Abstract
The chromogranin/secretogranin proteins are costored and coreleased with catecholamines from secretory vesicles in chromaffin cells and noradrenergic neurons. Chromogranin A (CHGA) regulates catecholamine storage and release through intracellular (vesiculogenic) and extracellular (catecholamine release–inhibitory) mechanisms. CHGA is a candidate gene for autonomic dysfunction syndromes, including intermediate phenotypes that contribute to human hypertension. Here, we show a surprising pattern of CHGA variants that alter the expression and function of this gene, both in vivo and in vitro. Functional variants include both common alleles that quantitatively alter gene expression and rare alleles that qualitatively change the encoded product to alter the signaling potency of CHGA-derived catecholamine release–inhibitory catestatin peptides.
Introduction
Chromogranin A (CHGA [MIM 118910]) is a central regulator of catecholamine vesicle storage and release that also plays key signaling roles in subsequent physiology (Taupenot et al. 2003). CHGA is targeted to the regulated secretory pathway (Taupenot et al. 2002), where it binds calcium and catecholamines (Videen et al. 1992). CHGA also binds the vesicle membrane, where it can influence the release of calcium from secretory granules to the cytosolic exocytotic machinery in secretory cells by modulating the inositol 1,4,5-trisphosphate receptor/Ca2+ channel (Yoo et al. 2002). CHGA is required for formation of catecholamine secretory vesicles in chromaffin-lineage PC12 cells and is sufficient to induce a regulated secretion system in nonsecretory cells (Kim et al. 2001). Proteolytic cleavage of CHGA at dibasic sites (Barbosa et al. 1991; Eskeland et al. 1996) produces, in secretory vesicles, several biologically active peptides, including catestatin (human CHGA352–372 and bovine CHGA344–364). Exocytosis of catecholamine vesicles is stimulated by cholinergic input. Catestatin inhibits subsequent catecholamine release by antagonizing nicotinic receptors, thus constituting an autocrine/paracrine feedback loop (Mahata et al. 1997). The formation (Taylor et al. 2000; Jiang et al. 2001), inhibitory mechanism (Mahata et al. 1997, 1999, 2000; Taupenot et al. 2000), and likely structure (Tsigelny et al. 1998) of catestatin have been recently characterized. Other biologically active CHGA fragments include the vascular smooth-muscle relaxant vasostatin (human CHGA1–76), the dysglycemic peptide pancreastatin (human CHGA250–301) (Cadman et al. 2002), and the antimicrobial peptides prochromacin (human CHGA79–439) and chromacin (human CHGA176–197), thought to play a role in the neuroendocrine stress response to systemic infection (Tasiemski et al. 2002).
We have previously proposed CHGA expression and activity as intermediate phenotypes in human hypertension (O’Connor et al. 2000). Hypertension shows substantial heritability in family studies, but the precise genes controlling blood pressure variability in the population remain poorly understood (Miall and Oldham 1963; O’Connor et al. 2000; Timberlake et al. 2001). Intermediate phenotypes—simpler, often monogenic, traits associated with multifactorial disease—offer an enticing approach to the genetics of common disorders with complex inheritance. CHGA is a likely regulator of intermediate phenotypes that contribute to hypertension (O’Connor et al. 2002). CHGA is overexpressed by chromaffin cells in rodent models of both genetic (spontaneously hypertensive rat [Schober et al. 1989; O’Connor et al. 1999]) and acquired (renovascular [Takiyyuddin et al. 1993]) hypertension, and twin studies have demonstrated heritability of plasma CHGA concentration in humans (Takiyyuddin et al. 1995). Conversely, plasma concentration of the catestatin fragment is diminished both in established hypertension and in still-normotensive offspring at genetic risk of developing the disease (Kennedy et al. 1998; Mahata et al. 2000; O’Connor et al. 2002). This suggests that expression levels of CHGA and its peptides act in the pathogenesis of hypertension and that steady-state levels of different fragments may be uncoupled from each other. To identify genetic variants in CHGA that might alter its function, we resequenced all eight exons and adjacent intronic regions, ∼1.2 kbp of 5′ promoter, and two intronic conserved noncoding regions from 180 ethnically diverse human subjects. Here, we report identification of both quantitative variation in expression level mediated by common promoter haplotypes and qualitative variation in catestatin peptide mediated by rare polymorphisms that alter its inhibitory potency.
Material and Methods
Subjects and Clinical Characterization
A series of 180 individuals was studied, selected to span a diverse range of ethnicities. Ethnicity was established by self-identification. None of the subjects had a history of renal failure. Subject characteristics are defined as in previous articles (e.g., O’Connor et al. 2002). Subjects were volunteers from urban southern California (San Diego), and each subject gave informed, written consent; the protocol was approved by the University of California San Diego institutional review board (see table A [online only]).
Table A.
Characterization of Resequenced Subjects in the Study
|
Population |
|||||
| Variable | Asian(n=44) | Black(n=57) | Hispanic(n=28) | White(n=51) | Global(n=180) |
| Sex (M/F) | 23/21 | 37/20 | 24/4 | 39/13 | 122/58 |
| Age (in years) | 33±14 | 44±11 | 48±10 | 50±11 | 45±13 |
| Hypertension (Y/N) | 8/36 | 14/43 | 8/20 | 12/40 | 44/136 |
| Height (in m) | 1.67±.09 | 1.73±.11 | 1.71±.08 | 1.74±.08 | 1.72±.09 |
| Weight (in kg) | 67±13 | 85±18 | 87±20 | 82±19 | 81±19 |
| Body surface area (in m2) | 1.75±.2 | 2.01±.24 | 2.02±.25 | 1.98±.25 | 1.96±.25 |
| BMI (in kg/m2) | 24±4 | 29±6 | 30±6 | 27±6 | 27±6 |
| Blood pressure (in mmHg): | |||||
| Systolic | 134±24 | 123±14 | 132±16 | 129±18 | 128±18 |
| Diastolic | 74±8 | 71±9 | 74±9 | 75±10 | 73±9 |
Molecular Genetics
Genomic DNA was prepared from leukocytes in EDTA-anticoagulated blood, through use of PureGene extraction columns (Gentra Biosystems), as described by Herrmann et al. (2000). Public draft human (Lander et al. 2001) and mouse (Waterston et al. 2002) genome sequence was obtained from the University of California Santa Cruz (UCSC) Genome Browser (UCSC Genome Bioinformatics Web site). Promoter positions were numbered with respect to the mRNA cap (transcriptional initiation) site. PCR primers were designed by Primer3 (Rozen and Skaletsky 2000; Primer3 Web site) to span each of the eight exons, as well as to include 50–100 bp of flanking intronic sequence. Target sequences were amplified by PCR from 16 ng genomic DNA in a final volume of 20 μl. Products were treated with exonuclease I and shrimp alkaline phosphatase to remove primers and dNTPs prior to cycle sequencing with BigDye terminators (Applied Biosystems). Sequence was determined on an ABI 3100 automated sequencer and analyzed using the Phred/Phrap/Consed suite of software to provide base quality scores (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998). Polymorphism and heterozygosity were detected using Polyphred (Nickerson et al. 1997; Rieder et al. 1998) and were manually confirmed. A subset of these data was cross-validated manually through use of base calls from Applied Biosystems software and visual inspection of trace files to identify heterozygotes. Rare SNPs were confirmed by resequencing in multiple individuals and from the reverse direction.
Statistical Analysis
Haplotypes were estimated from unphased genotypes by use of the PHASE program (Stephens et al. 2001). Haplotype homozygosities were confirmed by visual inspection. Pairwise linkage disequilibrium (LD) between each common SNP was quantified as D′ and Δ2 by use of the GOLD software package (Abecasis and Cookson 2000). Neutrality tests, including Tajima’s D, Fu and Li’s D, and the H test were performed using phased haplotype data in DnaSP (Rozas and Rozas 1999). Similar results were also obtained with Arlequin (Schneider et al. 2000), MEGA2 (Kumar et al. 2001), and hand calculations of Tajima’s D. We used the FST statistic as a relative measure of population differentiation and calculated FST for the CHGA promoter through use of the Arlequin package (Schneider et al. 2000). CHGA promoter haplotype networks were constructed in Arlequin, which computed a minimum spanning tree (MST) from the matrix of pairwise distances calculated between all pairs of haplotypes. Similar trees were also constructed using the neighbor-joining, parsimony, and maximum-likelihood methods. One-way post hoc ANOVA with a Bonferroni correction was performed using the SPSS software package to check the significance of the in vivo association study and in vitro haplotype-specific CHGA promoter activity.
Molecular Modeling
Models were based on native, linear, human catestatin (hCHGA352–372) nuclear magnetic resonance (NMR) structure in the Protein Data Bank. Point mutants were aligned to the wild-type backbone template and then subjected to energy-minimization/molecular mechanics (Mahata et al. 1998; Tsigelny et al. 1998).
Assays (CHGA Peptide Radioimmunoassays in Human Plasma)
EDTA-anticoagulated plasma was frozen and stored at −70°C prior to assay. CHGA region-specific radioimmunoassays for CHGA17–38 (hydrophobic disulfide loop), CHGA116–437 (large fragment), CHGA284–301 (pancreastatin), and CHGA361–372 (catestatin) were based on synthetic peptides and polyclonal rabbit antisera, as described elsewhere (Stridsberg 2000). 125I-radiolabeling of each peptide was enabled by either an endogenous or adventitious (terminal) Tyr residue. Several of the CHGA region-specific radioimmunoassays have been described in detail elsewhere (Stridsberg 2000; Stridsberg et al. 1995, 2000).
Synthetic Peptides
Particular CHGA regions, including linear, human catestatin CHGA352–372 (S352SMKLSFRARAYGFRGPGPQL372) or its point mutants (Gly364Ser; Pro370Leu), were synthesized by the solid-phase method, with 9-fluorenylmethoxycarbonyl (Fmoc) protection chemistry, as described elsewhere (Mahata et al. 2000). Peptides were purified to >95% homogeneity by preparative reverse-phase high-performance liquid chromatography (RP-HPLC) on C-18 silica columns. Authenticity and purity of peptides were further verified by analytical chromatography (RP-HPLC) and electrospray-ionization or matrix-assisted laser desorption/ionization (MALDI) mass spectrometry.
Catecholamine Secretion (Effect of Human Catestatin or Its Variants)
Rat PC12 pheochromocytoma cells were grown at 37°C with 6% CO2, in 10-cm dishes or six-well plates, in Dulbecco's minimum essential/high glucose medium supplemented with fetal bovine and horse serum and penicillin/streptomycin. Norepinephrine secretion from PC12 cells was monitored as described elsewhere (Mahata et al. 2000). In brief, cells were labeled for 3 h with 1 μCi 3H-L-norepinephrine (71.7 Ci/mmol) (DuPont/NEN) and then were incubated for 30 min, with or without the secretagogue nicotine (60 μM), in the presence or absence of peptide antagonists (0.1–10 μM). Release medium and cell lysates were assayed for 3H-L-norepinephrine by liquid scintillation counting. Net secretion is calculated as nicotine-stimulated release minus basal catecholamine release.
CHGA Promoter Haplotype/Reporter Activity Assays
Human CHGA promoter/reporter plasmids were constructed essentially as described by Rozansky et al. (1994) and Wu et al. (1994). Haplotype-specific promoter fragments corresponding to CHGA −1142/+54 bp were amplified from genomic DNA of known homozygotes (or heterozygotes for the two least common haplotypes), cloned into promoterless firefly luciferase reporter plasmid (pGL3-Basic [Promega]). Synthetic replacements were made by site-directed mutagenesis (QuickChange [Stratagene]). All promoter fragments were sequence verified before use. Promoter positions are numbered upstream (−) or downstream (+) of the cap site. Plasmids were purified on columns (Qiagen) prior to transfection. PC12 pheochromocytoma cells were transfected (at 50%–60% confluence, 1 d after 1:4 splitting) with 1 μg of supercoiled promoter haplotype-firefly luciferase reporter plasmid and 10 ng of the Renilla luciferase expression plasmid pRL-TK (Promega) as an internal control per well, by the liposome method (Superfect [Qiagen]). The firefly and Renilla luciferase activities in the cell lysates were measured 16–21 h after transfection, and the results were expressed as the ratio of firefly/Renilla luciferase activity (Stop & Glo [Promega]). Each experiment was repeated a minimum of three times.
Results
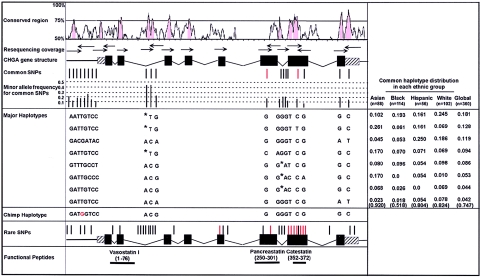
To identify genetic variants in CHGA that might alter its function, we resequenced all eight exons and adjacent intronic regions, the proximal promoter, and two intronic conserved noncoding regions (⩾75% conserved between human and mouse) from 180 ethnically diverse human subjects collected as part of our ongoing phenotypic studies (fig. 1). We identified 53 SNPs and 2 single-base insertion/deletions in this 5,725-bp footprint. Of these, 20 SNPs were common (minor allele frequency ⩾5%) in our initial sample. It is surprising that 8 of the common SNPs (and 13 total) occur within 1,175 bp in the proximal promoter. The estimated nucleotide diversity (π) (Tajima 1989) in the promoter region (0.0021) is two- to threefold higher than in other regions of CHGA, including introns (table 1), and is also several-fold higher than promoter regions of other genes we have resequenced from the same subjects (0.0007, 0.0001, and 0.0004 for similar-sized segments of PYY, PMX2B, and DBH, respectively, and 0.0008 for combined smaller segments of NPY2R and PNMT). This excess variation is even more extreme in samples with African and European ancestry, since Asian samples in our data show no increase in nucleotide diversity for the CHGA promoter. Among 17 coding-region polymorphisms, 11 encode amino acid substitutions; two of these, heterozygous Gly364Ser and Pro370Leu (positions in the mature CHGA protein), are relatively unusual (minor allele frequency 0.6%–3.1%) variants in the catestatin peptide that seem to influence its function in inhibiting vesicular catecholamine release (table B [online only]). We completely resequenced CHGA from two chimpanzees to determine the likely ancestral alleles at polymorphic sites and verified this ancestry for sites of interest with additional sequence from bonobo and gorilla.
Figure 1.
Resequencing strategy and identified variants. Sequences conserved between mouse and human CHGA were visualized with VISTA (Mayor et al. 2000). Location of common (upper) and rare (lower) SNPs relative to exons and conserved noncoding sequences is indicated by position. Red rods represent nonsynonymous SNPs, and black rods represent synonymous SNPs. Nucleotides in red in the chimpanzee haplotype indicate the minor allele in the human sequence. Computationally reconstructed haplotypes are indicated, along with their relative frequencies in ethnogeographic groups within our sample population. Nucleotide deletions in haplotype sequences are indicated by an asterisk (*).
Table 1.
Nucleotide Diversity (π) in Each Region of CHGA among Four Populations
| π in Population (Total No. of Segregating Sites)a |
|||||
| SequencedRegion (No.of BasesSequenced) | Asian(n=88) | Black(n=114) | Hispanic(n=56) | White(n=102) | Global(n=360) |
| Coding (1,371) | .0009 (8) | .0009 (10) | .0009 (8) | .0007 (7) | .0009 (17) |
| Promoter (1,175) | .0012 (8) | .0022 (11) | .0023 (10) | .0023 (9) | .0021 (13) |
| 5′UTR (213) | .0000 (0) | .0000 (0) | .0000 (0) | .0000 (0) | .0000 (0) |
| Intron (2,556) | .0011 (9) | .0012 (19) | .0009 (11) | .0011 (12) | .0011 (22) |
| 3′UTR (410) | .0003 (1) | .0015 (3) | .0011 (1) | .001 (1) | .0008 (3) |
| Total (5,725) | .0007 (26) | .0009 (43) | .0008 (30) | .0008 (29) | .0008 (55) |
π was calculated as described by Tajima (1989).
Table B.
Summary of CHGA SNP Discovery
|
Minor-Allele Frequency in Population |
||||||||
| SNPNumber | SNPa | SNP Positionb | Encoded Amino Acidc | Asian(n=44) | Black(n=57) | Hispanic(n=28) | White(n=51) | Global(n=180) |
| 1 | g/A | Promoter, −1106 | None | .102 | .277 | .196 | .255 | .215 |
| 2 | a/T | Promoter, −1018 | None | .125 | .107 | .107 | .173 | .130 |
| 3 | t/C | Promoter, −1014 | None | .045 | .237 | .303 | .21 | .195 |
| 4 | g/T | Promoter, −988 | None | .045 | .237 | .303 | .21 | .195 |
| 5 | g/A | Promoter, −722 | None | 0 | 0 | 0 | .01 | .003 |
| 6 | t/C | Promoter, −638 | None | 0 | .009 | .018 | 0 | .006 |
| 7 | g/A | Promoter, −462 | None | .045 | .237 | .303 | .21 | .195 |
| 8 | t/C | Promoter, −415 | None | .384 | .291 | .179 | .281 | .290 |
| 9 | g/A | Promoter, −372 | None | 0 | 0 | .018 | 0 | .003 |
| 10 | g/A | Promoter, −100 | None | 0 | .009 | 0 | 0 | .003 |
| 11 | c/A | Promoter, −89 | None | .023 | .127 | .232 | .198 | .148 |
| 12 | c/A | Promoter, −60 | None | 0 | .009 | 0 | 0 | .006 |
| 13 | c/T | Promoter, −57 | None | .105 | .127 | .107 | .198 | .136 |
| 14 | c/T | 295, intron 1 | None | 0 | 0 | 0 | .01 | .006 |
| 15 | g/C | 1060, exon 2 | A-18-A | .011 | 0 | 0 | 0 | .003 |
| 16 | g/A | 1286, intron 2 | None | 0 | .009 | 0 | 0 | .003 |
| 17 | g/A | 1314, intron 2 | None | 0 | .009 | 0 | .02 | .018 |
| 18 | t/C | 1422, intron 2 | None | 0 | 0 | 0 | .01 | .003 |
| 19 | a/* | 2341, intron 2 | None | .453 | .412 | .446 | .43 | .467 |
| 20 | g/A | 2399, intron 2 | None | .023 | .035 | .071 | .1 | .064 |
| 21 | g/A | 2479, intron 2 | None | 0 | .009 | 0 | 0 | .003 |
| 22 | g/A | 2768, intron 2 | None | 0 | .018 | 0 | 0 | .006 |
| 23 | c/T | 2796, intron 2 | None | .453 | .412 | .446 | .43 | .467 |
| 24 | c/T | 2818, intron 2 | None | 0 | .027 | 0 | 0 | .008 |
| 25 | g/A | 2942, intron 2 | None | .071 | .254 | .346 | .27 | .222 |
| 26 | g/A | 2944, intron 2 | None | 0 | .018 | 0 | 0 | .006 |
| 27 | t/C | 3611, intron 3 | None | 0 | .018 | 0 | .01 | .008 |
| 28 | a/G | 6580, exon 5 | E-89-G | 0 | .009 | 0 | 0 | .003 |
| 29 | g/A | 6596, exon 5 | Q-94-Q | .01 | 0 | 0 | 0 | .003 |
| 30 | c/G | 6838, intron 5 | None | 0 | 0 | .018 | 0 | .003 |
| 31 | t/C | 8132, exon 6 | D-128-D | 0 | .009 | .036 | 0 | .009 |
| 32 | g/A | 8274, exon 6 | E-176-K | 0 | .045 | 0 | 0 | .015 |
| 33 | g/C | 8540, exon 6 | E-264-D | .197 | .236 | .071 | .11 | .173 |
| 34 | g/A | 8587, intron 6 | None | .197 | .236 | .071 | .11 | .173 |
| 35 | g/A | 8687, intron 6 | None | .01 | .009 | .053 | 0 | .015 |
| 36 | g/* | 8757, intron 6 | None | .395 | .202 | .107 | .239 | .242 |
| 37 | g/A | 8759, intron 6 | None | .395 | .202 | .107 | .239 | .242 |
| 38 | c/T | 8955, intron 6 | None | 0 | .018 | .018 | 0 | .014 |
| 39 | c/T | 9105, intron 6 | None | 0 | .009 | 0 | 0 | .003 |
| 40 | t/C | 9179, intron 6 | None | .238 | .037 | .056 | .08 | .101 |
| 41 | c/T | 9226, exon 7 | R-271-W | .018 | 0 | .01 | 0 | .006 |
| 42 | c/G | 9236, exon 7 | A-274-G | 0 | .009 | .018 | 0 | .011 |
| 43 | g/A | 9358, exon 7 | G-315-S | 0 | 0 | 0 | .02 | .006 |
| 44 | t/C | 9410, exon 7 | L-332-P | 0 | .045 | 0 | 0 | .014 |
| 45 | c/T | 9558, exon7 | Y-381-Y | 0 | .009 | 0 | 0 | .003 |
| 46 | g/A | 9559, exon 7 | G-382-S | .018 | 0 | .059 | .049 | .031 |
| 47 | c/T | 9578, exon 7 | P-388-L | 0 | .018 | 0 | 0 | .006 |
| 48 | g/A | 9590, exon 7 | R-392-Q | 0 | 0 | 0 | .01 | .003 |
| 49 | c/T | 9610, exon 7 | R-399-W | .285 | .046 | .231 | .107 | .147 |
| 50 | g/A | 9678, exon 7 | E-421-E | .183 | 0 | .056 | .01 | .057 |
| 51 | g/A | 11583, intron 7 | None | 0 | .109 | 0 | 0 | .034 |
| 52 | g/A | 11687, exon 8 | E-441-E | .07 | .342 | .339 | .275 | .227 |
| 53 | c/T | 11825, exon 8, UTR | None | .07 | .096 | .339 | .274 | .177 |
| 54 | c/T | 11834, exon 8, UTR | None | 0 | .018 | 0 | 0 | .006 |
| 55 | c/T | 12012, exon 8, UTR | None | 0 | .018 | 0 | 0 | .006 |
Nucleotide deletions are indicated by an asterisk (*).
Positions are numbered upstream (−) or downstream (+) of the cap (transcription initiation) site.
Amino acid positions are numbered in premature CHGA protein.
To identify variants that are inherited together, we inferred haplotypes from the 20 most common SNPs in CHGA, through use of PHASE (Stephens et al. 2001). We identified eight major haplotypes, accounting for 74.7% of chromosomes examined (fig. 1). These eight haplotypes explicitly include the common variation in coding and likely regulatory regions and serve as surrogates (by LD) for any unexamined regions that might influence CHGA gene function in our clinically focused samples. Consistent with African origins for modern humans, the eight major haplotypes account for only 51.8% of all haplotypes in African Americans but account for 92% in the Asian sample. (The FST for the CHGA proximal promoter among four populations is 0.035, which is significantly different from 0 [P<.0001]. This is attributable to the difference between the Asian sample and the other three populations: pairwise differences between populations for Asian versus black, Hispanic, and white are independently significant: FST=0.062, 0.125, and 0.057, respectively [P<.0001], but differences among black, Hispanic, and white populations are not significant.)
The unusually high nucleotide diversity in the proximal promoter allowed us to examine this interval in greater detail. PHASE inferred eight haplotypes in the CHGA promoter region (table 2), six of which appeared to be relatively common (>4%). Four promoter SNPs in tight LD with each other (−1014, −988, −462, and −89; see table B [online only]) were common in the general population (although they were found in <5% of Asians), including one SNP (−988) for which the ancestral allele (in common with chimpanzee, bonobo, and gorilla) is the minor allele in all populations. Three of these SNPs (−1014, −988, and −462) are in absolute LD with each other (D′=1; Δ2=1), whereas the fourth (−89) shows complete but not absolute LD with the other three SNPs (D′=1; Δ2=0.744). To illustrate relationships among the promoter haplotypes, we constructed haplotype networks through use of Arlequin (fig. 2A). The haplotype MST structure showed that the four linked SNPs divided CHGA promoter haplotypes into two clusters. The haplotype clusters that differ at these linked sites define a deep division in the human lineage. It is interesting that the chimp promoter haplotype was located between the two clusters, one step closer to the cluster that did not contain minor alleles of the four SNPs. It is surprising that haplotypes that retain the ancestral SNP allele at −988 occur at only a 20% frequency in the current general population. The high π value and divergent haplotype structure in the CHGA promoter could suggest effects of balancing selection, as was shown for the cis-regulatory region of CCR5 (π=0.0021) (Bamshad et al. 2002).
Table 2.
Haplotype Distribution in the CHGA Promoter Region among Four Populations
|
Nucleotide at Position |
Frequency (No. of Chromosomes) in Population |
||||||||||||
| PromotorHaplotypeNumber | −1106 | −1018 | −1014 | −988 | −462 | −415 | −89 | −57 | Asian(n=88) | Black(n=114) | Hispanic(n=56) | White(n=102) | Total(n=360) |
| 1 | G | A | T | T | G | T | C | C | .466 (41) | .211 (24) | .321 (18) | .265 (27) | .306 (110) |
| 2 | A | A | T | T | G | T | C | C | .102 (9) | .272 (31) | .196 (11) | .255 (26) | .214 (77) |
| 3 | G | A | C | G | A | T | A | C | .045 (4) | .123 (14) | .268 (15) | .206 (21) | .15 (54) |
| 4 | G | A | T | T | G | C | C | C | .261 (23) | .158 (18) | .071 (4) | .078 (8) | .147 (53) |
| 5 | G | T | T | T | G | C | C | T | .102 (9) | .105 (12) | .107 (6) | .176 (18) | .125 (45) |
| 6 | G | A | C | G | A | T | C | C | 0 (0) | .114 (13) | .036 (2) | 0 (0) | .042 (15) |
| 7 | G | A | T | T | G | C | C | T | 0 (0) | .018 (2) | 0 (0) | .02 (2) | .011 (4) |
| 8 | G | T | T | T | G | C | C | C | .023 (2) | 0 (0) | 0 (0) | 0 (0) | .006 (2) |
Figure 2.
Functional variation in the CHGA promoter. A, Promoter haplotypes, each represented by a circle whose area represents the overall frequency of that haplotype in the sample. Each haplotype number corresponds to haplotype numbers in table 2. Each circle is subdivided to show the proportion of the individual haplotype frequency found in each of the four populations as represented by the indicated colors. Dashed lines indicate alternative topologies of equal length. Lines connecting haplotypes represent one nucleotide substitution, except where noted in parentheses. B, Association of CHGA proximal promoter SNP genotype (G-988-T) with in vivo plasma CHGA peptide levels in 102 subjects. All CHGA peptides levels are expressed as mean±SEM. Significant differences could be observed in two peptide fragments (large fragment, CHGA116–457; and pancreastatin, CHGA284–301) between minor-allele homozygotes and the other two groups. N = number of subjects for each genotype group. The allele frequencies were 22.5% for G and 77.5% for T. The genotypes were in Hardy-Weinberg equilibrium (χ2=0.011; P=.91). C, In vitro haplotype-specific CHGA promoter activity assay. Two haplotypes (3 and 6) showed a marked decrease in promoter activity compared with the other four common promoter haplotypes, two rare haplotypes, and chimp haplotype. Haplotype numbers correspond to haplotype numbers in table 2. There are also significant differences in promoter activity between haplotypes 3 and 6. *P<.0001 between haplotype 6 and other haplotypes. **P<.001 between haplotype 3 and the other haplotypes except haplotype 6. D, In vitro mutated haplotype activity assay in the CHGA promoter. Each mutated promoter haplotype was derived from either haplotype 1 or haplotype 6 (see table 2).
To determine whether the population structure of CHGA promoter variants is significantly different from expectation under selective neutrality, we applied several well-described tests, including Tajima’s D (Tajima 1989); Fu’s Fs, Fu and Li’s D* and F*, Fu and Li’s D and F, and Fay and Wu’s H test (Fu 1996; Fay and Wu 2000); and the HKA test (Hudson et al. 1987). The resulting statistics do not reach the .05 significance level for derived P values but are close in several cases (table 3), including a positive Tajima’s D and Fu’s Fs in white subjects. This is consistent with—but does not prove—selection acting on the promoter variants we identified. In protein coding sequence, we observed 11 replacement polymorphisms in the four populations but no fixed replacements between human and chimpanzee. This is significant by the McDonald-Kreitman test (McDonald and Kreitman 1991) with P<.001 (the test is independently significant at the .01 level for each ethnic group).
Table 3.
Neutral Selection Tests in the CHGA Promoter Region among Four Populations[Note]
| Population | Tajima’s D | Fu’s Fs | Fu and Li’s D* | Fu and Li’s F* | Fu and Li’s D | Fu and Li’s F | Fay and Wu’s H |
| Asian (n=88) | −.252 (.44) | .644 (.19) | 1.268 (.09) | .891 (.19) | 1.292 (.08) | .906 (.16) | −4.486 (.016) |
| Black (n=114) | .647 (.22) | .472 (.148) | .061 (.36) | .321 (.35) | .050 (.36) | .318 (.38) | −1.322 (.14) |
| Hispanic (n=56) | .766 (.16) | .953 (.159) | .136 (.39) | .408 (.38) | .118 (.34) | .403 (.36) | −.529 (.23) |
| White (n=102) | 1.460 (.074) | 2.757 (.068) | .564 (.45) | 1.037 (.14) | .568 (.16) | 1.049 (.13) | −1.594 (.1) |
| Global (n=360) | .498 (.24) | .059 (.125) | −.703 (.15) | −.293 (.38) | −.715 (.15) | −.300 (.40) | −1.867 (.09) |
Note.— Nominal P values are given in parentheses for each test.
To test the functional significance of common variations in CHGA, we first examined the influence of individual common polymorphisms on the expression of CHGA gene products in vivo (fig. 2B). We looked at associations of SNPs across CHGA with four plasma CHGA peptide fragments in 102 subjects for whom we had both physiologic and gene resequencing data. One-way ANOVA with Bonferroni correction for multiple comparisons identified significant association with plasma CHGA for only the three SNPs (−1014, −988, and −462) that presented absolute LD with each other in the promoter region, suggesting a haplotype-specific effect on plasma levels. Minor-allele homozygotes had significantly elevated levels of the two most abundant plasma CHGA peptides, compared with heterozygotes or major-allele homozygotes.
To test further the functional significance of common variation in the promoter region, we assayed expression of CHGA promoter haplotype-specific reporter constructs (fig. 2C). We placed the eight inferred promoter haplotypes upstream of a luciferase reporter and assayed expression in PC12 cells, which model a normal site of CHGA expression. It is interesting that the two common haplotypes containing minor alleles at the four complete-LD sites showed significantly lower expression than all the others, including chimpanzee. Haplotypes 3 (GACGATAC) and 6 (GACGATCC) are also significantly different from each other in expression level but differ in sequence only at −89. These results indicate that SNP −89 and at least one of the three SNPs in absolute LD are functionally significant.
To determine which of the three SNPs in absolute LD affect promoter activity, we mutated each of these sites in the promoter-reporter constructs for high-expressing haplotype 1 and low-expressing haplotype 6 (fig. 2D). For replacements on haplotype 1, only mutation of −462 (1−462A) shows significant decrease of promoter activity. For replacements on low-expressing haplotype 6, mutation of either −462 (6−462G) or −988 (6−988T) significantly increases expression, with −462G conferring the largest increase.
To test the functional significance of the rare amino acid replacements we found in the catestatin peptide sequence, we synthesized wild-type and variant peptides and assayed their potency for inhibition of nicotinic cholinergic-stimulated catecholamine release from chromaffin (PC12) cells (fig. 3). The Gly364Ser substitution alters a site that is otherwise absolutely conserved among seven mammalian species for which sequence is available but occurs in 11 of 180 subjects. This substitution in synthetic catestatin results in a 4.7-fold loss of potency in our assay. Pro370Leu occurred in 2 of 180 subjects; it is interesting that 370Leu is the normal allele in all reported nonprimate mammals. This substitution results in a 2.3-fold gain of potency of the synthetic peptide (fig. 3A and 3B). It is interesting that the Gly364Ser variant is distributed across other ethnic groups (five are Asian samples, five are white samples, and one is a Hispanic sample) but is absent from the African American sample (P<.01; χ2 test), whereas the Pro370Leu variant occurred only in African American samples.
Figure 3.
Catestatin peptide variants altering cholinergic inhibition and predicted structure. A, Peptide sequence alignment from several species (and the corresponding catestatin region), showing the extent of sequence conservation at Gly364Ser and Pro370Leu (red, second and third rows). Note that 370Leu is found in all nonprimate species available. Phylogenetic relationships and bootstrap values are taken from a published multiple gene comparison (Murphy et al. 2001). B, Altered efficacy of nicotinic inhibition by variant peptides in dose response from 0.1 to 10 μM, showing functional significance to each change, but in opposite directions. C, Homology modeling, predicting altered three-dimensional structure of Gly364Ser (left) and Pro370Leu (right) variants. Point mutants were aligned to the wild-type backbone template and then subjected to energy-minimization/homology-modeling using SWISS-MODEL at the ExPASy Web site and Swiss-PDBviewer (“DeepView”) for visualization and manipulation (Peitsch 1995).
To determine how these variants could influence peptide potency, we modeled the impact of amino acid substitutions on the NMR structure (PDB 1lv4) of human catestatin that we had recently determined. Homology modeling suggests that Gly364Ser distorts the peptide backbone in two locations, by 0.36 Å in the middle loop near the site of substitution and by 1.48 Å in the carboxy-terminal strand, which is adjacent in the three-dimensional structure (fig. 3C, left). By contrast, the Pro370Leu substitution model predicts a 0.59-Å shift in the middle loop but no shift in the carboxy-terminal strand near the site of substitution (fig. 3C, right).
Discussion
CHGA plays crucial roles in the sympathoadrenal system, both in the formation of catecholamine secretory vesicles and in the regulation of catecholamine release by nicotinic cholinergic stimuli (Taupenot et al. 2003). Here, we find both quantitative variation in expression level mediated by common promoter haplotypes and qualitative variation in rare catestatin peptide polymorphisms that alter its inhibitory potency several fold. We find 13 promoter SNPs that resolve into six relatively common haplotypes (frequencies >4%). These 5′ proximal haplotypes differ quantitatively in the ability to promote transcription, and one of the functionally variant SNP alleles (G-988-T) in the low-expression haplotype is an ancestral allele that has diminished in allele frequency in a region of likely natural selection. The functional variation we observed could be a substrate for environmental selective pressures dependent on catecholaminergic function.
Tests of selective neutrality provide modest support for recent selection on CHGA promoter haplotypes in white and Asian subjects. Tajima’s D and Fu’s Fs tests show positive statistics just above traditional significance thresholds in white subjects (with P values of .074 and .068, respectively), whereas Fu and Li’s D and D* tests are nearly significant and Fay and Wu’s H test is significant in Asian subjects (with P values of .08, .09, and .016, respectively). The difference between populations probably reflects the near absence of haplotypes 3 and 6 in Asian samples, increasing the proportion of chromosomes containing the nonancestral allele at position −988. We do not see significant deviation from neutrality in the other populations, but the statistical methods are relatively conservative and have power to detect selection only under limited circumstances.
Our results suggest that selection in modern humans favors haplotypes that impart a moderate range of CHGA expression. It is interesting to note that the most extreme promoter haplotypes for functional expression (6 and 7) are infrequent and are intermediates that link more-moderate and more-common promoter haplotypes in the minimum spanning network. In the coding sequence, the McDonald-Kreitman test showed significant departure from neutrality, because of the lack of fixed replacements. Consistent with purifying selection against replacements, we find that two relatively infrequent replacement polymorphisms in the catestatin region alter peptide potency several fold. Taken together with our functional studies, our molecular evolutionary data suggest the importance of maintaining CHGA expression within an approximately twofold range, but more-sensitive and more-powerful approaches will likely be needed to confirm the influence of selection in shaping modern haplotypes.
In vitro promoter studies indicate that three of the promoter SNPs we identified have functional consequences. The −89 SNP is the only difference between promoter haplotypes 3 and 6, accounting for ∼30% change in expression level. The −462 SNP accounts for another 30%–40% change, including most of the difference between haplotypes 1 and 6. It is interesting that the SNP at −988 had no effect on haplotype 1 but accounted for a slight increase in expression from haplotype 6. This may suggest an epistatic interaction among SNPs within the variant promoter haplotypes. The −462 and −89 polymorphisms each sit within consensus sites for known transcription factor families, identified computationally by rVista (Loots et al. 2002) or TESS (Schug and Overton 1997). These include TCF1α and GATA sites at −462 and one of several potential AP-2 sites at −89. However, occupancy and differentiation of these sites need to be demonstrated in vivo before a precise transcriptional mechanism for the altered expression phenotypes can be confirmed.
Two relatively infrequent amino acid replacement cSNPs in the catestatin peptide (CHGA352–372) produce quite different effects on catestatin potency, ultimately differing by 10.8-fold in their potency to inhibit nicotinic stimulation of catecholamine release from chromaffin cells. The Gly364Ser variant occurs at a highly conserved site among mammalian catestatins. The 364Ser variant we discovered reduces catestatin activity ∼4.7-fold. In contrast, the 370Leu variant is the normal allele in all mammalian species sequenced to date, except human and chimpanzee; thus, the Pro370Leu allele that increases catestatin activity ∼2.3-fold is a reversion to the usual amino acid seen among mammals.
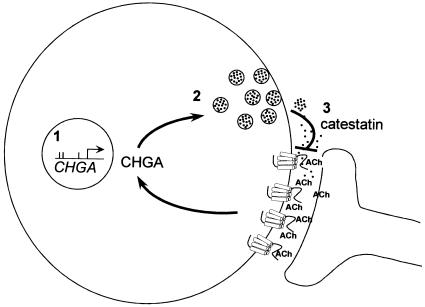
Our data demonstrate that both common and rare variants contribute to functional polymorphism at CHGA and suggest a model (fig. 4) for feedback regulation of catecholamine and chromogranin release mediated by catestatin. Correlations between quantitative functional alterations and allele frequencies suggest that the level of CHGA and catestatin is under selection but that different activities of CHGA may be under different selective pressures. Although larger cohort sizes will need to be examined to determine the impact of these alleles on complex phenotypes such as blood pressure, our results clearly reinforce a likely role for CHGA variation in intermediate phenotypes governing catecholamine physiology and suggest that current haplotype diversity at CHGA has been shaped by selective pressure in the modern human lineage.
Figure 4.
1-2-3 model for influence of CHGA polymorphisms on catecholamine storage and release. 1, Common promoter haplotypes affect the transcriptional level of the CHGA gene in chromaffin cells. 2, CHGA protein levels quantitatively affect pool size of catecholamine-chromogranin vesicles. 3, Variant catestatin peptides alter the release-dependent feedback inhibition of nicotinic-stimulated release. This feedback loop could explain the divergent effects of promoter polymorphisms on chromaffin cell CHGA expression and plasma levels.
Acknowledgments
The authors thank Brinda K. Rana for assistance and helpful discussion. This work was funded by program project grant P01 HL58120 from the National Heart, Lung and Blood Institute. B.A.H. is a Pew Scholar in the Biomedical Sciences.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CHGA) [PubMed]
- Primer3, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi (for design of PCR primers)
- Protein Data Bank, http://www.rcsb.org/pdb/ (for the NMR structure of catestatin, entry “1lv4”)
- SWISS-MODEL at ExPASy, http://www.expasy.org/swissmod/SWISS-MODEL.html
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/
References
- Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 10.1093/bioinformatics/16.2.182 [DOI] [PubMed] [Google Scholar]
- Bamshad MJ, Mummidi S, Gonzalez E, Ahuja SS, Dunn DM, Watkins WS, Wooding S, Stone AC, Jorde LB, Weiss RB, Ahuja SK (2002) A strong signature of balancing selection in the 5′ cis-regulatory region of CCR5. PNAS 99:10539–10544 10.1073/pnas.162046399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa JA, Gill BM, Takiyyuddin MA, O’Connor DT (1991) Chromogranin A: posttranslational modifications in secretory granules. Endocrinology 128:174–190 [DOI] [PubMed] [Google Scholar]
- Cadman PE, Rao F, Mahata SK, O’Connor DT (2002) Studies of the dysglycemic peptide pancreastatin, using a human forearm model. Ann NY Acad Sci 971:528–529 [DOI] [PubMed] [Google Scholar]
- Eskeland NL, Zhou A, Dinh TQ, Wu H, Parmer RJ, Mains RE, O’Connor DT (1996) Chromogranin A processing and secretion: specific role of endogenous and exogenous prohormone convertases in the regulated secretory pathway. J Clin Invest 98:148–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194 [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185 [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu C-I (2000) Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX (1996) New statistical tests of neutrality for DNA samples from a population. Genetics 143:557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202 [DOI] [PubMed] [Google Scholar]
- Herrmann V, Buscher R, Go MM, Ring KM, Hofer JK, Kailasam MT, O’Connor DT, Parmer RJ, Insel PA (2000) β2-adrenergic receptor polymorphisms at codon 16, cardiovascular phenotypes and essential hypertension in whites and African Americans. Am J Hypertens 13:1021–1026 10.1016/S0895-7061(00)01188-2 [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Taupenot L, Mahata SK, Mahata M, O’Connor DT, Miles LA, Parmer RJ (2001) Proteolytic cleavage of chromogranin A (CgA) by plasmin: selective liberation of a specific bioactive CgA fragment that regulates catecholamine release. J Biol Chem 276:25022–25029 10.1074/jbc.M101545200 [DOI] [PubMed] [Google Scholar]
- Kennedy BP, Mahata SK, O’Connor DT, Ziegler MG (1998) Mechanism of cardiovascular actions of the chromogranin A fragment catestatin in vivo. Peptides 19:1241–1248 10.1016/S0196-9781(98)00086-2 [DOI] [PubMed] [Google Scholar]
- Kim T, Tao-Cheng JH, Eiden LE, Loh YP (2001) Chromogranin A, an “on/off” switch controlling dense-core secretory granule biogenesis. Cell 106:499–509 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245 10.1093/bioinformatics/17.12.1244 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Loots GG, Ovcharenko I, Pachter L, Dubchak I, Rubin EM (2002) rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res 12:832–839 10.1101/gr.225502. Article published online before print in April 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahata M, Mahata SK, Parmer RJ, O’Connor DT (1998) Proadrenomedullin N-terminal 20 peptide: minimal active region to regulate nicotinic receptors. Hypertension 32:907–916 [DOI] [PubMed] [Google Scholar]
- ——— (1999) Desensitization of catecholamine release. The novel catecholamine release-inhibitory peptide catestatin (chromogranin A344–364) acts at the receptor to prevent nicotinic cholinergic tolerance. J Biol Chem 274:2920–2928 10.1074/jbc.274.5.2920 [DOI] [PubMed] [Google Scholar]
- Mahata SK, Mahata M, Wakade AR, O’Connor DT (2000) Primary structure and function of the catecholamine release inhibitory peptide catestatin (chromogranin A344–364): identification of amino acid residues crucial for activity. Mol Endocrinol 14:1525–1535 [DOI] [PubMed] [Google Scholar]
- Mahata SK, O’Connor DT, Mahata M, Yoo SH, Taupenot L, Wu H, Gill BM, Parmer RJ (1997) Novel autocrine feedback control of catecholamine release: a discrete chromogranin a fragment is a noncompetitive nicotinic cholinergic antagonist. J Clin Invest 100:1623–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046–1047 10.1093/bioinformatics/16.11.1046 [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654 10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- Miall WE, Oldham PD (1963) The hereditary factor in arterial blood pressure. Brit Med J 19:75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, O’Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW, Springer MS (2001) Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294:2348–2351 10.1126/science.1067179 [DOI] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751 10.1093/nar/25.14.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor DT, Insel PA, Ziegler MG, Hook VY, Smith DW, Hamilton BA, Taylor PW, Parmer RJ (2000) Heredity and the autonomic nervous system in human hypertension. Curr Hypertens Rep 2:16–22 [DOI] [PubMed] [Google Scholar]
- O’Connor DT, Kailasam MT, Kennedy BP, Ziegler MG, Yanaihara N, Parmer RJ (2002) Early decline in the catecholamine release-inhibitory peptide catestatin in humans at genetic risk of hypertension. J Hypertens 20:1335–1345 10.1097/00004872-200207000-00020 [DOI] [PubMed] [Google Scholar]
- O’Connor DT, Takiyyuddin MA, Printz MP, Dinh TQ, Barbosa JA, Rozansky DJ, Mahata SK, Wu H, Kennedy BP, Ziegler MG, Wright FA, Schlager G, Parmer RJ (1999) Catecholamine storage vesicle protein expression in genetic hypertension. Blood Press 8:285–295 10.1080/080370599439508 [DOI] [PubMed] [Google Scholar]
- Peitsch MC (1995) Protein modeling by e-mail. Bio/Technology 13:658–660 [Google Scholar]
- Rieder MJ, Taylor SL, Tobe VO, Nickerson DA (1998) Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res 26:967–973 10.1093/nar/26.4.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozansky DJ, Wu H, Tang K, Parmer RJ, O’Connor DT (1994) Glucocorticoid activation of chromogranin A gene expression: identification and characterization of a novel glucocorticoid response element. J Clin Invest 94:2357–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175 10.1093/bioinformatics/15.2.174 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN: a software for population genetics data analysis. University of Geneva, Geneva [Google Scholar]
- Schober M, Howe PR, Sperk G, Fischer-Colbrie R, Winkler H (1989) An increased pool of secretory hormones and peptides in adrenal medulla of stroke-prone spontaneously hypertensive rats. Hypertension 13:469–474 [DOI] [PubMed] [Google Scholar]
- Schug J, Overton GC (1997) TESS: transcription element search software on the WWW. University of Pennsylvania School of Medicine, Philadelphia [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stridsberg M (2000) Measurements of chromogranins and chromogranin-related peptides by immunological methods. In: Helle KB, Aunis D (eds) Chromogranins: functional and clinical aspects. Kluwer Academic/Plenum Publications, New York, pp 319–327 [DOI] [PubMed] [Google Scholar]
- Stridsberg M, Angeletti RH, Helle KB (2000) Characterisation of N-terminal chromogranin A and chromogranin B in mammals by region-specific radioimmunoassys and chromatographic separation methods. J Endocrinol 165:703–714 [DOI] [PubMed] [Google Scholar]
- Stridsberg M, Oberg K, Li Q, Engstrom U, Lundqvist G (1995) Measurements of chromogranin A, chromogranin B (secretogranin I), chromogranin C (secretogranin II) and pancreastatin in plasma and urine from patients with carcinoid tumors and endocrine pancreatic tumors. J Endocrinol 144:49–59 [DOI] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiyyuddin MA, De Nicola L, Gabbai FB, Dinh TQ, Kennedy B, Ziegler MG, Sabban EL, Parmer RJ, O’Connor DT (1993) Catecholamine secretory vesicles. Augmented chromogranins and amines in secondary hypertension. Hypertension 21:674–679 [DOI] [PubMed] [Google Scholar]
- Takiyyuddin MA, Parmer RJ, Kailasam MT, Cervenka JH, Kennedy B, Ziegler MG, Lin MC, Li J, Grim CE, Wright FA, O’Connor DT (1995) Chromogranin A in human hypertension: influence of heredity. Hypertension 26:213–220 [DOI] [PubMed] [Google Scholar]
- Tasiemski A, Hammad H, Vandenbulcke F, Breton C, Bilfinger TJ, Pestel J, Salzet M (2002) Presence of chromogranin-derived antimicrobial peptides in plasma during coronary artery bypass surgery and evidence of an immune origin of these peptides. Blood 100:553–559 10.1182/blood.V100.2.553 [DOI] [PubMed] [Google Scholar]
- Taupenot L, Harper KL, Mahapatra NR, Parmer RJ, O’Connor DT (2002) Intracellular protein trafficking into catecholamine storage vesicles: novel chimeric photoproteins visualized by deconvolution fluorescence microscopy. Ann NY Acad Sci 971:262–265 [DOI] [PubMed] [Google Scholar]
- Taupenot L, Harper KL, O’Connor DT (2003) The chromogranin-secretogranin family. N Engl J Med 348:1134–1149 10.1056/NEJMra021405 [DOI] [PubMed] [Google Scholar]
- Taupenot L, Mahata SK, Mahata M, Parmer RJ, O’Connor DT (2000) Interaction of the catecholamine release-inhibitory peptide catestatin (human chromogranin A352–372) with the chromaffin cell surface and Torpedo electroplax: implications for nicotinic cholinergic antagonism. Regul Pept 95:9–17 10.1016/S0167-0115(00)00135-X [DOI] [PubMed] [Google Scholar]
- Taylor CV, Taupenot L, Mahata SK, Mahata M, Wu H, Yasothornsrikul S, Toneff T, Caporale C, Jiang Q, Parmer RJ, Hook VY, O’Connor DT (2000) Formation of the catecholamine release-inhibitory peptide catestatin from chromogranin A: determination of proteolytic cleavage sites in hormone storage granules. J Biol Chem 275:22905–22915 10.1074/jbc.M001232200 [DOI] [PubMed] [Google Scholar]
- Timberlake DS, O’Connor DT, Parmer RJ (2001) Molecular genetics of essential hypertension: recent results and emerging strategies. Curr Opin Nephrol Hypertens 10:71–79 [DOI] [PubMed] [Google Scholar]
- Tsigelny I, Mahata SK, Taupenot L, Preece NE, Mahata M, Khan I, Parmer RJ, O’Connor DT (1998) Mechanism of action of chromogranin A on catecholamine release: molecular modeling of the catestatin region reveals a beta-strand/loop/beta-strand structure secured by hydrophobic interactions and predictive of activity. Regul Pept 77:43–53 10.1016/S0167-0115(98)00040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videen JS, Mezger MS, Chang YM, O’Connor DT (1992) Calcium and catecholamine interactions with adrenal chromogranins. Comparison of driving forces in binding and aggregation. J Biol Chem 267:3066–3073 [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, et al (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562 10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- Wu H, Rozansky DJ, Webster NJ, O’Connor DT (1994) Cell type-specific gene expression in the neuroendocrine system. A neuroendocrine-specific regulatory element in the promoter of chromogranin A, a ubiquitous secretory granule core protein. J Clin Invest 94:118–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, So SH, Huh YH, Park HY (2002) Inositol 1,4,5-trisphosphate receptor/Ca2+ channel modulatory role of chromogranins A and B. Ann NY Acad Sci 971:300–310 [DOI] [PubMed] [Google Scholar]