Abstract
Most dicot-infecting geminiviruses encode a replication enhancer protein (C3, AL3, or REn) that is required for optimal replication of their small, single-stranded DNA genomes. C3 interacts with C1, the essential viral replication protein that initiates rolling circle replication. C3 also homo-oligomerizes and interacts with at least two host-encoded proteins, proliferating cell nuclear antigen (PCNA) and the retinoblastoma-related protein (pRBR). It has been proposed that protein interactions contribute to C3 function. Using the C3 protein of Tomato yellow leaf curl virus, we examined the impact of mutations to amino acids that are conserved across the C3 protein family on replication enhancement and protein interactions. Surprisingly, many of the mutations did not affect replication enhancement activity of C3 in tobacco protoplasts. Other mutations either enhanced or were detrimental to C3 replication activity. Analysis of mutated proteins in yeast two-hybrid assays indicated that mutations that inactivate C3 replication enhancement activity also reduce or inactivate C3 oligomerization and interaction with C1 and PCNA. In contrast, mutated C3 proteins impaired for pRBR binding are fully functional in replication assays. Hydrophobic residues in the middle of the C3 protein were implicated in C3 interaction with itself, C1, and PCNA, while polar resides at both the N and C termini of the protein are important for C3-pRBR interaction. These experiments established the importance of C3-C3, C3-C1, and C3-PCNA interactions in geminivirus replication. While C3-pRBR interaction is not required for viral replication in cycling cells, it may play a role during infection of differentiated cells in intact plants.
Geminiviruses are single-stranded DNA viruses that infect plants and amplify their genomes by rolling circle and recombination-mediated replication in plant nuclei. Tomato yellow leaf curl virus (TYLCV) is a geminivirus that causes a debilitating disease in cultivated tomato (Solanum lycopersicum), resulting in extensive crop losses in the tropics and subtropics around the world (for a review, see reference 26). It is a member of the Begomovirus genus, which is characterized by its dicotyledonous hosts and whitefly (Bemisia tabaci) vector. There are many related species of TYLCV, including Tomato yellow leaf curl Sardinia virus (TYLCSV) and TYLCV, which were first identified in Italy and Israel, respectively.
Like other begomoviruses, TYLCV infection is initiated by whitefly-mediated transmission of virions from an infected plant to a recipient plant. The single-stranded DNA is converted to a double-stranded form, which then serves as a transcription template for the production of the viral replication proteins C1 (also designated AL1, AC1, and Rep) and C3 (also named AL3, AC3, and REn). The C1 protein acts as a rolling circle initiator to catalyze a site-specific cleavage and rejoining reaction in a conserved hairpin loop in the viral replication origin (22). The C3 protein greatly enhances viral DNA accumulation and symptoms in infected plants (6, 16, 27, 42, 43, 44). Begomoviruses do not encode their own DNA polymerases and, instead, rely on host DNA replication machinery to amplify their genomes in the nuclei of infected plant cells.
Several studies have shown that the begomovirus C1 and C3 proteins bind to viral and host proteins in yeast two-hybrid assays and by copurification of recombinant proteins. Both TYLCSV C3 and C1 proteins interact with proliferating cell nuclear antigen (PCNA) (2), an essential component of the DNA replisome (24). C1 and C3 homologs from Tomato golden mosaic virus (TGMV), also known as AL1 and AL3, interact with each other and themselves and independently interact with the host protein pRBR, the plant retinoblastoma homolog (reviewed in reference 14). TGMV AL1 also binds histone H3, a mitotic kinesin, a novel protein kinase (GRIK) (18), and Ubc9, a component of the sumoylation pathway (3). Tomato leaf curl virus C3 and TGMV AL3 were recently shown to interact with a transcription factor in the NAC family (38).
The functional importance of protein interactions has been demonstrated for TGMV AL1 binding to itself and pRBR. AL1 oligomerization is required for its replication activity and is intimately tied to its site-specific DNA binding activity and interactions with other proteins, such as AL3 and pRBR (19, 33, 40). AL1-pRBR binding is required for infection of differentiated plant cells, which must be reprogrammed for DNA replication (19). In contrast, the importance of C3 protein interactions in viral replication and infection has not been established. To better understand C3 function and the significance of its protein interactions in vivo, we generated a series of 30 mutations encompassing a total of 50 residues across TYLCV C3. We examined the impact of the mutations on C3 replication-enhancing activity and C3 protein interactions with itself, C1, PCNA, and pRBR.
MATERIALS AND METHODS
Plasmids.
A wild-type replicon containing 1.2 copies of TYLCV was constructed in two steps by inserting the 627-bp XmnI-SacI fragment from TYLCV (a Dominican Republic isolate, GenBank accession number AF024715 [28]) into pUC119 linearized with SmaI and SacI to make a partial virus copy containing the 5′ region. TYLCV digested with SacI was then cloned into the SacI site of the partial copy vector to make pTYLC2, a wild-type replicon plasmid. The plasmid pTYLC8, a mutant replicon with a disrupted C3 coding region, was constructed by site-directed mutagenesis with the primer sequence 5′-CTATTTCTATGATTCAAATATCAAATTAGTACTAAATACTCTTAAGAAACGACCAGT to loop out and remove the C3 sequence corresponding to TYLCV positions 1083 and 1229 while leaving the C2 open reading frame (ORF) intact. This construction resulted in a frameshift in the V2 ORF 3′ end, such that the last 3 amino acids of V2 were mutated from ISN to NIK. TYLCV V2 mutants replicate with a preference for double-stranded over single-stranded DNA accumulation (47).
The C3 DNA sequence used for plant expression and mutagenesis was a fragment generated by PCR using pTYLC2 as a template and the primers 5′-CGCATCTATTTCTATGATTCAATATC and 5′-CTGCAATAACCATGGATTCACGC. The PCR product was digested with NcoI, repaired with Klenow, and cloned into pUC119 previously linearized with SmaI. The C3 sequence was subsequently modified by site-directed mutagenesis (20) with the oligonucleotide 5′-CTCAACTTCCGGATTTGGACGAC to generate a silent BspEI site at position TYLCV 1283 to give pTYLC12, which was verified by DNA sequencing. The plasmid pTYLC13 contains the C3 coding sequence from pTYLC12 subcloned into pMON10018, a plant expression cassette with the Figwort mosaic virus 34S promoter (37) and the nopaline synthase 3′ end (10).
The plasmid pTYLC12 was used as a template for C3 site-directed mutagenesis using the primers listed in Table 1. After DNA sequencing, the fragment containing the desired mutation was subcloned into pTYLC13 using the engineered BspEI site in combination with either an NcoI or BamHI site to give the expression cassettes in Table 1.
TABLE 1.
TYLCV C3 mutantsa
| Mutant | Mutated residue(s) | Mutagenesis primer | Mutagenesis plasmid/expression cassette | Yeast AD/BD plasmid |
|---|---|---|---|---|
| a3 | D2, R4 | GTACCCATGGcTagcgcCACAGGGGAAC | pTYLC14/15 | pTYLC116/120 |
| a10 | E7, T10, Q13 | CGCACGGGcACTCATCgCTGCTCCTgcaGCAGAGAATGG | pTYLC32/33 | pTYLC118/122 |
| a20 | F19, W21 | GAATGGCGTTgcaATTgcGGAGATAAAC | pTYLC34/35 | pTYLC119/123 |
| a29 | YFK30 | AACAATCCCgcggccgcCAAGATAACAGACC | pTYLC16/17 | pTYLC104/109 |
| a38 | QR37, F39 | CCACAGCCACgcGgCcgcTCTAATGAACCAC | pTYLC36/37 | |
| a50 | Q49, RF52 | CATCATTTCCATTgcGATcgcAgcCAACCACAACATC | pTYLC38/39 | |
| a53 | QIRFNHN55 | GACATCATTTCCATTgcGgcAgcAgcCgcggcCgcCATCAGGGAAGGTAATG | pTYLC66/67 | pTYLC105/110 |
| a54 | NHN55 | GATAAGATTCgcggcCgcCATAGGAAGGTAATG | pTYLC40/41 | pTYLC126/129 |
| a58 | RK58 | CCACAACATCgcCgccGTAATGGGGA | pTYLC18/19 | |
| a64 | HK64 | GTAATGGGGATTgcCgcATGcTTTCTCCAACTTC | pTYLC42/43 | pTYLC127/130 |
| a68 | F66, NF69 | GATATGAAGTTCTTgcGgcTTTAgcTAGcCTTGGTGTAATTTC | pTYLC44/45 | pTYLC134/135 |
| a69 | F66, NF69, WT73 | Cloned from pTYLC21 and pTYLC45 | pTYLC68/69 | pTYLC106/111 |
| a73 | WT73 | CCGGATTgcGgCGACATTgCAGCCTCAGAC | pTYLC20/21 | pTYLC138/139 |
| a80 | WT73, FR87 | CCGGATTgcGgCGACATTgCAGCCTCAGACTAAGAGTAgcTgcATATGAAGTTC | pTYLC30/31 | pTYLC117/121 |
| a86 | RVFR87 | GTCGTTTCTTAgcAGcaGCTgcATATGAAGTTC | pTYLC70/71 | pTYLC107/112 |
| a87 | FR87 | TAAGAGTAgcTgcATATGAAGTTC | pTYLC22/23 | pTYLC132/133 |
| a93 | VLKYLD95 | AGATATGAAGcTgcTgcGgcTgcAGcTAGTCTTGGTG | pTYLC72/73 | pTYLC108/113 |
| a94 | KY93, D95 | GATTCACAAATGTgcTCTggcCgTCCGGATTTGGAC | pTYLC46/47 | pTYLC136/137 |
| a104 | NN104 | ATTCCATTgcCgcgGTAATCgcAGCAGTGGATC | pTYLC24/25 | |
| a127 | ETHD128 | CAATAAATGTAACAGcAgCTgcTGCTATAAAATATAAATT | pTYLC48/49 | pTYLC146/147 |
| a131 | KY131 | CTCATGATATcgcAgcTAATTTTATTAATTTG | pTYLC26/27 | pTYLC142/143 |
| a133 | K132, Y134 | AAAATATgcATTTgcTTAATTTGAT | pTYLC28/29 | pTYLC144/145 |
| c4 | R4E | CCATGGATTCAgaaACAGGGGAACTCATCACcGCTCCTCAGGC | pTYLC50/51 | |
| c7 | E7K | CACGCACAGGGaAACTCATCACTGCcCCTCAGGCAGAG | pTYLC52/53 | |
| c51 | R51E | CCATTCAGATAgaATTCAACCACAAC | pTYLC54/55 | |
| c57 | R57E | CCACAACATCgaGAAGGTAATGGGaATTCACAAATG | pTYLC56/57 | |
| c58 | RK58EE | CCACAACATCgaGgAGGTAATGGGaATTCACAAATG | pTYLC74/75 | |
| c84 | R84E | GTCGTTTCTTAgaAGTATTTcGATATGAAGTTC | pTYLC58/59 | |
| c107 | R107E | ACAATGTAATCgaAGCAGTGGATC | pTYLC60/61 | |
| c128 | D128K | TAAATGTAACAaAAACTCATGATATcAAATATAAATTTTATTA | pTYLC62/63 | |
| c130 | K130E | CTCATGATATcgAATATAAATTTTATTA | pTYLC64/65 |
The mutants are designated by the amino acid position of the center residue in the mutated sequence and an “a” for alanine mutations or a “c” for charge mutations. Lower case bases indicate altered nucleotides in the mutant.
A plant expression plasmid (pNSB46) containing the wild-type TGMV AL3 open reading frame under the control of the cauliflower mosaic virus 35S promoter and a TGMV A replicon (pMON1565) have been described previously (7, 9). The ΔAL3 replicon is a site-directed mutant that has an 88-bp deletion and a 3-bp insertion in AL3 between positions 1123 and 1211 of TGMV A sequence (44). pNSB46 was modified by site-directed mutagenesis (20) with the primers in Table 2 to give the indicated plant expression cassettes coding for mutant versions of TGMV AL3. The baculovirus expression vectors (Table 2) were created by insertion of a BglII-SacI fragment from the plant expression cassettes into pMON27025 previously digested with BamHI and SacI.
TABLE 2.
TGMV AL3 mutantsa
| Mutant | Mutated residue(s) | Mutagenesis primer | Expression cassette
|
|
|---|---|---|---|---|
| Plant | Insect | |||
| t29 | YFK30 | CAAATCCCTCgcggcCgcGATAATCAGCG | pNSB724 | pNSB751 |
| t53 | QIRFNHN54 | ATATACCACTTAGCaGCcGCgGCCGCcGCCGCCTGAGGAGAGC | pNSB798 | pNSB752 |
| t69 | F66, NF69, WT73 | CCACAAGGCAgcTCTcgcagtaCAAGTAggtgCtACATCGACGA | pNSB799 | pNSB753 |
| t86 | NRFK87 | ACTTATTTAAATgcggccgcATACTTAGTGATG | pNSB800 | pNSB804 |
| t93 | VMLYLE95 | TTAAATACTTAGcGcGgcCgcTGctCAGTTAGGC | pNSB801 | pNSB805 |
Lowercase bases indicate altered nucleotides in the mutant.
Yeast expression cassettes were generated using the pAS1-1 and pACT2 vectors from Clontech (Palo Alto, CA). The NcoI-BamHI C3 fragment of pTYLC13 was cloned into the same sites of pACT2 or pAS1-1 to give pTYLC100 or pTYLC101. pTYLC100 expresses TYLCV C3 fused to the GAL4 activation domain (AD), while pTYLC101 expresses C3 fused to the GAL4 DNA binding domain (DBD). The C1 ORF was a cloned and sequenced PCR fragment from pTYLC2 generated with the primers 5′-CCACAGTCGAATTCCCCGGGCTT and 5′-GGACACCGATTGGATCCAGCATG. The C1 fragment was cloned as a BamHI (trimmed)-SmaI fragment into the SmaI site of pACT2 or pAS1-1 to give pTYLC102 or pTYLC103. pTYLC102 expresses TYLCV C1 as an AD fusion, while pTYLC103 expresses C1 as a DBD fusion. The ZmRBR1 coding sequence fused to the GAL4-DBD of pAS1 has been described before (19). The PCNA clone is an activated domain fusion with PCNA from Solanum lycopersicum designated AD-LePCNA55-255 as described (2).
Replication assays.
Transient replication assays were performed with protoplasts prepared from Nicotiana tabacum suspension cells as described previously (8). Protoplasts were cotransfected with 10 μg of either the TYLCV or TGMV A wild-type replicon or with the mutant ΔC3 or ΔAL3 replicon and 10 μg of the indicated plant expression cassette for C3 or AL3. Total DNA was isolated 72 h after transfection and digested with SacI and DpnI for TYLCV or with BglII and DpnI for TGMV A. DNA was resolved on agarose gels and transferred to nylon membranes. A TGMV A-specific probe was prepared from a 1.8-kb EcoRI-XhoI fragment of TGMV A. A 2.8-kb SacI fragment of pTYLC2 was used to generate a TYLCV-specific probe. Probes were radiolabeled using [α-32P]dATP as described previously (8). Viral DNA was quantified by phosphorimager analysis. Assays were performed a minimum of three times.
Yeast two-hybrid assays.
Interaction between GAL4 fusion proteins was evaluated in Saccharomyces cerevisiae strain Y190 in growth assays after selection of transformed colonies on minimal media lacking leucine and tryptophan (−LW). C3 binding activity with itself was monitored with wild-type C3 fused to the GAL4-DBD and either wild-type or mutant C3 proteins fused to the GAL4-AD. Equivalent results were obtained with wild-type protein fused to the GAL4-AD and mutant proteins fused to the GAL4-DBD. Initial experiments indicated that C1-C3 interaction could be measured only with wild-type or mutant C3 protein fused to the GAL4-DBD and C1 fused to GAL4-AD. pRBR interaction assays gave highest values with AD-C3 and DBD-pRBR fusions. PCNA interaction assays were conducted with AD-PCNA and DBD-C3 protein fusions.
Protein interactions were quantified by measuring yeast growth (17). A 4- or 5-day-old yeast colony (0.1 to 0.2 cm) from either freshly streaked or newly transformed cells was resuspended in 100 μl of sterile water and diluted in three 10-fold serial dilutions. A quantity of yeast suspension (4 μl) from each dilution was inoculated onto −LW medium and onto medium lacking adenine, histidine, leucine, and tryptophan (−AHLW). After incubating plates at 30°C for 6 days, the yeast cell density resulting from each inoculation was determined by counting resuspended cells with a hemacytometer. The number of cells growing on −AHLW medium divided by the number of cells growing on −LW medium gave a measure of relative interaction strength. The growth assays were performed with at least four independent transformants for each two-hybrid combination.
Baculovirus protein expression.
Recombinant proteins were produced in Spodoptera frugiperda SF9 cells using a baculovirus expression system according to published protocols (23, 41). Crude extracts were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunoblotted with anti-AL3 serum (41), and visualized with the ECL detection system (Amersham Pharmacia Biotech, Piscataway, NJ).
RESULTS
TYLCV C3 is required for efficient viral replication and is interchangeable with TGMV AL3.
The TYLCV genome contains six overlapping open reading frames arranged as two divergent transcription units separated by a 300-bp 5′ intergenic region, which contains signals for viral replication and transcription (Fig. 1A). The viral replication proteins are expressed from the complementary sense transcription unit, which is highly conserved across all begomoviruses and resembles the left side of the DNA A component of TGMV (Fig. 1A). Here we use the C1 and C3 designations for the TYLCV replication proteins and the AL1 and AL3 nomenclature to distinguish the corresponding TGMV homologs.
FIG. 1.
TYLCV C3 and TGMV AL3 are functional homologs. (A) The entire genome of TYLCV and the replication-competent A component of TGMV are shown. TYLCV is a 2.8-kb DNA with six open reading frames, while TGMV A is a 2.5-kb DNA with five open reading frames (arrows). Both DNAs have a 5′ intergenic region (white box), which contains the rolling circle replication origin. (B) A linear diagram of TYLCV or TGMV A complementary sense open reading frames shows the region deleted in the ΔC3 and ΔAL3 replicons. (C) Tobacco protoplasts were electroporated with the indicated replicon and expression cassette, and double-stranded and single-stranded viral DNA accumulation were monitored and quantified on DNA gel blots at 72 h posttransfection by phosphorimage analysis. The replicons were wild-type TYLCV (TY, lane 1), ΔC3 (lanes 2 to 4), wild-type TGMV A (TG, lane 5), and ΔAL3 (lanes 6 to 8). The expression cassettes were an empty 35S cassette (lanes 1, 2, 5, and 6), a TYLCV C3 cassette (lanes 3 and 8), and a TGMV AL3 cassette (lanes 4 and 7).
TGMV AL3 is required for high levels of viral DNA accumulation in infected plants and transient replication assays (44). We asked if TYLCV C3 is also necessary for efficient viral DNA replication by comparing the accumulation of a wild-type TYLCV replicon and a ΔC3 replicon lacking the last 49 amino acids of the C3 open reading frame (Fig. 1A). The ΔC3 replicon is equivalent to a mutant TGMV A replicon (ΔAL3) (Fig. 1B) previously shown to accumulate DNA at significantly reduced levels compared to wild-type TGMV A (44). Tobacco protoplasts were transfected with either the wild-type TYLCV or TGMV replicons or the ΔC3 or ΔAL3 mutants, and viral replication was assessed on DNA gel blots 72 h posttransfection. Wild-type TYLCV (Fig. 1C, lane 1) and TGMV A (Fig. 1C, lane 5) replicated efficiently, accumulating both single-stranded and double-stranded DNA forms. The ΔC3 replicon accumulated to a greatly reduced level (Fig. 1C, lane 2), while ΔAL3 replication (Fig. 1C, lane 6) was undetectable at the shown level of sensitivity.
We then asked if the ΔC3 mutation could be complemented by cotransfection of plant expression cassettes corresponding to TYLCV C3 (Fig. 1C, lane 3) or TGMV AL3 (Fig. 1C, lane 4). The levels of double-stranded ΔC3 DNA increased ca. 50-fold in the presence of either expression cassette, to a level similar to the 50-fold dependency previously reported for TGMV but different from the three- to fivefold dependence on C3 seen for Beet curly top virus replication in tobacco cells (16, 44). Accumulation of the ΔAL3 replicon was also enhanced by the presence of an AL3 (Fig. 1C, lane 7) or a C3 (Fig. 1C, lane 8) expression cassette. These results established that, like other begomoviruses, efficient TYLCV DNA accumulation depends on the C3 protein and that the C3 and AL3 proteins of TYLCV and TGMV can substitute for each other in replication complementation assays. These results are similar to earlier studies showing that C3 proteins from diverse geminiviruses are functionally interchangeable (16, 45).
C3 sequence conservation.
TYLCV C3 and TGMV AL3 are small proteins of 134 and 132 residues, respectively, that include an unusually high number of hydrophobic amino acids. C3 and AL3 share 56% sequence identity and 67% similarity (Fig. 2). This level of conservation is consistent with geographical separation of the two viruses during evolution, with TYLCV of Old World lineage and TGMV of New World lineage. C3 protein family members display no significant homology to other proteins, and it has not been possible to infer how C3 enhances geminivirus DNA replication based on similarity to proteins of known activity. To better understand how TYLCV C3 acts as a replication enhancer, we performed comprehensive mutagenesis across the protein to delineate functional regions.
FIG. 2.
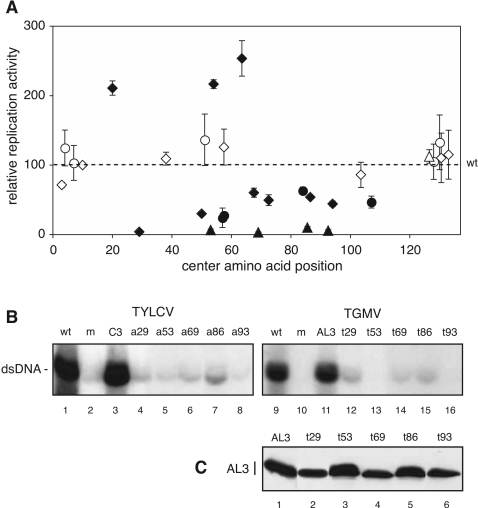
A consensus sequence for the geminivirus C3 protein family. The amino acid sequences of 113 C3 proteins were compared and scored for identical or functionally homologous residues. Amino acid positions that displayed ≥90% conservation are shown as bold capital letters. The uppercase letters designate conservation at the 70 to 90% level, while the lowercase letters indicate 50 to 70% conservation. Positions displaying <50% conservation are marked by an “x.” The sequences of TYLCV C3 and TGMV AL3 are shown below the consensus, with identical or similar residues designated by white letters on a black background. Each of the 30 mutated proteins is shown and labeled below the amino acid sequences. Mutations to alanine are prefixed with an “a,” followed by a center amino acid number within the mutated group. Mutations of two or three amino acids to alanine are indicated as diamonds. Extended mutations with at least four alanine substitutions are indicated by the triangles. Mutations that reversed an amino acid charge are prefixed with a “c” and designated by a circle.
C3 protein sequences from 76 Old World and 33 New World begomoviruses, 4 curtoviruses, and 1 topocuvirus were globally aligned with Clustal V Multiple Sequence Alignment software using a BLOSUM62 matrix (15). The alignment was used to derive a consensus sequence (Fig. 2), in which 6 amino acids display identity across all C3 proteins, 31 residues are conserved in >90% of the sequences, and an additional 23 positions show >90% conservation when similar replacements are considered. The conserved residues are in clusters, with the N terminus displaying stronger overall conservation than the C terminus.
C3 replication enhancement activity accommodates mutagenesis.
Based on the premise that the most conserved amino acids are likely to contribute to function, we used the alignment information to design three types of TYLCV C3 site-directed mutated proteins. The design also considered the annealing stability of the oligonucleotide used for mutagenesis and the creation of a silent restriction site for screening purposes. The first group of mutated C3 proteins contained two or three alanine substitutions in adjacent conserved residues (Fig. 2, diamonds). Some mutated proteins, like a29, contained two or three adjacent amino acids, while others, like a3, contained alanines surrounding unaltered residues. The second type of mutated C3 proteins, like a53, contained four or more alanine substitutions (Fig. 2, triangles). The first and second classes of mutated proteins are designated with an “a” prefix, for alanine, followed by a number which represents the number of the central amino acid within the mutated region. Alanine was chosen because it is a structure-neutral amino acid that is not likely to impact protein folding or result in unstable, poorly expressed proteins. The third class of mutated proteins, like c4, were charge reversals (Fig. 2, circles) and are designated with the prefix “c” for charged. Like the alanine mutations, the number designation in the charge reversal mutations is the central amino acid position of the mutated segment.
The impact of the mutations on C3 function was tested in replication complementation assays using the ΔC3 TYLCV replicon in combination with an expression cassette for each mutated C3 protein. The accumulation of double-stranded viral DNA was quantified on DNA gel blots and compared to DNA accumulation using a wild-type C3 expression cassette (Fig. 3). Each mutated C3 expression cassette was also tested in complementation assays with the TGMV ΔAL3 replicon. In all cases, the mutated C3 proteins displayed equivalent complementation phenotypes with the ΔC3 and ΔAL3 replicons (data not shown).
FIG. 3.
Replication enhancement activities of mutated C3 proteins. (A) Tobacco protoplasts were transfected with the TYLCV ΔC3 replicon and wild-type or mutated C3 expression cassettes. Double-stranded viral DNA accumulation was quantified on DNA gel blots by phosphorimage analysis. Relative replication activity was calculated as the ratio of viral DNA in the presence of the mutated expression cassette to that in the presence of the wild-type C3 expression cassettes and plotted against the central position of the mutated motif. Open symbols indicate proteins that displayed wild-type activity. Filled symbols above the dashed line represent proteins that complemented ΔC3 replication significantly better than wild-type C3, while filled symbols below the dashed line represent mutants that showed significantly reduced complementation. Each data point represents at least three independent experiments, with the bars showing 2 standard errors. The statistical significance of each sample relative to the wild type was determined using Student's t test and a cutoff of P < 0.05. The diamonds, triangles, and circles are defined in the Fig. 2 legend. (B) DNA gel blots showing replication complementation for severely impaired C3/AL3 mutants. Wild-type (wt) TYLCV and TGMV A accumulation are shown in lanes 1 and 9, respectively. Lanes 2 to 8 correspond to the ΔC3 replicon, and lanes 10 to 16 contain the ΔAL3 replicon. The expression cassettes in the first panel were an empty 35S cassette (lanes 1 and 2), a wild-type C3 cassette (lane 3), or the mutant C3 cassettes a29 (lane 4), a53 (lane 5), a69 (lane 6), a86 (lane 7), and a93 (lane 8). The expression cassettes in the second panel were an empty 35S cassette (lanes 9 and 10), a wild-type AL3 cassette (lane 11), or the mutant AL3 cassettes t29 (lane 12), t53 (lane 13), t69 (lane 14), t86 (lane 15), and t93 (lane 16). (C) Immunoblot showing TGMV AL3 protein expression in baculovirus-infected SF9 cells. Wild-type AL3 (lane 1), t29 (lane 2), t53 (lane 3), t69 (lane 4), t86 (lane 5), and t93 (lane 6) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected using an anti-AL3 antibody.
Of the 16 mutated proteins with two to three alanine substitutions, 7 behaved like wild-type C3 (Fig. 3A, open diamonds). Mutated proteins a20, a54, and a64 performed significantly better than wild-type C3, and mutated proteins a50, a68, a73, a87, and a94 were significantly impaired in the replication complementation assay (Fig. 3A, filled diamonds). Only the mutated protein a29, in which YFK28-30 was changed to three alanines, abolished C3 replication enhancement activity, suggesting that the YFK residues play a pivotal role in C3 function or regulation of its activity.
Because many of the alanine substitutions in conserved residues had no effect or caused only a moderate reduction in C3 activity in replication complementation assays, we asked if more severe mutations would cause phenotypic effects. Our first approach was to combine or extend the five mutations (a50, a68, a73, a87, and a94) that reduced C3 function to generate a53, a69, a86, and a93 (Fig. 2). The proteins a53, a69, and a93 exhibited negligible activity in replication complementation assays (Fig. 3A, filled triangles, and B, lanes 4 to 6 and 8). Although the a86 protein was also greatly impaired in C3 activity, it reproducibly supported more viral DNA accumulation than the other proteins (Fig. 3B, lane 7). The reduced activity of the mutated proteins did not reflect simply an increased number of alanine substitutions, because protein a127, which contains four alanine changes, displayed wild-type activity in the replication complementation assays. These results indicated that the region between positions 49 and 96 contains residues important for C3 replication enhancement.
We also generated nine mutated C3 proteins that contain charge reversals (E to K, R to E, or K to E) at conserved charged residues. The charge reversals (in c57, c58, c84, and c107) that impaired C3 function were changes to amino acids that are predicted to be exposed to the solvent in the middle of a hydrophobic cluster in the central region of C3 (Fig. 3A, filled circles). (Solvent accessibility was predicted by PHDacc analysis [36].) In contrast, charge reversals (in c4, c7, c128, and c130) in solvent-accessible residues in the N and C termini had no impact on C3 function (Fig. 3A, open circles).
The R57E mutation contained in c57 and c58 resulted in proteins with reduced capacities to complement ΔC3 replication. This arginine is 100% conserved among all C3 sequences, and amino acid 58 is always a positive charge. An R57A substitution (a58) had no effect on C3 function, indicating that charge reversal but not loss of charge is inhibitory. This suggests that the side group may be part of a salt bridge with another amino acid in C3 or in a different protein. Consistent with this idea, we observed no effect of the charge reversal in c51, which is modified at an arginine that is predicted to be buried.
The same residues are important for C3 and AL3 activity.
There were five highly impaired alanine-scanning mutated C3 proteins that complemented ΔC3 minimally or not at all (Fig. 3B). We mutated the equivalent amino acids in TGMV AL3 and tested the mutated AL3 proteins in complementation assays using the ΔAL3 replicon (Fig. 3B). (The TGMV AL3 versions were designated t29, t53, t69, t86, and t93 in agreement with the TYLCV nomenclature.) In these assays, wild-type TGMV AL3 complemented the defect in ΔAL3, resulting in wild-type levels of viral DNA accumulation (Fig. 3B, lane 11). In contrast, the mutated AL3 proteins were severely impaired, resulting in greatly reduced levels of viral DNA (Fig. 3B, lanes 12 to 16). TGMV AL3 proteins t29, t69, and t86 supported a very low but detectable amount of viral DNA accumulation (Fig. 3B, lanes 12, 14, and 15), whereas t53 and t93 had no detectable activity in the complementation assays (Fig. 3B, lanes 13 and 16).
To verify that the loss of the AL3 activity was not due to poor protein production, we monitored the accumulation of the mutated proteins in a baculovirus expression system on immunoblots using an anti-AL3 antibody (41). Wild-type TGMV AL3 (Fig. 3C, lane 1) and all of the mutated proteins (Fig. 3C, lanes 2 to 6) were expressed at similar levels in insect cells, indicating that the loss of activity most likely was due to impaired AL3 function and probably did not reflect reduced expression or instability of the mutated proteins. We were unable to perform equivalent experiments in tobacco protoplasts because of the low transfection efficiency of the C3 expression cassettes.
We also asked if the mutated AL3 proteins act in a trans-dominant negative manner to inhibit the replication of wild-type TGMV. The five mutated AL3 expression cassettes (Fig. 3B) were tested in tobacco protoplasts cotransfected with a TGMV A replicon that produced wild-type AL3. A similar strategy was used previously to identify AL1 trans-dominant mutants (32). However, even with a 40-fold excess of expression cassette to input replicon DNA, we were unable to detect inhibition of TGMV replication by any of the mutant AL3 proteins (data not shown). The same results were seen in protoplast assays containing mutated C3 expression cassettes and a wild-type TYLCV replicon (data not shown).
TYLCV C3 protein interactions.
TGMV AL3 participates in protein interactions with itself, AL1, and the host factor pRBR (40). PCNA was retrieved in a library screen for TYLCSV C3 partners (2). We used yeast two-hybrid assays to ask if TYLCV C3 also forms oligomers and interacts with C1, pRBR, and PCNA. Positive interactions were indicated by yeast growth in the absence of histidine and adenine. None of the protein fusions was able to activate histidine/adenine auxotrophy alone. Cells transformed with plasmids encoding AD-C3 and DBD-C3 or DBD-pRBR fusions grew in the absence of adenine and histidine, indicative of positive protein interactions (Fig. 4A). Similarly, cells cotransformed with a DBD-C3 cassette and either AD-C1 or AD-PCNA cassettes grew on medium lacking histidine and adenine. The specificity of the different interactions was established by the failure of selected mutated TYLCV C3 proteins fused to the GAL4-AD to support yeast growth in the presence of DBD cassettes for wild-type C3 or pRBR. Likewise, C3-PCNA and C3-C1 binding specificity were shown by the lack of yeast growth with certain mutated DBD-C3 fusions. The replication-inactive protein a29 did not bind to C3, C1, or PCNA, while the replication-competent protein a133 failed to bind to pRBR. Together, these results established that, like other begomovirus AL3/C3 proteins, TYLCV C3 interacts with itself, C1, PCNA, and pRBR and that the interactions are differentially impacted by the C3 mutations.
FIG. 4.
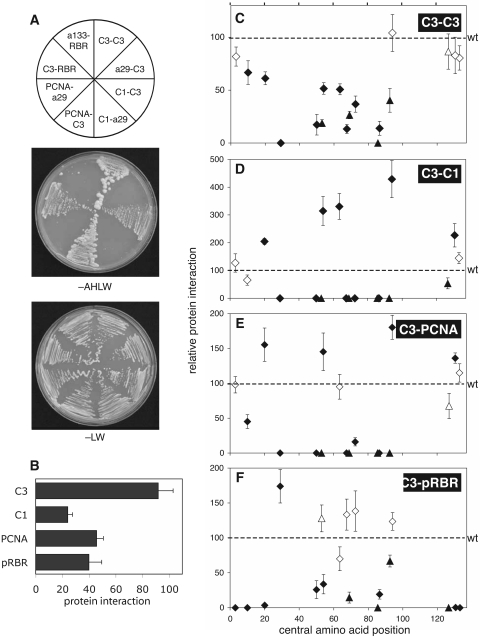
TYLCV C3 protein interactions. (A) C3 protein interactions were measured in yeast two-hybrid assays. Wild-type or mutated C3 sequence was fused to the GAL4 activation domain (indicated by the first protein listed in each protein pair) or the GAL4 DNA binding domain (second protein listed). Transformants were selected for protein interactions on medium lacking adenine, histidine, leucine, and tryptophan (−AHLW). Medium lacking leucine and tryptophan (−LW) selected for the input plasmids only. Yeast cotransfected with a wild-type C3 cassette and a cassette corresponding to C3, C1, PCNA, or pRBR grew on both −AHLW and −LW media. Yeast cotransfected with certain mutant C3 cassettes and a C3, C1, PCNA, or pRBR cassette grew only on −LW medium. (B) The strength of the interactions shown for wild-type C3 in panel A was quantified in growth assays. Yeast growth on −AHLW medium was normalized to growth on −LW medium as a measure of strength. (C to F) Growth assays for yeast cotransfected with indicated C3 mutants and (C) wild-type C3, (D) C1, (E) PCNA, and (F) pRBR. Growth in the presence of mutated C3 proteins was normalized to growth in the presence of wild-type C3, which was set at 100. Proteins that were not significantly different than wild type, determined as in Fig. 3, are shown as open symbols. Significantly different proteins are marked with filled symbols. Each data point reflects at least four independent experiments, with the bars representing 2 standard errors. The diamonds and triangles are defined in the Fig. 2 legend.
We adapted a quantitative yeast growth assay to measure the strengths of the various C3 protein interactions (17). The more commonly used β-galactosidase assay lacked the sensitivity required to measure C3 interactions (unpublished data). In the growth assay, interaction strength is quantified as the ratio of the cell density of transformants grown on adenine- and histidine-deficient medium (−AHLW) to growth on medium selecting for only the two-hybrid plasmids (−LW). With wild-type C3-C3 interaction, nearly as many cells grew without adenine and histidine as with the added nutrients, indicative of a strong interaction (Fig. 4B). The other interactions were weaker, demonstrated by smaller colony size and slower growth on −AHLW medium (Fig. 4B).
C3 oligomerization is necessary for replication enhancement.
We used the growth assay to ask if the inability of certain alanine substitutions to fully complement ΔC3 can be attributed to defects in protein interactions. We also tested alanine replacements clustered in the N and C termini that were wild-type for replication enhancement and mutated proteins with enhanced replication complementation phenotypes (Table 3).
TABLE 3.
Functional properties of mutant TYLCV C3 proteinsa
| Mutant | Replication | C3-C3 | C3-C1 | C3-PCNA | C3-pRBR |
|---|---|---|---|---|---|
| a3 | wt | wt | wt | wt | Inactive |
| c4 | wt | ||||
| c7 | wt | ||||
| a10 | wt | Reduced | wt | Reduced | inactive |
| a20 | Enhanced | Reduced | Enhanced | Enhanced | inactive |
| a29 | Inactive | Inactive | Inactive | inactive | Enhanced |
| a38 | wt | ||||
| a50 | Reduced | Reduced | Inactive | inactive | Reduced |
| c51 | wt | ||||
| a53 | inactive | Reduced | Inactive | inactive | wt |
| a54 | Enhanced | Reduced | Enhanced | Enhanced | Reduced |
| c57 | Reduced | ||||
| a58 | wt | ||||
| c58 | Reduced | ||||
| a64 | Enhanced | Reduced | Enhanced | wt | wt |
| a68 | Reduced | Reduced | Inactive | inactive | wt |
| a69 | Inactive | Reduced | Inactive | Inactive | Reduced |
| a73 | Reduced | Reduced | Inactive | Reduced | wt |
| c84 | Reduced | ||||
| a86 | Trace | Inactive | Inactive | Inactive | Inactive |
| a87 | Reduced | Reduced | Inactive | Inactive | Reduced |
| a93 | Inactive | Reduced | Inactive | Inactive | Reduced |
| a94 | Reduced | wt | Enhanced | Enhanced | wt |
| a104 | wt | ||||
| c107 | Reduced | ||||
| a127 | wt | wt | Reduced | wt | Inactive |
| c128 | wt | ||||
| c130 | wt | ||||
| a131 | wt | wt | Enhanced | Enhanced | Inactive |
| a133 | wt | wt | wt | wt | Inactive |
All activities classified as reduced or enhanced showed P < 0.05 when compared to wild-type C3 using a Students t test. wt, wild type.
Thirteen of the 18 tested mutated proteins were significantly impaired for C3 oligomerization (Fig. 4C, filled symbols), with 6 displaying less than 50% of wild-type activity and 2 with no detectable activity. The mutations that reduced C3 oligomerization by at least 50% were also deleterious to C3 function in complementation assays (cf. Fig. 3A and 4C). The two proteins, a29 and a86, that were inactive for C3 oligomerization were also inactive or retained only trace replication enhancement activity (Table 3). These results suggested that although the efficiency of C3-C3 interactions can be reduced, a wild-type level of oligomerization is necessary for optimal replication enhancement.
All of the mutations between residues 27 and 91 that impaired C3 function also reduced C3 oligomerization (Fig. 4C). This region mirrors the previously demonstrated TGMV AL3 oligomerization region between amino acids 35 and 112 (40), indicating that C3 and AL3 utilize similar oligomerization mechanisms. Because the alanine substitutions involved two or more amino acids, it is not possible to define precisely which of the targeted amino acids is most important in C3 self-interaction. However, our data indicated that phenylalanine replacements were favored in disrupting C3 oligomerization. Phenylalanine was modified at one or more positions in 6 of the 11 impaired proteins with mutations in the central region, representing 25% of the modified residues. In contrast, phenylalanine accounts for only 6% of the total amino acids in the C3 protein.
C3 interactions with PCNA and C1 but not with pRBR are required for replication enhancement.
Ten of the mutated C3 proteins were significantly impaired for C1 binding, with nine displaying no detectable activity (Fig. 4D, filled symbols below the dashed line). Interestingly, five mutated proteins were more active for C1 binding than wild-type C3 (Fig. 4D, filled symbols above the dashed line). Our ability to detect enhanced C1 binding probably reflected that C3-C1 binding is weaker than C3 oligomerization. Three proteins (a20, a54, and a64) with enhanced C3-C1 binding also enhanced replication significantly more than wild-type C3 (Table 3). These mutated proteins showed reduced C3 oligomerization. Two mutants (a94 and a131) that had wild-type C3 oligomerization and increased C3-C1 binding were reduced and wild type, respectively, for replication enhancement, indicating that enhanced C1 binding in combination with wild-type C3 oligomerization is not sufficient for optimal C3 function. All of the mutated C3 proteins that were inactive in replication assays were also inactive for C1 interaction. However, four proteins (a50, a68, a73, and a87) were inactive for C1 binding in the yeast two-hybrid assays but retained replication activity as high as 50% of wild type (cf. Fig. 3A and 4D).
Two regions containing overlapping mutations may have uncovered elements that modulate C3-C1 binding. The first cluster contains mutations made in the a50, a54, and a53 proteins (Fig. 2). The protein a53 is an expanded version of a50 and a54. C3-C1 interaction was impaired when amino acids 49, 51, and 52 were mutated in either a50 or a53, but when three immediately adjacent amino acids, NHN54-56, were depolarized to alanines in a54, C1 interaction was enhanced. A similar situation occurred with a87, a86, a93, and a94, which are located within a putative α-helical domain. Mutation of amino acids FR86,87 inactivated C1 interaction whether alone in a87 or as part of the extended mutation contained in a86. Mutation of LKYLD91-95 abolished C1 interaction, while modification of only KY92,93 and K95 in the protein a94 enhanced C1 interaction (Fig. 2 and Table 3).
Ten of the mutated C3 proteins were impaired for PCNA binding (Fig. 4E, filled symbols below the dashed line), with eight displaying no detectable activity. Three mutated proteins were more active for PCNA binding than wild-type C3 (Fig. 4E, filled symbols above the dashed line). Even though wild-type C3 binding to PCNA was slightly stronger than to C1 (Fig. 4B), the correlation between the PCNA and C1 data sets was 0.91. Only mutant a10 had a reduced C3-PCNA interaction without an impact on C3-C1 binding. Conversely, only the mutation within a127 reduced C3-C1 interaction with no effect on C3-PCNA binding. Replication-inactive proteins were also inactive for C3-PCNA binding. However, like the situation with C1, several mutated versions of C3 appeared to be inactive for PCNA binding but could still support replication enhancement (Table 3).
Twelve of the mutated C3 proteins were impaired for pRBR binding, with seven displaying no detectable activity (Fig. 4F, filled symbols below the dashed line). One altered protein was significantly more active for pRBR binding than wild-type C3 (Fig. 4F, filled symbol above the dashed line). pRBR binding was most susceptible to mutations in the N and C termini of C3 (Fig. 4F) that alter charged amino acids, with the exception of a20 (Fig. 2). These mutations had no impact on replication enhancement and were functional for C3-C3 interactions (Table 3), indicating that the inhibitory effects on pRBR binding were specific. Two replication-inactive proteins, a29 and a53, were enhanced and wild type, respectively, for pRBR interaction. These results showed that unlike C1 and PCNA, pRBR binding is not necessary for C3 replication enhancement activity in cultured cells.
DISCUSSION
The C3 protein acts as a replication enhancer during geminivirus DNA accumulation. Mutant C3 replicons can replicate, albeit to low levels, in single cells and plants. C3 is not required for C1 DNA binding, cleavage/ligation, or topoisomerase activities in vitro (4, 22, 33, 34) or for C1-mediated release of unit-length viral DNA from replicating bacterial plasmids (35, 39). Members of the Mastrevirus genus and Nanovirus family, which also replicate via a rolling circle mechanism, do not encode C3 homologs (12, 13). Together, these observations indicate that C3 is not essential for C1-mediated initiation or host-catalyzed DNA synthesis during rolling circle replication but do not provide insight into how C3 influences the efficiency of begomovirus and curtovirus replication. The C3 family displays no significant homology to proteins with known functions (29), and no catalytic activity has been associated with C3 proteins. As a consequence, the mechanism whereby C3 enhances viral DNA accumulation has been elusive. The studies reported here demonstrate that C3 interactions with itself, C1, and PCNA contribute to replication enhancement.
Two lines of evidence support the hypothesis that C3 acts primarily through protein interactions. First, C3 replication enhancement activity is highly tolerant to mutation, while C1 activity is very susceptible to modification. This difference probably reflects different modes of action for the two viral replication proteins. C1 has enzymatic functions that are very sensitive to mutation (30-33). In contrast, C3 protein interactions are likely to be difficult to abolish by site-directed mutation because of the involvement of multiple contacts and nonspecific stabilization by residues outside of the binding site (32, 46). Second, mutated C3 proteins that are impaired for replication enhancement are also impaired for interaction with wild-type C3, C1, and/or PCNA. C1 and PCNA may be part of the viral replisome, and it is likely that interactions with other proteins, like C3, will influence their activities during viral replication. However, there were several cases where negative protein interactions did not bring about a total loss of replication competency. One possibility is that the yeast assay does not detect weak interaction with some C3 mutants. Alternatively, protein interactions might occur through interactions with a bridging protein, like pRBR. This idea is supported by the observation that the proteins that could not bind C1 or PCNA and were competent for replication enhancement retained both pRBR binding and C3-C3 oligomerization, while the mutants that had impaired C1 and PCNA binding and were impaired for replication enhancement lost either C3 or pRBR binding activity or both. Viral proteins are commonly multifunctional; therefore, it is likely that C3 enhances replication through multiple mechanisms, some of which are independent of C1, PCNA, or even C3 oligomerization.
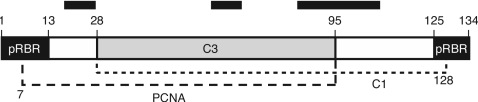
C3 oligomerization was affected by changes in the center of the protein between amino acids 28 and 95 (Fig. 5). Many of the mutations were pleiotropic for C3 oligomerization and binding to C1 and PCNA, suggesting that oligomerization might be a prerequisite for C1 and PCNA interactions. However, TGMV AL1 binds monomeric AL3 (40), and one C3 mutation, contained in a10, reduced C3 oligomerization without affecting C1 binding. Another C3 mutation, contained in a64, reduced C3-C3 interactions but not C3-PCNA binding. There were several mutated C3 proteins that were reduced in C3 oligomerization but retained pRBR binding. Hence, it is not likely that C3 oligomerization is essential for binding to any of the partners tested here. However, our data suggest that C3 oligomerization is an important component of its replication enhancement activity. Multimerization provides multiple sites for protein binding when two or more protein binding sites overlap as in the case of C3 binding to C1 and PCNA. Multimerization can also lead to the assembly of large multicomponent complexes with different activities dependent upon composition (25). One can envision a high-molecular-weight complex containing C3, C1, PCNA, and other host factors required for viral DNA replication. The existence of a multimeric C3 complex is supported by gel filtration analysis of native TGMV AL3, which fractionated with a complex of ≥100 kDa (data not shown).
FIG. 5.
Model of C3 protein interactions. TYLCV C3 consists of 134 amino acids containing three hydrophobic clusters shown as filled boxes above the rectangular diagram. pRBR interaction with C3 was decreased by mutations clustered in the first 13 and final 9 amino acids of C3 (shown as solid regions). C3 homo-oligomerization was adversely affected by mutations in the middle of the protein, shown shaded within the rectangular region. PCNA interaction with C3 (dashed line) was impacted by mutations near the beginning of C3 to amino acid 95. C1 interaction with C3 (dotted line) was sensitive to mutations between amino acids 28 and 128.
C1 and PCNA binding were impacted by mutations in overlapping regions between amino acids 28 and 128 and 7 and 95, respectively (Fig. 5). For both PCNA and C1, conversion of phenylalanine, leucine, or tryptophan to alanine abolished C3 binding, while conversion of polar residues to alanine enhanced interaction. These data suggest that C3 interacts with C1 and PCNA through nonpolar and aromatic contacts. The combined PCNA and C1 binding region contains three hydrophobic areas (Fig. 5) that would be shielded from water by protein interactions (21). Neither of the known canonical PCNA binding sequences (24) is present in C3, but there are numerous aromatic residues in C3 that could interact with the four aromatic amino acids near the beginning of the minimal domain in PCNA required for TYLSCV C3 binding (2). The region of TGMV AL1 (residues 101 to 119) that specifically interacts with AL3 contains a tryptophan and phenylalanine. The requirement for AL1 multimerization for AL3 interaction (40) may reflect the need for multiple aromatic residues for stable binding.
C3 contacts with pRBR map to the N and C termini and minimally overlap those involved in oligomerization and interactions with C1 and PCNA (Fig. 5). C3-pRBR binding also differs from the other protein interactions in that it is not essential for replication enhancement in cultured plant cells. The high level of conservation of the C3 amino acids that alter pRBR binding suggests that these residues play an essential function not uncovered in the protoplast assays. The requirements for viral replication in actively cycling suspension cells are likely to only partially reflect those in mature cells of infected plants. This idea is consistent with the proposed role for pRBR in modulating plant development as well as the cell division cycle. In plants, C3 could impact pRBR function by direct binding and/or by modulating its interaction with C1, which is necessary to induce mature plant cells to produce the requisite host replication machinery for viral DNA synthesis (5). The hypothesis can be addressed only in whole-plant infectivity assays using viral replicons carrying C3-pRBR binding mutations. However, these studies are complicated by the overlap of the ends of the C3 gene with the essential C2 gene and the bidirectional polyadenylation site in the viral replicon.
There are some differences in the predicted binding regions for TYLCV C3 reported here and those characterized earlier for TGMV AL3 (40). However, there are two important differences between the TYLCV C3 and TGMV AL3 studies. The C3 analysis involved the characterization of site-directed mutants in yeast dihybrid assays, whereas the AL3 analysis involved truncated proteins expressed in insect cells. Alanine substitution may be more detrimental to protein interactions because alanine is rarely found at protein-protein interfaces (11). Earlier experiments with AL1 showed that the yeast system is more effective at detecting impaired protein interactions (32), because the high level of mutated protein expression in insect cells can stabilize weak interactions or drive nonspecific binding. This may explain why the earlier study showed TGMV AL1 and pRBR binding to an AL3 truncation corresponding to amino acids 1 to 37 while the present work indicates that C3-C1 and C3-pRBR binding involves amino acids outside of that region (40).
A recent study proposed that TLCV C3 binding to a transcription factor, SlNAC1, and induction of its expression is the mechanism whereby C3 enhances geminivirus replication (38). However, it is unlikely that C3-SlNAC1 interactions are sufficient for C3 replication enhancement activity. C3 mutant replicons typically are constructed by deleting 3′ sequences that do not overlap with the C2 open reading frame to yield truncated proteins of 83 to 111 amino acids (16, 27, 42, 43, 44). This region of C3 retains the capacity to bind to SlNAC1 (38), but the C3 mutant replicons support little if any detectable replication in protoplasts and are severely attenuated in infectivity assays in planta. Instead, our data suggest that the loss of replication enhancement activity by the truncated C3 proteins reflects their inability to form oligomers with themselves, C1, and/or PCNA. In addition, C3 activity is required for efficient geminivirus replication in suspension cells, which already contain abundant SlNAC1 orthologues (48). Therefore, induction and binding of an SlNAC1 orthologue is not necessary or sufficient for C3 replication enhancement activity.
One application of the inactive mutated C3 proteins would be to express the inactivated C3 proteins in transgenic plants to interfere with geminivirus infection. The virus-nonspecific nature of C3 replication-enhancing activity suggests that inactivated C3 proteins have the potential to confer broad-based geminivirus resistance. This is in contrast to mutated or truncated C1 proteins, which are most effective against closely related viruses, consistent with the strong specificity of C1 proteins for their cognate viral genomes (1, 9). However, because none of the inactive C3 proteins displayed trans-dominant negative activity in cultured cells, it will be necessary to evaluate their interference potential directly in plants.
Acknowledgments
We thank the Monsanto Corporation for the gift of TYLCV DNA and pMON10018. The PCNA clone AD-LePCNA55-255 was a gift from Eduardo Bejarono. Carla Mattos contributed helpful suggestions in the writing of the manuscript. Dominique Robertson provided critical reading of the manuscript.
The research was supported by grants to L.H.-B. from USDA-NRI (2001-35319-10856), the Monsanto Corporation, and the North Carolina Biotechnology Association (2000-ARG-0021).
REFERENCES
- 1.Brunetti, A., R. Tavazza, E. Noris, A. Lucioli, G. P. Accotto, and M. Tavazza. 2001. Transgenically expressed T-Rep of tomato yellow leaf curl Sardinia virus acts as a trans-dominant-negative mutant, inhibiting viral transcription and replication. J. Virol. 75:10573-10581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castillo, A. G., D. Collinet, S. Deret, A. Kashoggi, and E. R. Bejarano. 2003. Dual interaction of plant PCNA with geminivirus replication accessory protein (REn) and viral replication protein (Rep). Virology 312:381-394. [DOI] [PubMed] [Google Scholar]
- 3.Castillo, A. G., L. J. Kong, L. Hanley-Bowdoin, and E. R. Bejarano. 2004. Interaction between a geminivirus replication protein and the plant sumoylation system. J. Virol. 78:2758-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desbiez, C., C. David, A. Mettouchi, J. Laufs, and B. Gronenborn. 1995. Rep protein of tomato yellow leaf curl geminivirus has an ATPase activity required for viral DNA replication. Proc. Natl. Acad. Sci. USA 92:5640-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egelkrout, E. M., D. Robertson, and L. Hanley-Bowdoin. 2001. Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13:1437-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elmer, J. S., L. Brand, G. Sunter, W. E. Gardiner, D. M. Bisaro, and S. G. Rogers. 1988. Genetic analysis of the tomato golden mosaic virus. II. The product of the AL1 coding sequence for is required for replication. Nucleic Acids Res. 16:7043-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elmer, J. S., G. Sunter, W. E. Gardiner, L. Brand, C. K. Browning, D. M. Bisaro, and S. G. Rogers. 1988. Agrobacterium-mediated inoculation of plants with tomato golden mosaic virus DNAs. Plant Mol. Biol. 10:225-234. [DOI] [PubMed] [Google Scholar]
- 8.Fontes, E. P., P. A. Eagle, P. S. Sipe, V. A. Luckow, and L. Hanley-Bowdoin. 1994. Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J. Biol. Chem. 269:8459-8465. [PubMed] [Google Scholar]
- 9.Fontes, E. P., H. J. Gladfelter, R. L. Schaffer, I. T. Petty, and L. Hanley-Bowdoin. 1994. Geminivirus replication origins have a modular organization. Plant Cell 6:405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraley, R. T., S. G. Rogers, R. B. Horsch, E. A. Eichholtz, J. S. Flick, C. L. Fink, N. L. Hoffmann, and S. C. Woo. 1983. Expression of bacterial genes in plant cells. Proc. Natl. Acad. Sci. USA 80:4803-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaser, F., D. M. Steinberg, I. A. Vakser, and N. Ben-Tal. 2001. Residue frequencies and pairing preferences at protein-protein interfaces. Proteins 43:89-102. [PubMed] [Google Scholar]
- 12.Gronenborn, B. 2004. Nanoviruses: genome organisation and protein function. Vet. Microbiol. 98:103-109. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez, C. 1999. Geminivirus DNA replication. Cell. Mol. Life Sci. 56:313-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanley-Bowdoin, L., S. B. Settlage, and D. Robertson. 2004. Reprogramming plant gene expression: a prerequisite to geminivirus DNA replication. Mol. Plant Pathol. 5:149-156. [DOI] [PubMed] [Google Scholar]
- 15.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hormuzdi, S. G., and D. M. Bisaro. 1995. Genetic analysis of beet curly top virus: examination of the roles of L2 and L3 genes in viral pathogenesis. Virology 206:1044-1054. [DOI] [PubMed] [Google Scholar]
- 17.Humphrey, J. S., S. Salim, M. R. Erdos, F. S. Collins, L. C. Brody, and R. D. Klausner. 1997. Human BRCA1 inhibits growth in yeast: potential use in diagnostic testing. Proc. Natl. Acad. Sci. USA 94:5820-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong, L., and L. Hanley-Bowdoin. 2002. A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14:1817-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong, L. J., B. M. Orozco, J. L. Roe, S. Nagar, S. Ou, H. S. Feiler, T. Durfee, A. B. Miller, W. Gruissem, D. Robertson, and L. Hanley-Bowdoin. 2000. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:482-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 22.Laufs, J., W. Traut, F. Heyraud, V. Matzeit, S. G. Rogers, J. Schell, and B. Gronenborn. 1995. In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc. Natl. Acad. Sci. USA 92:3879-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maga, G. H. U. 2003. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 116:3051-3060. [DOI] [PubMed] [Google Scholar]
- 25.Marianayagam, N. J., and J. M. Matthews. 2004. The power of two: protein dimerization in biology. Trends Biochem. Sci. 29:618-625. [DOI] [PubMed] [Google Scholar]
- 26.Moriones, E., and J. Navas-Castillo. 2000. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 71:123-134. [DOI] [PubMed] [Google Scholar]
- 27.Morris, B., K. Richardson, P. Eddy, X. C. Zhan, A. Haley, and R. Gardner. 1991. Mutagenesis of the AC3 open reading frame of African cassava mosaic virus DNA A reduces DNA B replication and ameliorates disease symptoms. J. Gen. Virol. 72:1205-1213. [DOI] [PubMed] [Google Scholar]
- 28.Nakhla, M. K., D. P. Maxwell, M. G. Carvalho, and R. L. Gilbertson. 1994. Widespread occurrence of the eastern Mediterranean strain of tomato yellow leaf curl geminivirus in tomatoes in the Dominican Republic. Plant Dis. 78:926. [Google Scholar]
- 29.Obenauer, J. C., C. L. Cantley, and M. B. Yaffe. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31:3635-3641. [DOI] [PMC free article] [PubMed]
- 30.Orozco, B. M., and L. Hanley-Bowdoin. 1998. Conserved sequence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J. Biol. Chem. 273:24448-24456. [DOI] [PubMed] [Google Scholar]
- 31.Orozco, B. M., and L. Hanley-Bowdoin. 1996. A DNA structure is required for geminivirus origin function. J. Virol. 270:148-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orozco, B. M., L. J. Kong, L. A. Batts, S. Elledge, and L. Hanley-Bowdoin. 2000. The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J. Biol. Chem. 275:6114-6122. [DOI] [PubMed] [Google Scholar]
- 33.Orozco, B. M., A. B. Miller, S. B. Settlage, and L. Hanley-Bowdoin. 1997. Functional domains of a geminivirus replication protein. J. Biol. Chem. 272:9840-9846. [DOI] [PubMed] [Google Scholar]
- 34.Pant, V., D. Gupta, N. R. Choudhury, V. G. Malathi, A. Varma, and S. K. Mukherjee. 2001. Molecular characterization of the Rep protein of the blackgram isolate of Indian mungbean yellow mosaic virus. J. Gen. Virol. 82:2559-2567. [DOI] [PubMed] [Google Scholar]
- 35.Rigden, J. E., I. B. Dry, L. R. Krake, and M. A. Rezaian. 1996. Plant virus DNA replication processes in Agrobacterium: insight into the origins of geminiviruses? Proc. Natl. Acad. Sci. USA 93:10280-10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rost, B., and C. Sander. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19:55-72. [DOI] [PubMed] [Google Scholar]
- 37.Sanger, M., S. Daubert, and R. M. Goodman. 1990. Characteristics of a strong promoter from figwort mosaic virus: comparison with the analogous 35S promoter from cauliflower mosaic virus and the regulated mannopine synthase promoter. Plant Mol. Biol. 14:433-443. [DOI] [PubMed] [Google Scholar]
- 38.Selth, L. A., S. C. Dogra, M. S. Rasheed, H. Healy, J. W. Randles, and M. A. Rezaian. 2005. A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell 17:311-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selth, L. A., J. W. Randles, and M. A. Rezaian. 2002. Agrobacterium tumefaciens supports DNA replication of diverse geminivirus types. FEBS Lett. 516:179-182. [DOI] [PubMed] [Google Scholar]
- 40.Settlage, S. B., A. B. Miller, W. Gruissem, and L. Hanley-Bowdoin. 2001. Dual interaction of a geminivirus replication accessory factor with a viral replication protein and a plant cell cycle regulator. Virology 279:570-576. [DOI] [PubMed] [Google Scholar]
- 41.Settlage, S. B., A. B. Miller, and L. Hanley-Bowdoin. 1996. Interactions between geminivirus replication proteins. J. Virol. 70:6790-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stanley, J., J. R. Latham, M. S. Pinner, I. Bedford, and P. G. Markham. 1992. Mutational analysis of the monopartite geminivirus beet curly top virus. Virology 191:396-405. [DOI] [PubMed] [Google Scholar]
- 43.Sung, Y. K., and R. H. A. Coutts. 1995. Mutational analysis of potato yellow mosaic geminivirus. J. Gen. Virol. 76:1773-1780. [DOI] [PubMed] [Google Scholar]
- 44.Sunter, G., M. D. Hartitz, S. G. Hormuzdi, C. L. Brough, and D. M. Bisaro. 1990. Genetic analysis of tomato golden mosaic virus: ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology 179:69-77. [DOI] [PubMed] [Google Scholar]
- 45.Sunter, G., D. C. Stenger, and D. M. Bisaro. 1994. Heterologous complementation by geminivirus AL2 and AL3 genes. Virology 203:203-210. [DOI] [PubMed] [Google Scholar]
- 46.Tansey, W. P., S. Ruppert, R. Tjian, and W. Herr. 1994. Multiple regions of TBP participate in the response to transcriptional activators in vivo. Genes Dev. 8:2756-2769. [DOI] [PubMed] [Google Scholar]
- 47.Wartig, L., A. Kheyr-Pour, E. Noris, F. DeKouchkovsky, F. Jouanneau, B. Gronenborn, and I. Jupin. 1997. Genetic analysis of the monopartite tomato yellow leaf curl geminivirus: roles of V1, V2, and C2 ORFs in viral pathogenesis. Virology 228:132-140. [DOI] [PubMed] [Google Scholar]
- 48.Zimmerman, P., M. Hirsch-Hoffman, L. Hennig, and W. Gruissem. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136:2621-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]