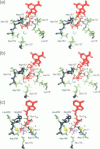
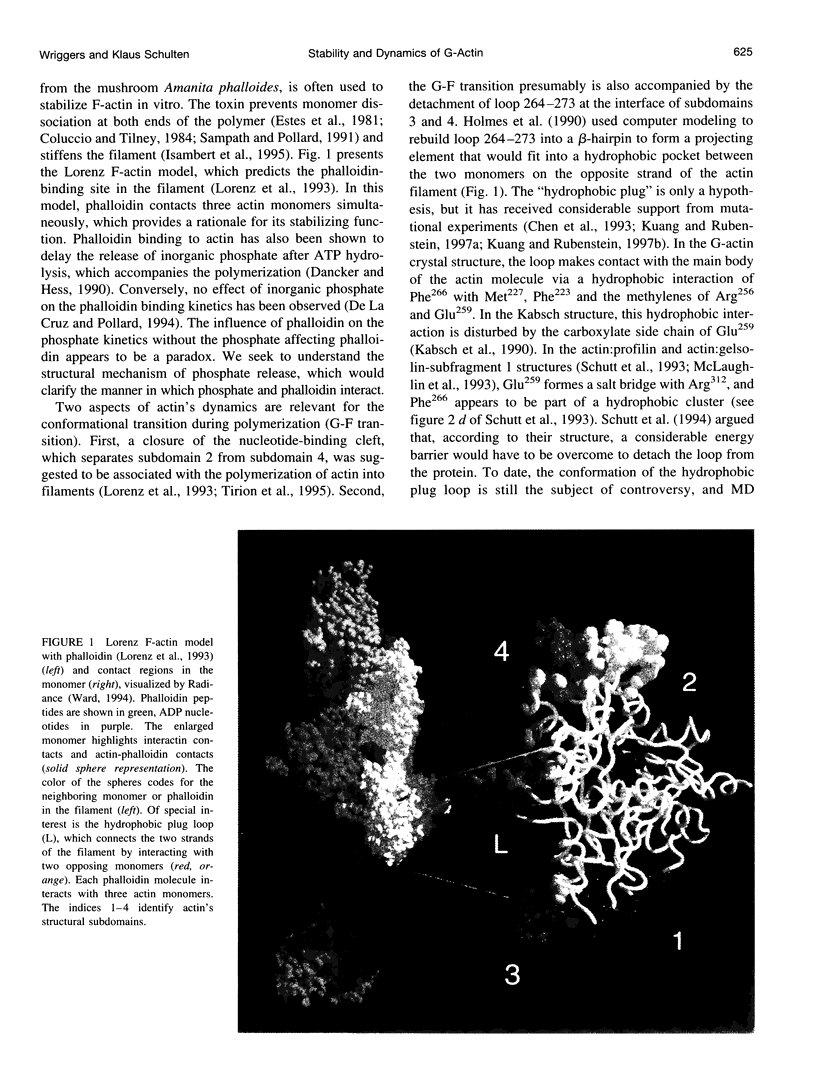
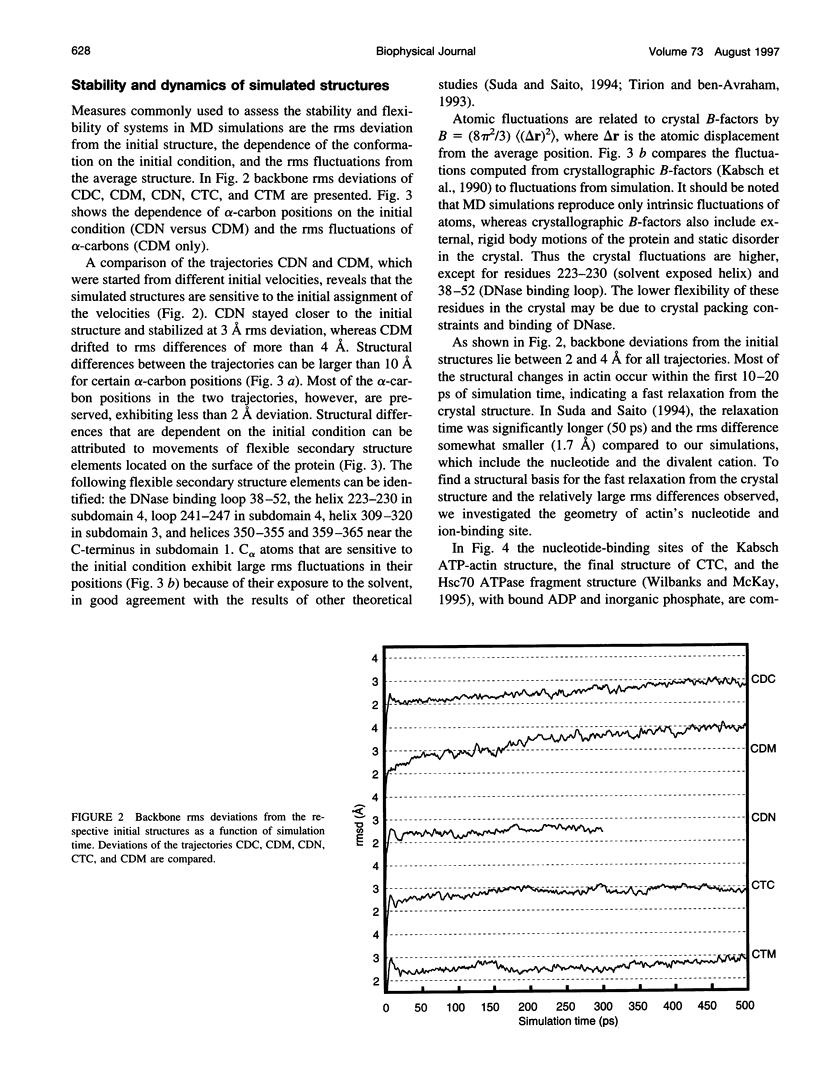
Abstract
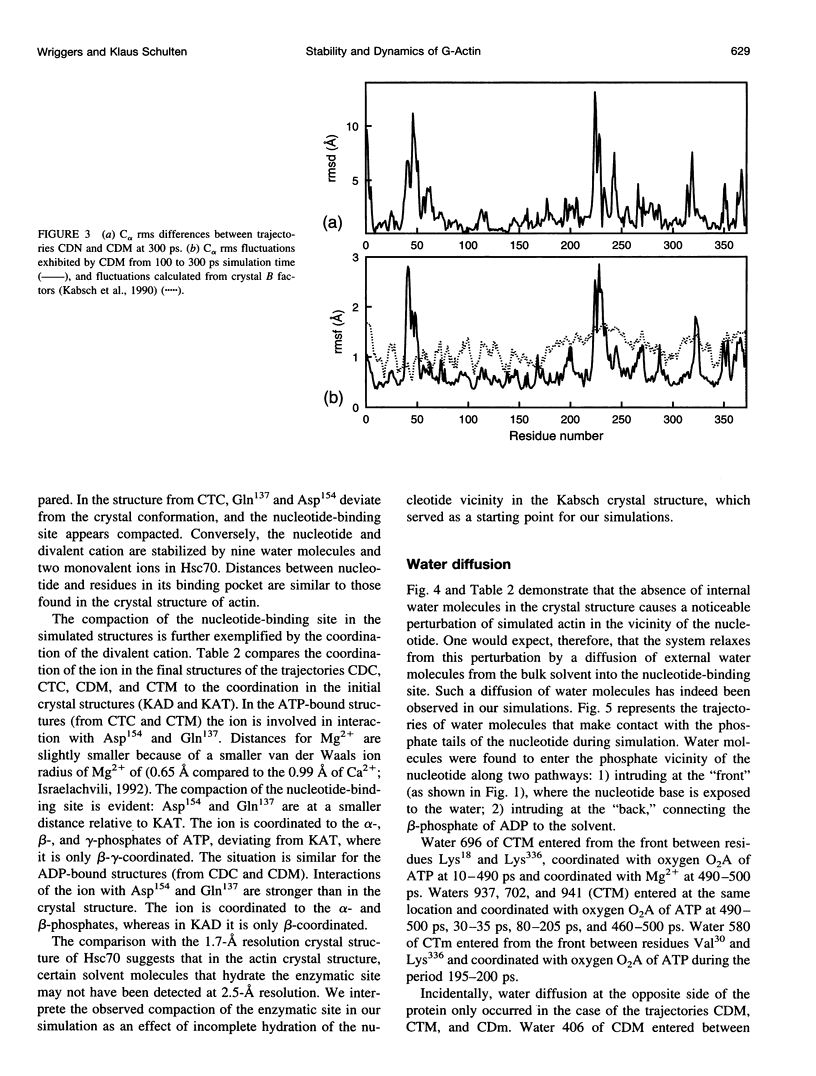
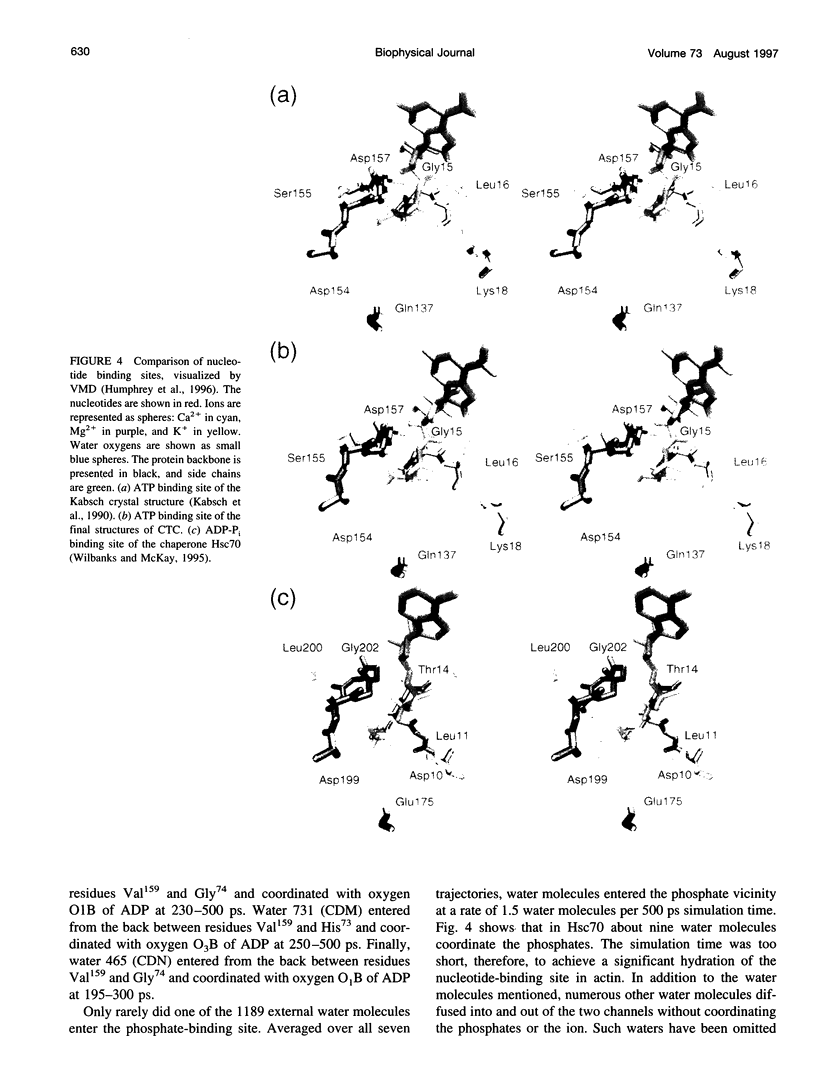
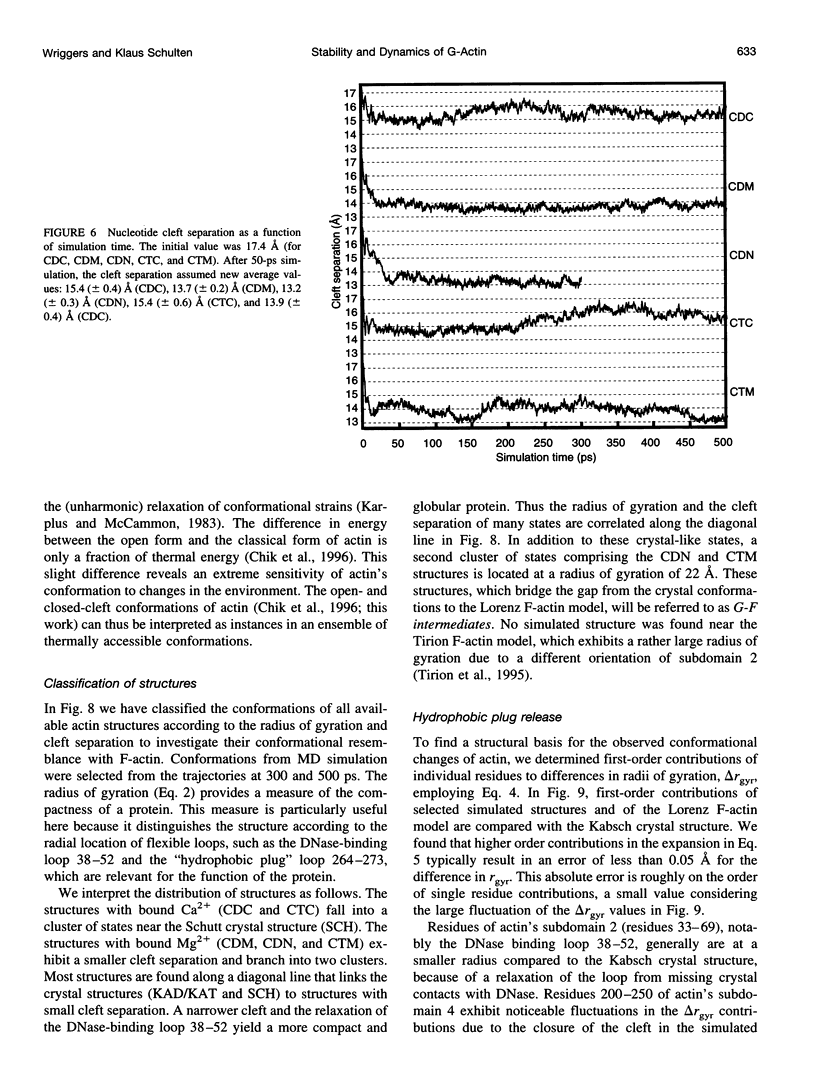
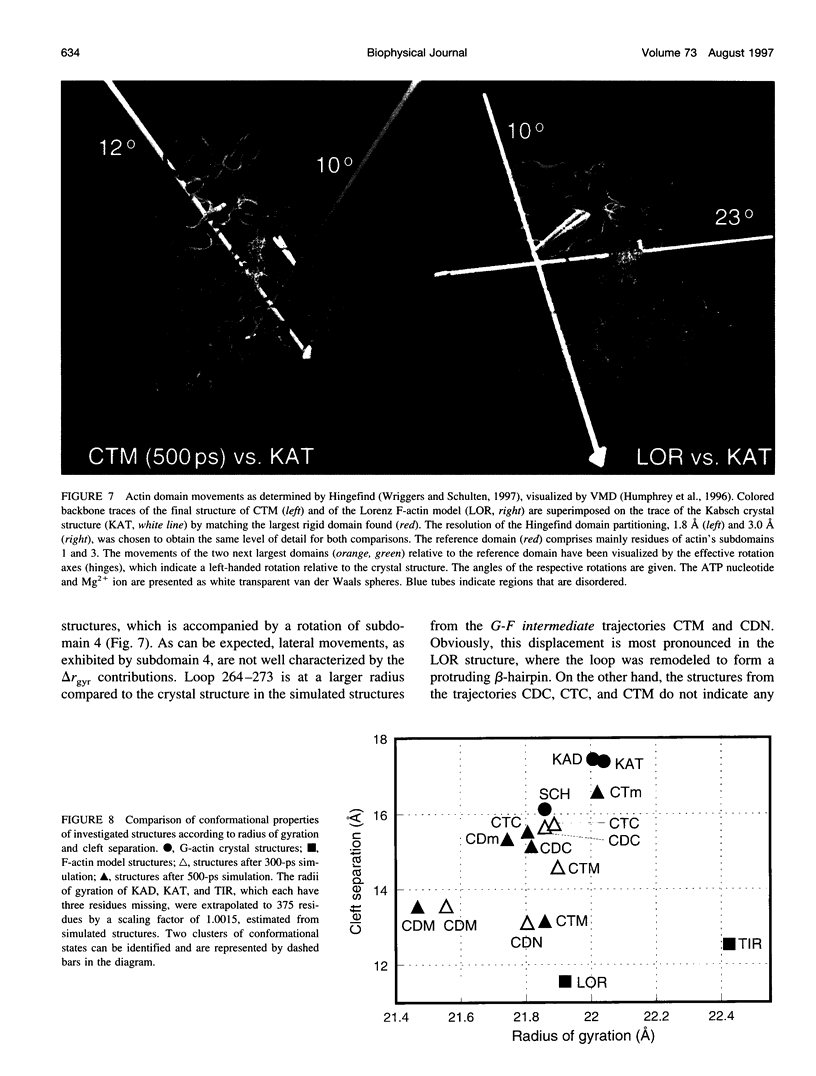
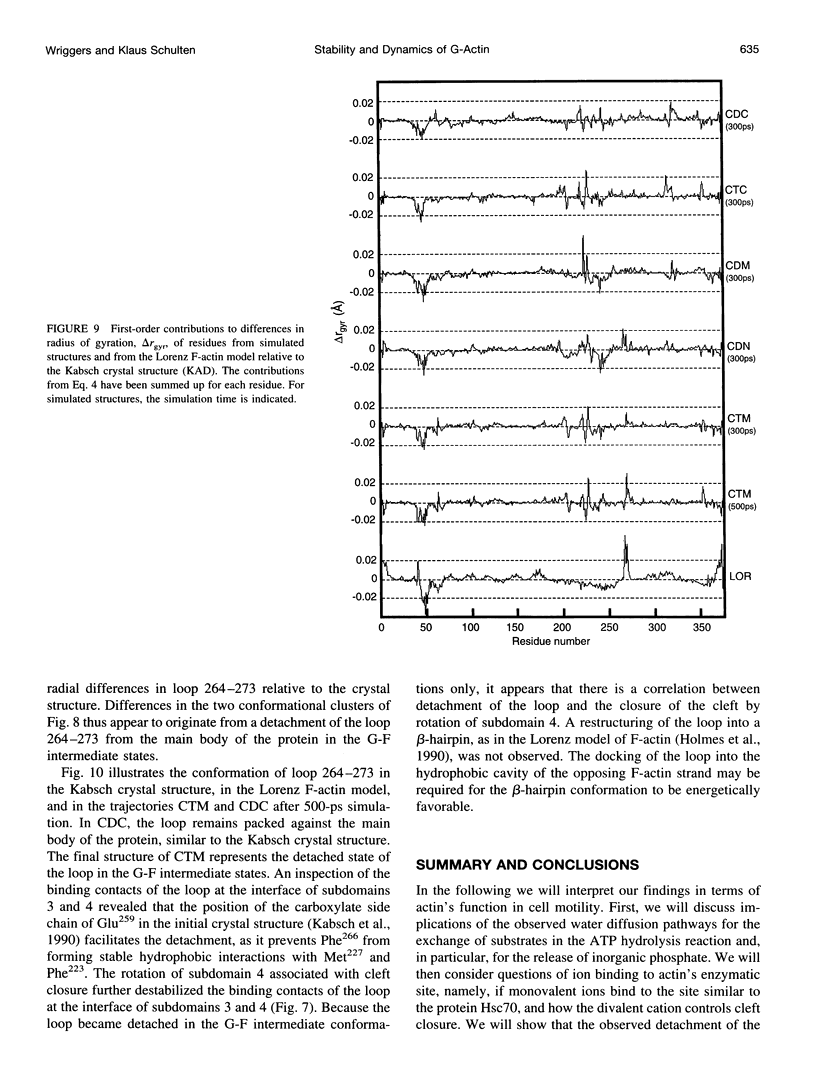
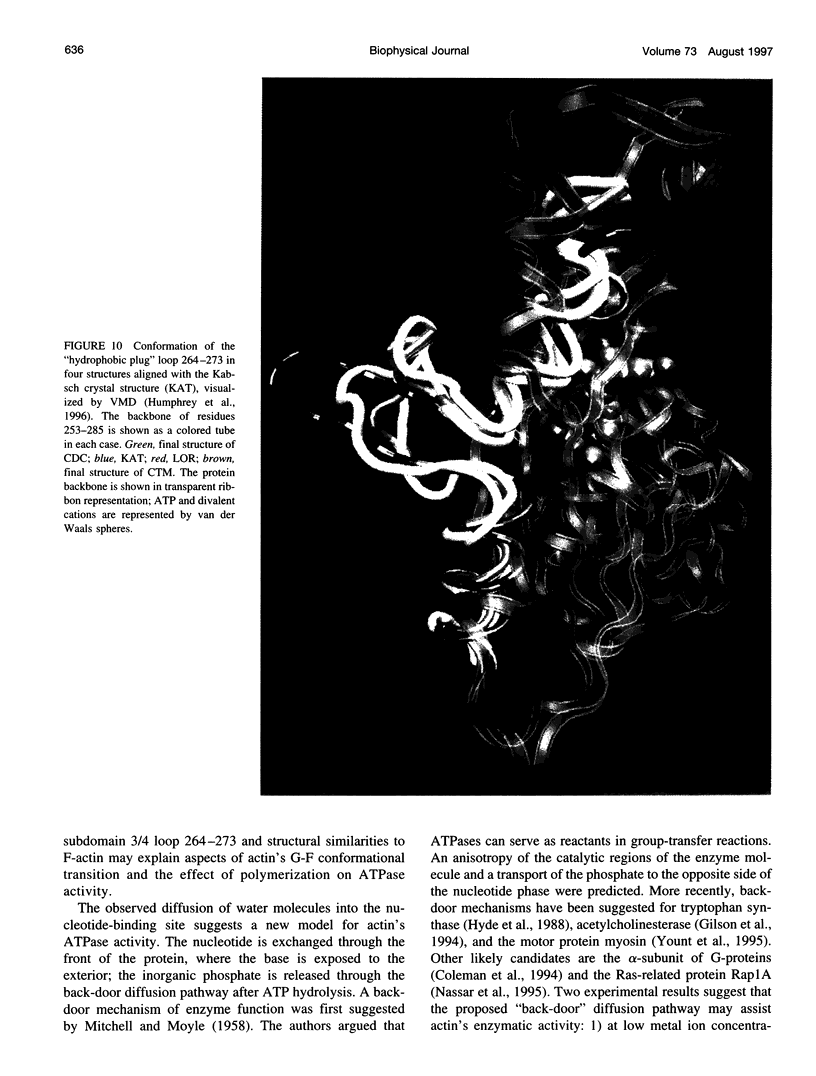
Molecular dynamics simulations have been performed on solvated G-actin bound to ADP and ATP, starting with the crystal structure of the actin-DNase 1 complex, including a Ca2+ or Mg2+ ion at the high-affinity divalent cation-binding site. Water molecules have been found to enter the nucleotide-binding site (phosphate vicinity) along two pathways, from the side where the nucleotide base is exposed to water, as well as from the opposite side. The water channels suggest a "back-door" mechanism for ATP hydrolysis in which the phosphate is released to a side opposite that of nucleotide binding and unbinding. The simulations also reveal a propensity of G-actin to alter its crystallographic structure toward the filamentous structure. Domain movement closes the nucleotide cleft, the movement being more pronounced for bound Mg2+. The conformational change is interpreted as a response of the system to missing water molecules in the crystal structure. The structures arising in the simulations, classified according to nucleotide cleft separation and radius of gyration of the protein, fall into two distinct clusters: a cluster of states that are similar to the G-actin crystal structure, and a cluster of states with small cleft separation and with the subdomain 3/4 loop 264-273 detached from the protein. The latter states resemble the putative filamentous structure of actin, in which the loop connects the two strands of the actin filament.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen P. G., Laham L. E., Way M., Janmey P. A. Binding of phosphate, aluminum fluoride, or beryllium fluoride to F-actin inhibits severing by gelsolin. J Biol Chem. 1996 Mar 1;271(9):4665–4670. doi: 10.1074/jbc.271.9.4665. [DOI] [PubMed] [Google Scholar]
- Breed J., Sankararamakrishnan R., Kerr I. D., Sansom M. S. Molecular dynamics simulations of water within models of ion channels. Biophys J. 1996 Apr;70(4):1643–1661. doi: 10.1016/S0006-3495(96)79727-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M. F. Actin: protein structure and filament dynamics. J Biol Chem. 1991 Jan 5;266(1):1–4. [PubMed] [Google Scholar]
- Chen X., Cook R. K., Rubenstein P. A. Yeast actin with a mutation in the "hydrophobic plug" between subdomains 3 and 4 (L266D) displays a cold-sensitive polymerization defect. J Cell Biol. 1993 Dec;123(5):1185–1195. doi: 10.1083/jcb.123.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chik J. K., Lindberg U., Schutt C. E. The structure of an open state of beta-actin at 2.65 A resolution. J Mol Biol. 1996 Nov 8;263(4):607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- Coleman D. E., Berghuis A. M., Lee E., Linder M. E., Gilman A. G., Sprang S. R. Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science. 1994 Sep 2;265(5177):1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- Coluccio L. M., Tilney L. G. Phalloidin enhances actin assembly by preventing monomer dissociation. J Cell Biol. 1984 Aug;99(2):529–535. doi: 10.1083/jcb.99.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daggett V., Levitt M. Realistic simulations of native-protein dynamics in solution and beyond. Annu Rev Biophys Biomol Struct. 1993;22:353–380. doi: 10.1146/annurev.bb.22.060193.002033. [DOI] [PubMed] [Google Scholar]
- Dancker P., Hess L. Phalloidin reduces the release of inorganic phosphate during actin polymerization. Biochim Biophys Acta. 1990 Aug 17;1035(2):197–200. doi: 10.1016/0304-4165(90)90116-e. [DOI] [PubMed] [Google Scholar]
- De La Cruz E. M., Pollard T. D. Transient kinetic analysis of rhodamine phalloidin binding to actin filaments. Biochemistry. 1994 Dec 6;33(48):14387–14392. doi: 10.1021/bi00252a003. [DOI] [PubMed] [Google Scholar]
- Devreotes P. N., Zigmond S. H. Chemotaxis in eukaryotic cells: a focus on leukocytes and Dictyostelium. Annu Rev Cell Biol. 1988;4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- Estes J. E., Selden L. A., Gershman L. C. Mechanism of action of phalloidin on the polymerization of muscle actin. Biochemistry. 1981 Feb 17;20(4):708–712. doi: 10.1021/bi00507a006. [DOI] [PubMed] [Google Scholar]
- Flaherty K. M., DeLuca-Flaherty C., McKay D. B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990 Aug 16;346(6285):623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Flaherty K. M., McKay D. B., Kabsch W., Holmes K. C. Similarity of the three-dimensional structures of actin and the ATPase fragment of a 70-kDa heat shock cognate protein. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):5041–5045. doi: 10.1073/pnas.88.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty K. M., Wilbanks S. M., DeLuca-Flaherty C., McKay D. B. Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment. J Biol Chem. 1994 Apr 29;269(17):12899–12907. [PubMed] [Google Scholar]
- Frieden C., Lieberman D., Gilbert H. R. A fluorescent probe for conformational changes in skeletal muscle G-actin. J Biol Chem. 1980 Oct 10;255(19):8991–8993. [PubMed] [Google Scholar]
- Gilson M. K., Straatsma T. P., McCammon J. A., Ripoll D. R., Faerman C. H., Axelsen P. H., Silman I., Sussman J. L. Open "back door" in a molecular dynamics simulation of acetylcholinesterase. Science. 1994 Mar 4;263(5151):1276–1278. doi: 10.1126/science.8122110. [DOI] [PubMed] [Google Scholar]
- Holmes K. C., Popp D., Gebhard W., Kabsch W. Atomic model of the actin filament. Nature. 1990 Sep 6;347(6288):44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996 Feb;14(1):33-8, 27-8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Hyde C. C., Ahmed S. A., Padlan E. A., Miles E. W., Davies D. R. Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988 Nov 25;263(33):17857–17871. [PubMed] [Google Scholar]
- Isambert H., Venier P., Maggs A. C., Fattoum A., Kassab R., Pantaloni D., Carlier M. F. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J Biol Chem. 1995 May 12;270(19):11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C. Atomic structure of the actin:DNase I complex. Nature. 1990 Sep 6;347(6288):37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. Dynamics of proteins: elements and function. Annu Rev Biochem. 1983;52:263–300. doi: 10.1146/annurev.bi.52.070183.001403. [DOI] [PubMed] [Google Scholar]
- Kinosian H. J., Selden L. A., Estes J. E., Gershman L. C. Nucleotide binding to actin. Cation dependence of nucleotide dissociation and exchange rates. J Biol Chem. 1993 Apr 25;268(12):8683–8691. [PubMed] [Google Scholar]
- Kuang B., Rubenstein P. A. Beryllium fluoride and phalloidin restore polymerizability of a mutant yeast actin (V266G,L267G) with severely decreased hydrophobicity in a subdomain 3/4 loop. J Biol Chem. 1997 Jan 10;272(2):1237–1247. doi: 10.1074/jbc.272.2.1237. [DOI] [PubMed] [Google Scholar]
- Kuang B., Rubenstein P. A. The effects of severely decreased hydrophobicity in a subdomain 3/4 loop on the dynamics and stability of yeast G-actin. J Biol Chem. 1997 Feb 14;272(7):4412–4418. doi: 10.1074/jbc.272.7.4412. [DOI] [PubMed] [Google Scholar]
- Levitt M. Molecular dynamics of native protein. I. Computer simulation of trajectories. J Mol Biol. 1983 Aug 15;168(3):595–617. doi: 10.1016/s0022-2836(83)80304-0. [DOI] [PubMed] [Google Scholar]
- Lorenz M., Popp D., Holmes K. C. Refinement of the F-actin model against X-ray fiber diffraction data by the use of a directed mutation algorithm. J Mol Biol. 1993 Dec 5;234(3):826–836. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- McLaughlin P. J., Gooch J. T., Mannherz H. G., Weeds A. G. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 1993 Aug 19;364(6439):685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- Nassar N., Horn G., Herrmann C., Scherer A., McCormick F., Wittinghofer A. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature. 1995 Jun 15;375(6532):554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Goldberg I., Schwarz W. H. Nucleotide exchange, structure, and mechanical properties of filaments assembled from ATP-actin and ADP-actin. J Biol Chem. 1992 Oct 5;267(28):20339–20345. [PubMed] [Google Scholar]
- Sampath P., Pollard T. D. Effects of cytochalasin, phalloidin, and pH on the elongation of actin filaments. Biochemistry. 1991 Feb 19;30(7):1973–1980. doi: 10.1021/bi00221a034. [DOI] [PubMed] [Google Scholar]
- Schutt C. E., Myslik J. C., Rozycki M. D., Goonesekere N. C., Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993 Oct 28;365(6449):810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- Schutt C. E., Rozycki M. D., Lindberg U. What's the matter with the ribbon? Curr Biol. 1994 Feb 1;4(2):185–186. doi: 10.1016/s0960-9822(94)00046-1. [DOI] [PubMed] [Google Scholar]
- Small J. V. Microfilament-based motility in non-muscle cells. Curr Opin Cell Biol. 1989 Feb;1(1):75–79. doi: 10.1016/s0955-0674(89)80040-7. [DOI] [PubMed] [Google Scholar]
- Steinbach P. J., Brooks B. R. Protein hydration elucidated by molecular dynamics simulation. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9135–9139. doi: 10.1073/pnas.90.19.9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka-Golaszewska H., Wozniak A., Hult T., Lindberg U. Effects of the type of divalent cation, Ca2+ or Mg2+, bound at the high-affinity site and of the ionic composition of the solution on the structure of F-actin. Biochem J. 1996 Jun 15;316(Pt 3):713–721. doi: 10.1042/bj3160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter M. M. Water-protein interactions: theory and experiment. Annu Rev Biophys Biophys Chem. 1991;20:577–600. doi: 10.1146/annurev.bb.20.060191.003045. [DOI] [PubMed] [Google Scholar]
- Theriot J. A., Mitchison T. J., Tilney L. G., Portnoy D. A. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature. 1992 May 21;357(6375):257–260. doi: 10.1038/357257a0. [DOI] [PubMed] [Google Scholar]
- Tirion M. M., ben-Avraham D., Lorenz M., Holmes K. C. Normal modes as refinement parameters for the F-actin model. Biophys J. 1995 Jan;68(1):5–12. doi: 10.1016/S0006-3495(95)80156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirion M. M., ben-Avraham D. Normal mode analysis of G-actin. J Mol Biol. 1993 Mar 5;230(1):186–195. doi: 10.1006/jmbi.1993.1135. [DOI] [PubMed] [Google Scholar]
- Valentin-Ranc C., Carlier M. F. Role of ATP-bound divalent metal ion in the conformation and function of actin. Comparison of Mg-ATP, Ca-ATP, and metal ion-free ATP-actin. J Biol Chem. 1991 Apr 25;266(12):7668–7675. [PubMed] [Google Scholar]
- Wilbanks S. M., McKay D. B. How potassium affects the activity of the molecular chaperone Hsc70. II. Potassium binds specifically in the ATPase active site. J Biol Chem. 1995 Feb 3;270(5):2251–2257. doi: 10.1074/jbc.270.5.2251. [DOI] [PubMed] [Google Scholar]
- Yount R. G., Lawson D., Rayment I. Is myosin a "back door" enzyme? Biophys J. 1995 Apr;68(4 Suppl):44S–49S. [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hermans J. Hydrophilicity of cavities in proteins. Proteins. 1996 Apr;24(4):433–438. doi: 10.1002/(SICI)1097-0134(199604)24:4<433::AID-PROT3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]