Abstract
Human genes use various mechanisms to generate different transcripts having different exon content, which in turn generate multiple protein isoforms having differential and even opposite biological activities. To understand the biological consequences of gene transcriptional activity modulation, it is necessary to integrate the capability of genes to generate distinct functional products, particularly because transcriptional stimuli also affect the exon content of their target gene products. For this purpose, we have developed a bioinformatics suite, FAST DB, which defines easily and accurately the exon content of all known transcripts produced by human genes. In addition, several tools have been developed, including a graphical presentation of all gene products, a sequence multi-alignment of all gene transcripts and an in silico PCR computer program. The FAST DB interface also offers extensive links to website resources for promoter analysis and transcription factor binding site prediction, splicing regulatory sequence prediction, as well as 5′- and 3′-untranslated region analysis. FAST DB has been designed to facilitate studies that integrate transcriptional and post-transcriptional events to investigate the expression regulation of human gene products.
INTRODUCTION
About 95% of human genes contain exons (between 7 and 12 in average) separated by introns. Exons contain the information necessary for the production of proteins, whereas introns are removed during the splicing process that gives rise to messenger RNAs (mRNAs). The mRNAs are then exported to the cytosol where they are translated. Owing to the presence of exons separated by introns, a single gene can produce different mRNAs having various exon contents. At its 5′ end or within internal introns, a given gene can have different promoters driving the production of transcripts that have different 5′-untranslated regions (5′-UTRs) and that sometimes encode protein isoforms with different N-terminal domains (1,2). At its 3′ end or within internal exons and/or introns, a given gene can have different transcriptional termination sequences and/or polyadenylation sites allowing the production of different transcripts that have different 3′-UTRs and that eventually encode protein isoforms with different C-terminal domains (3–5). During the splicing process, different introns (or parts of introns) and different exons (or parts of exons) can be alternatively spliced (6–10). A given intron can be retained in an mRNA molecule (intron retention), whereas a given exon can be skipped (exon skipping or exon cassette). The 5′ or the 3′ end of a given intron can be differentially selected (alternative 5′- or 3′-splicing site, respectively), which modifies the size of the exons included in the mRNA. It is estimated that 75% of these events occur in the translated regions of mRNAs and have consequences at the protein level (6–9,11). Alternative splicing events either generate splice variants encoding truncated proteins by the introduction of a stop codon, or yield protein isoforms that contain different domains. This allows a single gene to produce proteins with different properties regarding their stability or cellular localization, their ability to be regulated by post-translational modifications and to respond to signaling pathways, and their ability to interact with partners and/or to perform enzymatic reactions (6–9). The biological importance of such mechanisms is illustrated by genes involved in cell death as a single gene can produce different protein isoforms with either pro- or anti-apoptotic effects (12). Moreover, the human sequencing project and the cloning and sequencing of an increasing number of human transcripts reveal that most human genes (between 40 and 70%) generate different transcripts, which contributes to increase the human proteome diversity encoded by a limited number of genes (6–9,13,14).
Owing to the production of different translatable mRNAs from a given gene, it is not possible to predict the biological consequences resulting from gene transcriptional modulation only. This is particularly important because a transcriptional stimulus can ‘switch’ the promoter that drives the production of its gene products, and can also change the nature (exon content) of its target gene products. Indeed, the promoter identity driving the expression of a gene can affect the nature of the splice variants produced by this gene and, as we have shown, transcriptional stimuli, such as steroid hormones, simultaneously control the transcriptional rate of their target genes and the nature (exon content) of the spliced variant produced (15–18). Consistent with these observations, different transcription factors or transcriptional coregulators have different effects on splicing and 3′-end processing (17–23). In this context, studies of gene expression regulation need to account for the capability of human genes to produce different transcripts (24–26). For this reason, we developed a bioinformatics suite, named FAST DB (Friendly Alternative Splicing and Transcripts Database), that allows for defining easily and accurately the exon content of the different known transcripts produced by human genes based on a computerized analysis of human and mouse cDNAs and human expressed sequence tags (ESTs) libraries. In addition, a multi-alignment of all the transcript sequences of a given gene allows for visualizing the common and specific sequences of these transcripts. Therefore, it becomes very easy to design probes for downstream experimental applications, in particular PCR amplification. Thanks to FAST DB interface, users can design primers in a few minutes for PCR amplification of either all the gene products or specific variants, as well as for the co-amplification of splice variants giving rise to PCR products of different sizes. In addition, several links to various website resources are provided for the analysis of promoter regions and the analysis of 5′- and 3′-UTRs, as well as links to other splicing databases recently developed (27–32). Therefore, FAST DB is a bioinformatics tool designed for a rapid, extensive and accurate search to support integrated studies of gene transcriptional and post-transcriptional regulation.
MATERIALS AND METHODS
Filling FAST DB
All data contained in FAST DB were obtained through an informatics analysis of sequences available from public libraries. FAST DB was developed in PERL v5.8.5 (www.perl.org) using Bio::EnsEMBL, CGI, DBI, BioPerl, GD and PDF::API2 modules (www.cpan.org) on an AMD Athlon64 3000+ processor with 1.5 Go of RAM and with the Mandrake 10.1 Linux distribution (www.mandrakelinux.com). The FAST DB algorithm recovered all the exon sequences defined in EnsEMBL version 26 (homo_sapiens_core_26_35 database) and each of these exons was ‘blasted’ against two cDNA databanks using standalone BLAST v2.2.10. A full-length transcript databank was downloaded from the UCSC website (genome.ucsc.edu) and a partial mRNA databank was downloaded from the NCBI website (www.ncbi.nlm.nih.gov). These two databanks were formatted using Formatdb (‘formatdb -i downloaded_databank -pF -oT -sT’). All the recovered transcripts with an E-value < 10−40 (Blast was made using Bio::ToolsRun::StandAloneBlast and transcript sequences were recovered using fastacmd) were aligned against genomic sequences using sim4. By parsing the sim4 output and using strict criteria (see Supplementary Material), each transcript was then selected or excluded. The sim4 output also allowed for defining the different exons contained in each transcript. These exons were called ‘transcript exons’ and were clustered by genomic position. ‘Genomic exons’ were defined using the more frequent first and last position of the different clustered ‘transcript exon’.
To define alternative events generating the different products of a single gene, the FAST DB algorithm compared each ‘transcript exon’ with its corresponding ‘genomic exon’. FAST DB defined seven types of events (see Supplementary Material). To be defined as an alternative first exon, a ‘transcript exon’ had to be the first exon of at least one transcript and, if there were other internal exons at this genomic position, it had to start at least 10 nt upstream of the first position of the corresponding ‘genomic exon’. To be defined as an alternative last exon, a ‘transcript exon’ had to be the last exon of at least one transcript and, if there were other internal exons at this genomic position, it had to end at least 10 nt downstream of the last position of the corresponding ‘genomic exon’. The FAST DB algorithm also defined several splicing events. An ‘alternative 3′-splice site’ event was defined when the first position of a ‘transcript exon’ was different from the first position of the corresponding ‘genomic exon’. An ‘alternative 5′-splice site’ event was defined when the last position of a ‘transcript exon’ was different from the last position of the corresponding ‘genomic exon’. An ‘intron retention’ event was defined when a whole intronic sequence was included in at least one ‘transcript exon’ sequence. FAST DB also defined an ‘internal exon deletion’ (IED) event when a ‘transcript exon’ presented an internal sequence deletion compared with the corresponding ‘genomic exon’ sequence. Such deleted sequences correspond in fact to small introns that are frequently not spliced out (data not shown). Finally, an ‘exon skipping’ event was defined when at least one transcript had no defined ‘genomic exon’.
Once the ‘genomic exons’ and the alternative events were defined, the FAST DB algorithm filled a MySQL database (using DBI module for PERL). FAST DB used MySQL v4.0.20 (www.mysql.com). To decrease the loading time of its website, the FAST DB algorithm makes PNG files of gene and transcript representations (using GD module for PERL) and PDF files (using PDF::API2 module for PERL) and stores these files on a server. The same algorithm was used for human ESTs and mouse cDNAs analyses.
Multi-alignment and in silico PCR
FAST DB multi-alignment is available by using the FAST DB graphical interface. This interface was created using PERL (CGI module) on an APACHE server v2.0.50 (www.apache.org). FAST DB multi-alignment was performed using Clustalw and Partial Order Alignment. For a given gene, the sequences of ‘transcript exons’ corresponding to one ‘genomic exon’ were aligned using Clustalw. The results for each ‘genomic exon’ position were then assembled to present the multi-alignment of the different cDNAs. ‘Transcript exons’ containing a retained intron were each divided into two exons and one intron (or more intron/exon in case of multiple consecutive retained introns). Each exon was aligned with its corresponding ‘genomic exons’, and then the intron sequence was inserted between both exons. In case of a ‘genomic exon’ presenting an IED event, all the corresponding ‘transcript exons’ were aligned using Partial Order Alignment with the global alignment option (33,34). Nevertheless, owing to multi-alignment programming difficulties, in particular when ‘transcript exon’ sequences poorly overlap, some ‘transcript exons’ are not properly aligned (see Supplementary Material). To overcome this limitation, the sim4 alignment of each transcript sequence against the genomic sequence is made available (see below).
Housekeeping genes
We have selected a set of 707 housekeeping (HK) genes by compiling results from several reports defining HK genes based on the presence of their transcripts in a wide variety of tissues (35–38). We have linked all these genes with the FAST DB database. We have then excluded redundant genes, have assigned a unique ID to each of these genes and have set a new table of 707 ID to do statistical analyses.
RESULTS
By comparing the sequence of human genes with the sequence of their transcripts available from public libraries, the FAST DB algorithm provides the genomic organization and the exon content of transcripts corresponding to >12 000 human genes (see Materials and Methods and Supplementary Material). FAST DB contains genes defined by ‘good quality’ cDNAs only. Genes missing in FAST DB might not be defined by human cDNAs or ‘good quality’ cDNAs (see Supplementary Material).
More than 150 000 exons (10 exons per gene on average) have been defined in FAST DB using 80 000 transcripts (6 transcripts per gene on average). All the events defined by FAST DB and yielding different transcripts from single genes are based on a computerized analysis of known full-length and partial human cDNAs. For each gene, the analysis of the corresponding ESTs has been independently made available, which allows the prediction of additional events (see below). When possible, a similar analysis has been performed for mouse orthologous genes, allowing inter-species comparison (see below).
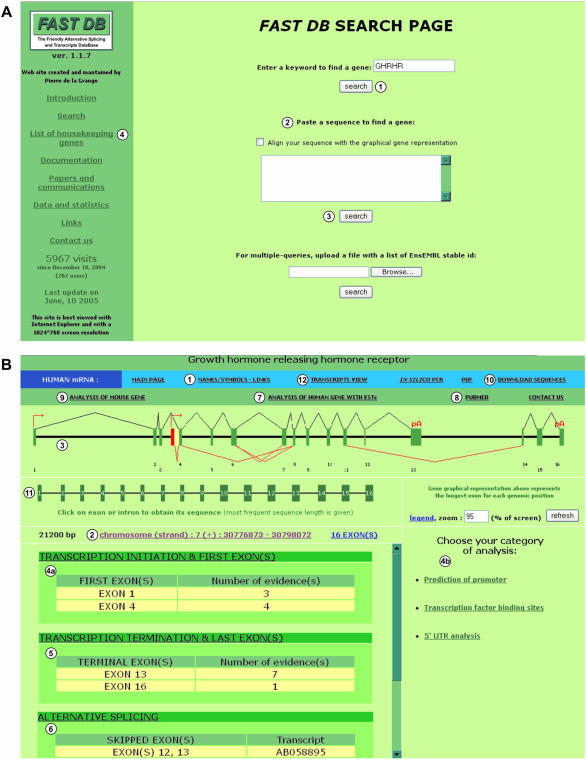
The FAST DB ‘SEARCH PAGE’ (http://193.48.40.18/fastdb/) offers different ways of accessing a given gene. FAST DB can be queried using either any name of the gene (see Supplementary Material and Figure 1A, item 1) or a partial sequence of the gene of interest, which will be identified by a ‘BLAST’ search (Figure 1A, item 2). Alternatively, multiple queries are possible by uploading files containing a list of genes of interest (ENsEMBL stable ID) (Figure 1A, item 3 and see User's guide). The list of FAST DB links to all the genes obtained by this query might be saved for further analysis. This multiple query interface is of interest to assist in designing custom made microarrays.
Figure 1.
FAST DB ‘SEARCH PAGE’ and ‘MAIN PAGE’ corresponding to human GHRHR gene. (A) FAST DB ‘SEARCH PAGE’. (1) Search by keyword; (2) BLAST search against FAST DB genes; (3) multiple query interface; (4) list of 707 HK genes with a link to FAST DB main page for each of them. (B) FAST DB ‘MAIN PAGE’ corresponding to human GHRHR gene. (1) Link on the navigation banner to display other names and symbols of GHRHR gene and several links to NCBI, ExPASy and EnsEMBL websites; (2) chromosomal localization of the gene (link to UCSC genome browser); (3) graphical representation of the gene; (4a) list of alternative first exons and links to several websites [displayed on (4b)] for promoter predictions, transcription factor binding sites and 5′-UTR analysis; (5) list of alternative terminal exons and links to several 3′-UTR analysis websites; (6) list of alternative splicing events and links to other alternative splicing databases; (7) link to FAST DB analysis that includes human ESTs; (8) link to PUBMED; (9) link to FAST DB analysis of the orthologous mouse gene; (10) link for downloading cDNA and genomic sequences; (11) on this graphical representation, each exon or intron is clickable to display its length and sequence; (12) link on the navigation banner to display the graphical representation of the transcripts.
After running the search engine, a gene or a list of genes is provided as a query result. By clicking on the requested gene, FAST DB opens the ‘MAIN PAGE’ of the gene. For example, Figure 1B shows the FAST DB analysis corresponding to the human gene of the growth hormone releasing hormone receptor (GHRHR).
Analysis of the exon content of human gene products
In addition to general information regarding a requested gene through links to different website resources, such as EnsEMBL, NCBI, ExPASy (Figure 1B, item 1) and the UCSC genome browser (Figure 1B, item 2), the ‘MAIN PAGE’ shows a graphical representation of the genomic organization of the requested gene (Figure 1B, item 3), with introns represented as horizontal lines and exons as filled rectangles. An arrow on top of exons indicates that these exons are potential alternative first transcript exons and define upstream promoters, whereas the symbol ‘pA’ on top of certain exons indicates that these exons are defined as potential alternative last transcript exons under several criteria (see Supplementary Material). It is important to underline that the ‘pA’ symbol does not indicate a polyadenylation site position but indicates only a potential terminal exon. Each represented exon corresponds to the longest exon at this genomic position identified among all the different gene transcripts. V-shaped lines above all exons join the most frequent last position of an exon with the most frequent first position of the next exon. V-shaped lines below certain exons indicate that these exons or part of them are differentially included in the final gene products.
As mentioned above, several mechanisms allow for differential selection of the exons (or part of them) that will be incorporated in the mature transcripts. For a requested gene, all the differentially selected exons defined by the FAST DB algorithm (see Materials and Methods and Supplementary Material) are listed at the bottom of the ‘MAIN PAGE’ (Figure 1B, items 4a, 5 and 6). When several promoters exist within a given gene, the transcripts generated from the different promoters differ in their first exon. When several transcription termination sequences and/or polyadenylation sites exist within a given gene, the transcripts generated differ in their last exon. Because the first and last exons within a given transcript could result from cloning and sequencing mistakes, strong criteria must be used in the computerized definition of these exons (see Supplementary Material). It is important to underline that we cannot exclude that upstream or downstream exons that are absent from the available transcripts actually exist. Therefore, to gain more confidence in candidate first and last exons, we have indicated the number of transcripts that include every single first and last exon [Figure 1B, items 4a and 5 ‘Number of evidence(s)’]. Moreover, users can perform computer-assisted analyses to look for other features of first exons (e.g. GC content) and last exons (e.g. polyadenylation sites). By clicking on ‘TRANSCRIPTION INITIATION & FIRST EXONS’ (Figure 1B, item 4a), users have access to several website resources (displayed on the right panel of Figure 1B, item 4b) for promoter and transcription factor binding site predictions (39–44). Links to 5′-UTR analysis website resources are also available, which provide further information on elements present within candidate first exons (45–47). Similarly, searches for last exons features are performed using different website resources accessible through the ‘TRANSCRIPTION TERMINATION & LAST EXONS’ link (Figure 1B, item 5) that is displayed on the right panel (3,45–48). Although some of these links direct users to databases containing pre-defined genes, other tools analyze a provided sequence. In this case, FAST DB creates a direct link to the website resource after the user has chosen the sequence of interest, which can correspond to exons or introns plus eventually upstream or downstream sequences of different sizes. In addition to providing more confidence in candidate first and last exons, these links let the user search for known sequences implicated in post-transcriptional regulation, such as transcript stability and translation (3,45–48).
The computerized analysis of the exon content of transcripts present in public libraries is based on transcript selection (elimination of bad quality sequences) and on exon definition. Because using different criteria has consequences on the final analysis result, clicking on ‘ALTERNATIVE SPLICING’ in FAST DB provides links to other website resources that contain potential complementary information on transcript exon content (Figure 1B, item 6) (27–32). Through this link, it is also possible to access website resources predicting splicing regulatory sequences within exons (ESEfinder and RESCUE-ESE) (49,50) as well as computer programs that score splicing sites (FSplice, Gene Splicer and BDGP) (51,52). FAST DB has created a link that directs the user to the website resource after choosing sequences corresponding to exons or exon/intron boundaries.
Because ESTs provide further information regarding differentially selected exons but are potentially of bad quality, an independent analysis of ESTs is available by clicking on ‘ANALYSIS OF HUMAN GENE WITH ESTs’ (Figure 1B, item 7). This analysis allows for ‘predicting’ other potential alternatively spliced exons. Additional information described in the literature on splicing events for the analyzed gene is available by clicking on the ‘PUBMED’ link (Figure 1B, item 8). If a transcript has not been analyzed by FAST DB or if users identified a new transcript, the sequence of such a transcript can be entered from the ‘SEARCH PAGE’ (Figure 1A, item 2). The transcript sequence must be at least 20 nt long. After clicking the ‘search’ button and checking the ‘Align your sequence with the graphical gene representation’ box, the input sequence localization is displayed under the graphical gene representation. The input sequence is also aligned against the genomic sequence in the ‘in silico PCR’ page (see below). Finally, for some genes, a similar analysis for the corresponding mouse orthologous gene is available by clicking on ‘ANALYSIS OF MOUSE GENE’ for inter-species comparison (Figure 1B, item 9).
In summary, FAST DB offers several ways of obtaining extensive information on the exons that are differentially selected within mature transcripts: analysis of full-length and partial human cDNAs and human ESTs, analysis of mouse cDNAs, links to other website resources and PUBMED, and analysis of transcripts entered by the users. Importantly, all the sequences used by FAST DB can be downloaded by clicking on the ‘DOWNLOAD SEQUENCES’ button (Figure 1B, item 10). From this link, it is also possible to obtain the exonic sequences underlined within the corresponding genomic sequence (data not shown). Alternatively, the sequence of individual exons or introns can be obtained by clicking on the graphical representation (Figure 1B, item 11).
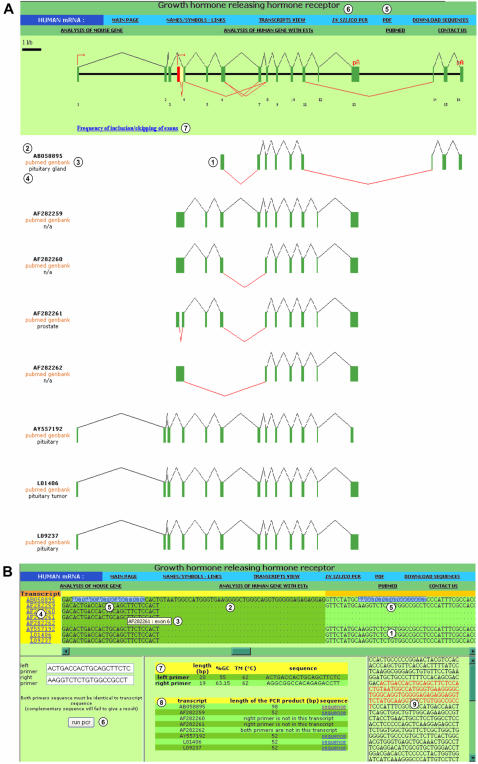
Graphical presentation of the transcripts and in silico PCR
To identify the transcripts used to establish the different events listed on the ‘MAIN PAGE’, users access the graphical representation (exon content) of each transcript by clicking on the ‘TRANSCRIPTS VIEW’ button (Figure 1B, item 12). This graphical representation provides users with an easy understanding of the exon content of all known transcripts produced by the requested gene (Figure 2A, item 1). The accession number (Figure 2A, item 2), a link to Pubmed and Genbank (Figure 2A, item 3), and information regarding the tissues from which the transcripts have been cloned (Figure 2A, item 4) are available. All the graphical and content information is easily printable by clicking on the ‘PDF’ button (Figure 2A, item 5). Altogether, these data provide an excellent tool for scientists designing an experimental approach to study the expression regulation of the products of a given gene. To facilitate such studies, FAST DB provides a specific tool accessed by clicking on the ‘IN SILICO PCR’ button (Figure 2A, item 6).
Figure 2.
Graphical representation of GHRHR transcripts, navigation banner and in silico PCR. (A) Graphical representation of GHRHR transcripts. (1) Graphical representation of transcript aligned with the gene graphical representation; (2) transcript accession number; (3) links to Genbank and Pubmed; (4) name of the tissue where the transcript was cloned; (5) link to the PDF version of the current GHRHR analysis; (6) link to the multi-alignment of the GHRHR transcript sequences and in silico PCR; (7) for each exon, number of transcripts that include/skip the exon. (B) In silico PCR. (1) Multi-alignment of all GHRHR transcripts performed exon by exon; (2) variable sequence found in one transcript due to an alternative 5′-splice site; (3) current exon and name of the current transcript displayed by pointing the cursor on the corresponding sequence for a few seconds; (4) transcript accession numbers linked to an alignment of the corresponding transcript sequence with the genomic sequence; (5) selection of sequences for PCR primers directly on the multi-alignment; (6) selected PCR primer sequences pasted in the corresponding boxes to run in silico PCR; (7) length, GC% content, Tm and 5′–3′ sequence of the selected primers; (8) length of the expected PCR product for each transcript and link to its sequence; (9) sequence of the PCR product within the sequence of the template transcript.
The ‘IN SILICO PCR’ link provides users with a multi-alignment (see Materials and Methods and Supplementary Material) of all transcript sequences of a given gene (Figure 2B, item 1). Thanks to this alignment, the specific sequences of certain transcripts are distinguished from the sequences shared by all transcripts (Figure 2B, item 2). By leaving the cursor a few seconds on the sequence of interest, the corresponding exon number is displayed and the graphical view of the transcripts printed from the PDF file easily compares with the multi-alignment result (Figure 2B, item 3). In case of a multi-alignment difficulty for one of the transcripts (see Supplementary Material), the alignment of each transcript sequence against the genomic sequence is made available by clicking on the accession number of the transcript on the left of the multi-alignment (Figure 2B, item 4).
Based on this multi-alignment, it becomes easy to design PCR primers. As shown in Figure 2B, the user selects sequences flanking a region that presents alternative splicing (Figure 2B, item 5). These selected sequences can be copied and pasted in the primer boxes (Figure 2B, item 6). Clicking on the ‘Run PCR’ button brings up information on the sequence, length, %GC content and Tm of the selected primers (Figure 2B, item 7). The sizes of the PCR products obtained from the different transcripts are also provided (Figure 2B, item 8). Clicking on the ‘sequence’ link on the right (Figure 2B, item 8) displays the sequence of the PCR product for the selected transcript (Figure 2B, item 9). This sequence can be selected and pasted in any computer program for prediction of restriction enzyme sites either for checking the nature of the PCR products experimentally obtained or for cloning purpose.
As described above, users can select primers that flank an alternative region of a gene to co-amplify different spliced variants and quantify the effect of a stimulus on the ratio of different spliced products. Primers can also be selected within sequences shared by all known transcripts to amplify all the gene products as a single PCR product and determine the impact of a transcriptional stimulus taking into account all the target gene products. Finally, primers can be selected to specifically amplify a variant by choosing primers within specific sequences of a splice variant. In addition, because the name of each exon appears on the multi-alignment, primers can be designed at the junction of exons to avoid amplification of genomic DNA.
Another application of the multi-alignment interface is to predict whether an alternative splicing event would have biological consequences at the protein level. Users can select a specific variable sequence and analyze it with Blastx (http://www.ncbi.nlm.nih.gov/BLAST/) to test if this sequence encodes a known protein and with Interpro (http://www.ebi.ac.uk/interpro/) to test if this sequence encodes a specific protein domain.
Analysis of HK genes
Because HK genes are widely expressed across tissues, studies of gene expression regulation are often performed using HK genes as internal control (35–38). Interestingly, the genomic organization of HK genes has recently been shown to be different from that of tissue-specific (TS) expressed genes (37,38). HK genes are usually more ‘compact’ than TS genes, mostly because of the smaller size of their introns and because HK genes have fewer exons/introns than TS genes. Nevertheless, to our knowledge, there is no general information available regarding the potential ability of HK genes to generate multiple transcripts. Because HK genes are essential in transcriptional studies, we have set up an analysis of HK gene products in FAST DB. For this purpose, we used 707 HK genes that had been defined in previous reports based on their wide expression in many tissues (35–38).
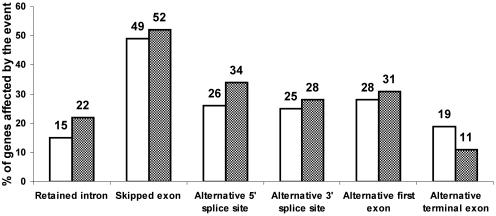
Analyzing all the genes (other that HK genes) present within FAST DB, we observed that 3458 genes (∼28%) out of 12 538 analyzed genes contain at least two alternative first exons and 2414 genes (∼19%) contain at least two different last exons (Figure 3). Our analysis did not take into account the different polyadenylation sites within a single exon, which would significantly increase the percentage of transcripts having differential 3′ end (3–5). The most frequently occurring alternative splicing event was ‘exon skipping’. Half of the analyzed genes contain at least one exon that was skipped. About 26% of the analyzed genes contain at least one exon with alternative 5′ end and 25% of the analyzed genes contain at least one exon with alternative 3′ end. About 15% of the analyzed genes contain at least one intron that was retained within a transcript. These results, which are consistent with those of other analyzes (6–9,12–14,27–32), demonstrate that the capability of human genes to generate different transcripts is a rule and not an exception because 66% of human genes generate at least two different transcripts having different exon content.
Figure 3.
Appearance of alternative events in producing different transcripts from HK versus TS genes. The horizontal axis defines categories of alternative events. The vertical bars represent the proportion (%) of TS genes (white bars) and HK genes (shaded bars) affected at least once by the alternative event.
Using the set of 707 HK genes, we confirmed previous findings that HK genes contain fewer exons than other genes (two introns fewer in average) and that small (<10 kb) HK genes are more frequent compared with small TS genes (20% versus 10%, respectively). Nevertheless, we observed that HK genes can generate multiple transcripts similarly to TS genes (Figure 3). Half of HK genes contain at least one exon cassette (Figure 3). About one-third of HK genes generate products with different 5′-alternative spliced sites and a similar proportion of these genes contains 3′-alternative spliced sites. About 22% of HK genes contain at least one intron retained in a mature transcript. Interestingly, although one-third of HK and TS genes contain multiple first exons, we observed that only 11% of HK genes contain multiple terminal exons compared with ∼19% of TS genes.
In conclusion, despite a different genomic organization, HK genes are able to generate transcript diversity at a similar level to that of TS genes. Therefore, caution is required in designing primers when HK genes are used as transcription internal controls because the exon content of HK gene products might vary depending on the biological conditions. To help users select primers and avoid alternative regions in these genes, the list of 707 HK genes is accessed by clicking the ‘List of housekeeping genes’ link on the FAST DB ‘SEARCH PAGE’ (Figure 1A, item 4).
DISCUSSION
FAST DB has been designed to facilitate the study of the expression regulation of the various transcripts produced by human genes. This goal was achieved by: (i) a clear and ‘intuitive’ presentation of the information. (ii) The most complete set of information on the nature of the transcripts produced by human genes based on the analysis of full-length and partial cDNAs, as well as human ESTs. Links to other public databases that contain potential complementary information on the nature of transcripts produced by human genes are included. The possibility for users to enter their own transcript sequences and a PUBMED link enriches the information available. The analysis of mouse orthologous genes is provided for inter-species comparison. (iii) A sequence multi-alignment of all transcripts produced by a single gene, which facilitates the design of probes for downstream experiments. (iv) A link to a ‘List of housekeeping genes’ to help users design primers that are used as internal controls. The rationale is based on our observation that HK genes generate transcripts of different exonic content. (v) Links to website resources for promoter analysis and transcriptional factor binding site predictions, splicing regulatory sequence prediction, as well as for 5′- and 3′-UTR analysis, which facilitate studies integrating transcriptional and post-transcriptional aspects.
Knowing the exon content of transcripts is required for understanding the biological consequences of transcriptional stimuli. Indeed, genes cannot be longer considered as ‘simple’ functional units. Genes are rather an ‘assemblage’ of exons that are differentially incorporated within the gene products that in turn generate protein isoforms with different biological activities or functions. The FAST DB analysis has been performed on 12 538 human genes defining 151 747 exons and we estimated that 31 318 exons are subject to regulation. This means that ∼20% of total human exons are differentially integrated within gene products. This proportion is probably underestimated because the statistical analysis was performed using only full or partial cDNAs present within public libraries. This large amount of exons differentially incorporated within gene products is in good agreement with the poor definition of exon and intron boundaries, which creates the right conditions for physiological regulation and evolution (8,10,53–55).
AVAILABILITY
FAST DB is freely available on the internet at http://193.48.40.18/fastdb/.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by the INSERM ‘AVENIR’ program (D.A.), Société Française d'Hématologie and Association Française contre les Myopathies (P.G.), INSERM ‘Poste Vert’ (N.M.) and Ligue Nationale Contre le Cancer (M.D.). Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Landry J.R., Mager D.L., Wilhelm B.T. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 2003;19:640–648. doi: 10.1016/j.tig.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 2.Zhang T., Haws P., Wu Q. Multiple variable first exons: a mechanism for cell- and tissue-specific gene regulation. Genome Res. 2004;14:79–89. doi: 10.1101/gr.1225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H., Hu J., Recce M., Tian B. PolyA_DB: a database for mammalian mRNA polyadenylation. Nucleic Acids Res. 2005;33:D116–D120. doi: 10.1093/nar/gki055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian B., Hu J., Zhang H., Lutz C.S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaudoing E., Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T.A., Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Kriventseva E.V., Koch I., Apweiler R., Vingron M., Bork P., Gelfand M.S., Sunyaev S. Increase of functional diversity by alternative splicing. Trends Genet. 2003;19:124–128. doi: 10.1016/S0168-9525(03)00023-4. [DOI] [PubMed] [Google Scholar]
- 8.Maniatis T., Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 9.Hastings M.L., Krainer A.R. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- 10.Smith C.W., Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 11.Sorek R., Shamir R., Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Wu J.Y., Tang H., Havlioglu N. Alternative pre-mRNA splicing and regulation of programmed cell death. Prog. Mol. Subcell. Biol. 2003;31:153–185. doi: 10.1007/978-3-662-09728-1_6. [DOI] [PubMed] [Google Scholar]
- 13.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 14.Grimwood J., Gordon L.A., Olsen A., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., et al. The DNA sequence and biology of human chromosome 19. Nature. 2004;428:529–535. doi: 10.1038/nature02399. [DOI] [PubMed] [Google Scholar]
- 15.Cramer P., Pesce C.G., Baralle F.E., Kornblihtt A.R. Functional association between promoter structure and transcript alternative splicing. Proc. Natl Acad. Sci. USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagani F., Stuani C., Zuccato E., Kornblihtt A.R., Baralle F.E. Promoter architecture modulates CFTR exon 9 skipping. J. Biol. Chem. 2003;278:1511–1517. doi: 10.1074/jbc.M209676200. [DOI] [PubMed] [Google Scholar]
- 17.Auboeuf D., Honig A., Berget S.M., O'Malley B.W. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- 18.Auboeuf D., Dowhan D.H., Kang Y.K., Larkin K., Lee J.W., Berget S.M., O'Malley B.W. Differential recruitment of nuclear receptor coactivators may determine alternative RNA splice site choice in target genes. Proc. Natl Acad. Sci. USA. 2004;101:2270–2274. doi: 10.1073/pnas.0308133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nogues G., Kadener S., Cramer P., Bentley D., Kornblihtt A.R. Transcriptional activators differ in their abilities to control alternative splicing. J. Biol. Chem. 2002;277:43110–43114. doi: 10.1074/jbc.M208418200. [DOI] [PubMed] [Google Scholar]
- 20.Rosonina E., Bakowski M.A., McCracken S., Blencowe B.J. Transcriptional activators control splicing and 3′-end cleavage levels. J. Biol. Chem. 2003;278:43034–43040. doi: 10.1074/jbc.M307289200. [DOI] [PubMed] [Google Scholar]
- 21.Auboeuf D., Dowhan D.H., Li X., Larkin K., Ko L., Berget S.M., O'Malley B.W. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol. Cell. Biol. 2004;24:442–453. doi: 10.1128/MCB.24.1.442-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monsalve M., Wu Z., Adelmant G., Puigserver P., Fan M., Spiegelman B.M. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 23.Nagai K., Yamaguchi T., Takami T., Kawasumi A., Aizawa M., Masuda N., Shimizu M., Tominaga S., Ito T., Tsukamoto T., et al. SKIP modifies gene expression by affecting both transcription and splicing. Biochem. Biophys. Res. Commun. 2004;316:512–517. doi: 10.1016/j.bbrc.2004.02.077. [DOI] [PubMed] [Google Scholar]
- 24.Yeakley J.M., Fan J.B., Doucet D., Luo L., Wickham E., Ye Z., Chee M.S., Fu X.D. Profiling alternative splicing on fiber-optic arrays. Nat. Biotechnol. 2002;20:353–358. doi: 10.1038/nbt0402-353. [DOI] [PubMed] [Google Scholar]
- 25.Relogio A., Ben-Dov C., Baum M., Ruggiu M., Gemund C., Benes V., Darnell R.B., Valcarcel J. Alternative splicing microarrays reveal functional expression of neuron-specific regulators in Hodgkin lymphoma cells. J. Biol. Chem. 2005;280:4779–4784. doi: 10.1074/jbc.M411976200. [DOI] [PubMed] [Google Scholar]
- 26.Pan Q., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Huang H.D., Horng J.T., Lee C.C., Liu B.J. ProSplicer: a database of putative alternative splicing information derived from protein, mRNA and expressed sequence tag sequence data. Genome Biol. 2003;4:R29. doi: 10.1186/gb-2003-4-4-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee C., Atanelov L., Modrek B., Xing Y. ASAP: the Alternative Splicing Annotation Project. Nucleic Acids Res. 2003;31:101–105. doi: 10.1093/nar/gkg029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thanaraj T.A., Stamm S., Clark F., Riethoven J.J., Le Texier V., Muilu J. ASD: the Alternative Splicing Database. Nucleic Acids Res. 2004;32:D64–D69. doi: 10.1093/nar/gkh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng C.L., Nair T.M., Gribskov M., Kwon Y.S., Li H.R., Fu X.D. A database designed to computationally aid an experimental approach to alternative splicing. Pac. Symp. Biocomput. 2004:78–88. doi: 10.1142/9789812704856_0008. [DOI] [PubMed] [Google Scholar]
- 31.Pospisil H., Herrmann A., Bortfeldt R.H., Reich J.G. EASED: Extended Alternatively Spliced EST Database. Nucleic Acids Res. 2004;32:D70–D74. doi: 10.1093/nar/gkh136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H.D., Horng J.T., Lin F.M., Chang Y.C., Huang C.C. SpliceInfo: an information repository for mRNA alternative splicing in human genome. Nucleic Acids Res. 2005;33:D80–D85. doi: 10.1093/nar/gki129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee C., Grasso C., Sharlow M.F. Multiple sequence alignment using partial order graphs. Bioinformatics. 2002;18:452–464. doi: 10.1093/bioinformatics/18.3.452. [DOI] [PubMed] [Google Scholar]
- 34.Grasso C., Lee C. Combining partial order alignment and progressive multiple sequence alignment increases alignment speed and scalability to very large alignment problems. Bioinformatics. 2004;20:1546–1556. doi: 10.1093/bioinformatics/bth126. [DOI] [PubMed] [Google Scholar]
- 35.Warrington J.A., Nair A., Mahadevappa M., Tsyganskaya M. Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol. Genomics. 2000;2:143–147. doi: 10.1152/physiolgenomics.2000.2.3.143. [DOI] [PubMed] [Google Scholar]
- 36.Hsiao L.L., Dangond F., Yoshida T., Hong R., Jensen R.V., Misra J., Dillon W., Lee K.F., Clark K.E., Haverty P., et al. A compendium of gene expression in normal human tissues. Physiol. Genomics. 2001;7:97–104. doi: 10.1152/physiolgenomics.00040.2001. [DOI] [PubMed] [Google Scholar]
- 37.Eisenberg E., Levanon E.Y. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- 38.Vinogradov A.E. Compactness of human housekeeping genes: selection for economy or genomic design? Trends Genet. 2004;20:248–253. doi: 10.1016/j.tig.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grabe N. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. 2002;2:S1–S15. [PubMed] [Google Scholar]
- 41.Boardman P.E., Oliver S.G., Hubbard S.J. SiteSeer: visualisation and analysis of transcription factor binding sites in nucleotide sequences. Nucleic Acids Res. 2003;31:3572–3575. doi: 10.1093/nar/gkg511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burden S., Lin Y.X., Zhang R. Improving promoter prediction for the NNPP2.2 algorithm: a case study using Escherichia coli DNA sequences. Bioinformatics. 2005;21:601–607. doi: 10.1093/bioinformatics/bti047. [DOI] [PubMed] [Google Scholar]
- 43.Prestridge D.S. Predicting Pol II promoter sequences using transcription factor binding sites. J. Mol. Biol. 1995;249:923–932. doi: 10.1006/jmbi.1995.0349. [DOI] [PubMed] [Google Scholar]
- 44.Bajic V.B., Seah S.H., Chong A., Zhang G., Koh J.L., Brusic V. Dragon Promoter Finder: recognition of vertebrate RNA polymerase II promoters. Bioinformatics. 2002;18:198–199. doi: 10.1093/bioinformatics/18.1.198. [DOI] [PubMed] [Google Scholar]
- 45.Jacobs G.H., Rackham O., Stockwell P.A., Tate W., Brown C.M. Transterm: a database of mRNAs and translational control elements. Nucleic Acids Res. 2002;30:310–311. doi: 10.1093/nar/30.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mignone F., Grillo G., Licciulli F., Iacono M., Liuni S., Kersey P.J., Duarte J., Saccone C., Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lambert A., Fontaine J.F., Legendre M., Leclerc F., Permal E., Major F., Putzer H., Delfour O., Michot B., Gautheret D. The ERPIN server: an interface to profile-based RNA motif identification. Nucleic Acids Res. 2004;32:W160–W165. doi: 10.1093/nar/gkh418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tabaska J.E., Zhang M.Q. Detection of polyadenylation signals in human DNA sequences. Gene. 1999;231:77–86. doi: 10.1016/s0378-1119(99)00104-3. [DOI] [PubMed] [Google Scholar]
- 49.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fairbrother W.G., Yeo G.W., Yeh R., Goldstein P., Mawson M., Sharp P.A., Burge C.B. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pertea M., Lin X., Salzberg S.L. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001;29:1185–1190. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reese M.G., Eeckman F.H., Kulp D., Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 53.Ast G. How did alternative splicing evolve? Nature Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 54.Yeo G., Hoon S., Venkatesh B., Burge C.B. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl Acad. Sci. USA. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeo G., Holste D., Kreiman G., Burge C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.