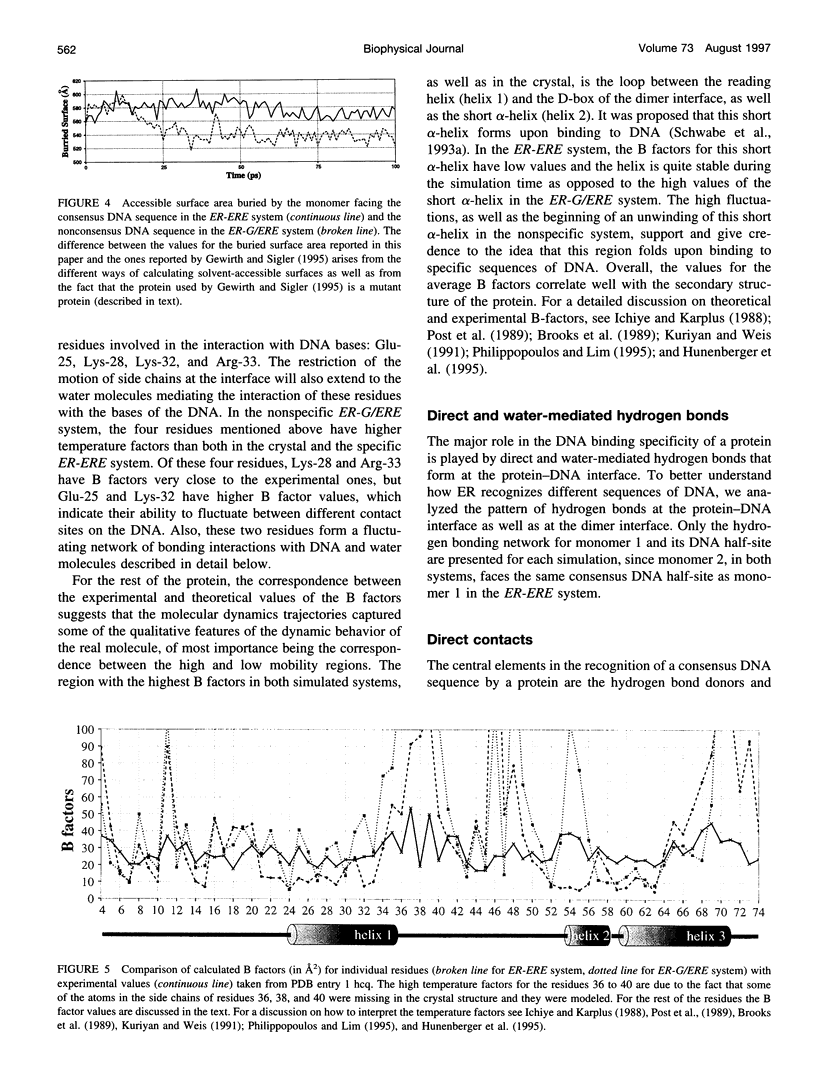
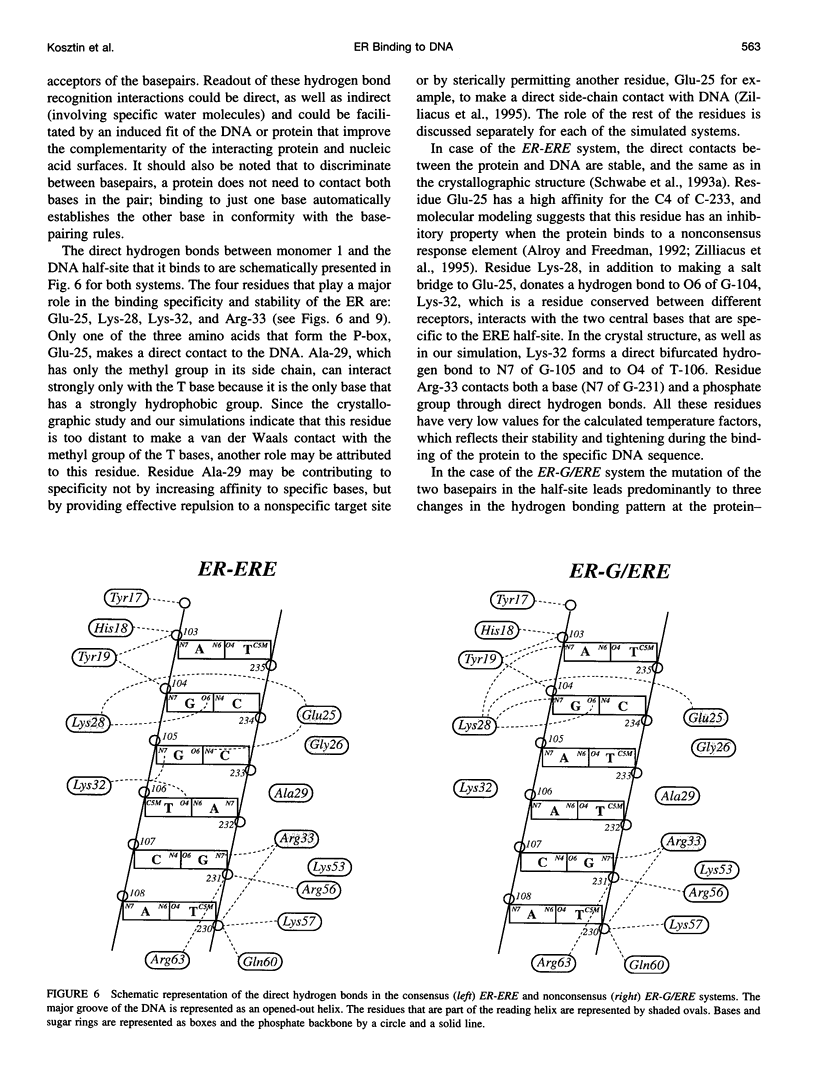
Molecular dynamics simulations are carried out to investigate the binding of the estrogen receptor, a member of the nuclear hormone receptor family, to specific and non-specific DNA. Two systems have been simulated, each based on the crystallographic structure of a complex of a dimer of the estrogen receptor DNA binding domain with DNA. One structure includes the dimer and a consensus segment of DNA, ds(CCAGGTCACAGTGACCTGG); the other structure includes the dimer and a nonconsensus segment of DNA, ds(CCAGAACACAGTGACCTGG). The simulations involve an atomic model of the protein-DNA complex, counterions, and a sphere of explicit water with a radius of 45 A. The molecular dynamics package NAMD was used to obtain 100 ps of dynamics for each system with complete long-range electrostatic interactions. Analysis of the simulations revealed differences in the protein-DNA interactions for consensus and nonconsensus sequences, a bending and unwinding of the DNA, a slight rearrangement of several amino acid side chains, and inclusion of water molecules at the protein-DNA interface region. Our results indicate that binding specificity and stability is conferred by a network of direct and water mediated protein-DNA hydrogen bonds. For the consensus sequence, the network involves three water molecules, residues Glu-25, Lys-28, Lys-32, Arg-33, and bases of the DNA. The binding differs for the nonconsensus DNA sequence in which case the fluctuating network of hydrogen bonds allows water molecules to enter the protein-DNA interface. We conclude that water plays a role in furnishing DNA binding specificity to nuclear hormone receptors.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alroy I., Freedman L. P. DNA binding analysis of glucocorticoid receptor specificity mutants. Nucleic Acids Res. 1992 Mar 11;20(5):1045–1052. doi: 10.1093/nar/20.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop T. C., Kosztin D., Schulten K. How hormone receptor-DNA binding affects nucleosomal DNA: the role of symmetry. Biophys J. 1997 May;72(5):2056–2067. doi: 10.1016/S0006-3495(97)78849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop T. C., Schulten K. Molecular dynamics study of glucocorticoid receptor-DNA binding. Proteins. 1996 Jan;24(1):115–133. doi: 10.1002/(SICI)1097-0134(199601)24:1<115::AID-PROT8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Danielsen M., Hinck L., Ringold G. M. Two amino acids within the knuckle of the first zinc finger specify DNA response element activation by the glucocorticoid receptor. Cell. 1989 Jun 30;57(7):1131–1138. doi: 10.1016/0092-8674(89)90050-0. [DOI] [PubMed] [Google Scholar]
- Eriksson M. A., Härd T., Nilsson L. Molecular dynamics simulations of the glucocorticoid receptor DNA-binding domain in complex with DNA and free in solution. Biophys J. 1995 Feb;68(2):402–426. doi: 10.1016/S0006-3495(95)80203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M. A., Nilsson L. Structure, thermodynamics and cooperativity of the glucocorticoid receptor DNA-binding domain in complex with different response elements. Molecular dynamics simulation and free energy perturbation studies. J Mol Biol. 1995 Oct 27;253(3):453–472. doi: 10.1006/jmbi.1995.0566. [DOI] [PubMed] [Google Scholar]
- Evans R. M. The steroid and thyroid hormone receptor superfamily. Science. 1988 May 13;240(4854):889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J. A., Johnson R. C., Dickerson R. E. Hin recombinase bound to DNA: the origin of specificity in major and minor groove interactions. Science. 1994 Jan 21;263(5145):348–355. doi: 10.1126/science.8278807. [DOI] [PubMed] [Google Scholar]
- Freedman L. P., Luisi B. F. On the mechanism of DNA binding by nuclear hormone receptors: a structural and functional perspective. J Cell Biochem. 1993 Feb;51(2):140–150. doi: 10.1002/jcb.240510205. [DOI] [PubMed] [Google Scholar]
- Gewirth D. T., Sigler P. B. The basis for half-site specificity explored through a non-cognate steroid receptor-DNA complex. Nat Struct Biol. 1995 May;2(5):386–394. doi: 10.1038/nsb0595-386. [DOI] [PubMed] [Google Scholar]
- Green S., Chambon P. Oestradiol induction of a glucocorticoid-responsive gene by a chimaeric receptor. Nature. 1987 Jan 1;325(6099):75–78. doi: 10.1038/325075a0. [DOI] [PubMed] [Google Scholar]
- Green S., Kumar V., Theulaz I., Wahli W., Chambon P. The N-terminal DNA-binding 'zinc finger' of the oestrogen and glucocorticoid receptors determines target gene specificity. EMBO J. 1988 Oct;7(10):3037–3044. doi: 10.1002/j.1460-2075.1988.tb03168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J. H., Capp M. W., Hohenwalter M. D., Baskerville M., Record M. T., Jr Thermodynamic stoichiometries of participation of water, cations and anions in specific and non-specific binding of lac repressor to DNA. Possible thermodynamic origins of the "glutamate effect" on protein-DNA interactions. J Mol Biol. 1992 Nov 5;228(1):252–264. doi: 10.1016/0022-2836(92)90504-d. [DOI] [PubMed] [Google Scholar]
- Ha J. H., Spolar R. S., Record M. T., Jr Role of the hydrophobic effect in stability of site-specific protein-DNA complexes. J Mol Biol. 1989 Oct 20;209(4):801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- Harris L. F., Sullivan M. R., Popken-Harris P. D., Hickok D. F. Molecular dynamics simulations in solvent of the glucocorticoid receptor protein in complex with a glucocorticoid response element DNA sequence. J Biomol Struct Dyn. 1994 Oct;12(2):249–270. doi: 10.1080/07391102.1994.10508740. [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996 Feb;14(1):33-8, 27-8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Ichiye T., Karplus M. Anisotropy and anharmonicity of atomic fluctuations in proteins: implications for X-ray analysis. Biochemistry. 1988 May 3;27(9):3487–3497. doi: 10.1021/bi00409a054. [DOI] [PubMed] [Google Scholar]
- Kim J. L., Nikolov D. B., Burley S. K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993 Oct 7;365(6446):520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Krust A., Green S., Argos P., Kumar V., Walter P., Bornert J. M., Chambon P. The chicken oestrogen receptor sequence: homology with v-erbA and the human oestrogen and glucocorticoid receptors. EMBO J. 1986 May;5(5):891–897. doi: 10.1002/j.1460-2075.1986.tb04300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988 Oct 7;55(1):145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- Kumar V., Green S., Stack G., Berry M., Jin J. R., Chambon P. Functional domains of the human estrogen receptor. Cell. 1987 Dec 24;51(6):941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- Kuriyan J., Weis W. I. Rigid protein motion as a model for crystallographic temperature factors. Proc Natl Acad Sci U S A. 1991 Apr 1;88(7):2773–2777. doi: 10.1073/pnas.88.7.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudet V., Hänni C., Coll J., Catzeflis F., Stéhelin D. Evolution of the nuclear receptor gene superfamily. EMBO J. 1992 Mar;11(3):1003–1013. doi: 10.1002/j.1460-2075.1992.tb05139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J Biomol Struct Dyn. 1988 Aug;6(1):63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- Livingstone J. R., Spolar R. S., Record M. T., Jr Contribution to the thermodynamics of protein folding from the reduction in water-accessible nonpolar surface area. Biochemistry. 1991 Apr 30;30(17):4237–4244. doi: 10.1021/bi00231a019. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991 Aug 8;352(6335):497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Lundbäck T., Cairns C., Gustafsson J. A., Carlstedt-Duke J., Härd T. Thermodynamics of the glucocorticoid receptor-DNA interaction: binding of wild-type GR DBD to different response elements. Biochemistry. 1993 May 18;32(19):5074–5082. doi: 10.1021/bi00070a015. [DOI] [PubMed] [Google Scholar]
- Lundbäck T., Zilliacus J., Gustafsson J. A., Carlstedt-Duke J., Härd T. Thermodynamics of sequence-specific glucocorticoid receptor-DNA interactions. Biochemistry. 1994 May 17;33(19):5955–5965. doi: 10.1021/bi00185a037. [DOI] [PubMed] [Google Scholar]
- Mader S., Kumar V., de Verneuil H., Chambon P. Three amino acids of the oestrogen receptor are essential to its ability to distinguish an oestrogen from a glucocorticoid-responsive element. Nature. 1989 Mar 16;338(6212):271–274. doi: 10.1038/338271a0. [DOI] [PubMed] [Google Scholar]
- Philippopoulos M., Lim C. Molecular dynamics simulation of E. coli ribonuclease H1 in solution: correlation with NMR and X-ray data and insights into biological function. J Mol Biol. 1995 Dec 8;254(4):771–792. doi: 10.1006/jmbi.1995.0654. [DOI] [PubMed] [Google Scholar]
- Post C. B., Dobson C. M., Karplus M. A molecular dynamics analysis of protein structural elements. Proteins. 1989;5(4):337–354. doi: 10.1002/prot.340050409. [DOI] [PubMed] [Google Scholar]
- Prévost C., Louise-May S., Ravishanker G., Lavery R., Beveridge D. L. Persistence analysis of the static and dynamical helix deformations of DNA oligonucleotides: application to the crystal structure and molecular dynamics simulation of d(CGCGAATTCGCG)2. Biopolymers. 1993 Mar;33(3):335–350. doi: 10.1002/bip.360330303. [DOI] [PubMed] [Google Scholar]
- Rastinejad F., Perlmann T., Evans R. M., Sigler P. B. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995 May 18;375(6528):203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- Ravishanker G., Swaminathan S., Beveridge D. L., Lavery R., Sklenar H. Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCGAATTCGCG) double helix: "curves", dials and windows. J Biomol Struct Dyn. 1989 Feb;6(4):669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- Rodgers D. W., Harrison S. C. The complex between phage 434 repressor DNA-binding domain and operator site OR3: structural differences between consensus and non-consensus half-sites. Structure. 1993 Dec 15;1(4):227–240. doi: 10.1016/0969-2126(93)90012-6. [DOI] [PubMed] [Google Scholar]
- Schneider B., Cohen D. M., Schleifer L., Srinivasan A. R., Olson W. K., Berman H. M. A systematic method for studying the spatial distribution of water molecules around nucleic acid bases. Biophys J. 1993 Dec;65(6):2291–2303. doi: 10.1016/S0006-3495(93)81306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe J. W., Chapman L., Finch J. T., Rhodes D., Neuhaus D. DNA recognition by the oestrogen receptor: from solution to the crystal. Structure. 1993 Nov 15;1(3):187–204. doi: 10.1016/0969-2126(93)90020-h. [DOI] [PubMed] [Google Scholar]
- Schwabe J. W., Chapman L., Finch J. T., Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993 Nov 5;75(3):567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- Schwabe J. W., Chapman L., Rhodes D. The oestrogen receptor recognizes an imperfectly palindromic response element through an alternative side-chain conformation. Structure. 1995 Feb 15;3(2):201–213. doi: 10.1016/s0969-2126(01)00150-2. [DOI] [PubMed] [Google Scholar]
- Schwabe J. W., Neuhaus D., Rhodes D. Solution structure of the DNA-binding domain of the oestrogen receptor. Nature. 1990 Nov 29;348(6300):458–461. doi: 10.1038/348458a0. [DOI] [PubMed] [Google Scholar]
- Seibel G. L., Singh U. C., Kollman P. A. A molecular dynamics simulation of double-helical B-DNA including counterions and water. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6537–6540. doi: 10.1073/pnas.82.19.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakked Z., Guzikevich-Guerstein G., Frolow F., Rabinovich D., Joachimiak A., Sigler P. B. Determinants of repressor/operator recognition from the structure of the trp operator binding site. Nature. 1994 Mar 31;368(6470):469–473. doi: 10.1038/368469a0. [DOI] [PubMed] [Google Scholar]
- Sharp K. A., Honig B. Salt effects on nucleic acids. Curr Opin Struct Biol. 1995 Jun;5(3):323–328. doi: 10.1016/0959-440x(95)80093-x. [DOI] [PubMed] [Google Scholar]
- Sklenar H., Etchebest C., Lavery R. Describing protein structure: a general algorithm yielding complete helicoidal parameters and a unique overall axis. Proteins. 1989;6(1):46–60. doi: 10.1002/prot.340060105. [DOI] [PubMed] [Google Scholar]
- Spolar R. S., Record M. T., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994 Feb 11;263(5148):777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Suzuki M. A framework for the DNA-protein recognition code of the probe helix in transcription factors: the chemical and stereochemical rules. Structure. 1994 Apr 15;2(4):317–326. doi: 10.1016/s0969-2126(00)00033-2. [DOI] [PubMed] [Google Scholar]
- Umesono K., Evans R. M. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989 Jun 30;57(7):1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- Zilliacus J., Wright A. P., Carlstedt-Duke J., Nilsson L., Gustafsson J. A. Modulation of DNA-binding specificity within the nuclear receptor family by substitutions at a single amino acid position. Proteins. 1995 Jan;21(1):57–67. doi: 10.1002/prot.340210107. [DOI] [PubMed] [Google Scholar]