Abstract
The ability to taste phenylthiocarbamide (PTC) is a classic phenotype that has long been known to vary in human populations. This phenotype is of genetic, epidemiologic, and evolutionary interest because the ability to taste PTC is correlated with the ability to taste other bitter substances, many of which are toxic. Thus, variation in PTC perception may reflect variation in dietary preferences throughout human history and could correlate with susceptibility to diet-related diseases in modern populations. To test R. A. Fisher’s long-standing hypothesis that variability in PTC perception has been maintained by balancing natural selection, we examined patterns of DNA sequence variation in the recently identified PTC gene, which accounts for up to 85% of phenotypic variance in the trait. We analyzed the entire coding region of PTC (1,002 bp) in a sample of 330 chromosomes collected from African (n=62), Asian (n=138), European (n=110), and North American (n=20) populations by use of new statistical tests for natural selection that take into account the potentially confounding effects of human population growth. Two intermediate-frequency haplotypes corresponding to “taster” and “nontaster” phenotypes were found. These haplotypes had similar frequencies across Africa, Asia, and Europe. Genetic differentiation between the continental population samples was low (FST=0.056) in comparison with estimates based on other genes. In addition, Tajima’s D and Fu and Li’s D and F statistics demonstrated a significant deviation from neutrality because of an excess of intermediate-frequency variants when human population growth was taken into account (P<.01). These results combine to suggest that balancing natural selection has acted to maintain “taster” and “nontaster” alleles at the PTC locus in humans.
Introduction
Variation in the ability to taste the synthetic compound phenylthiocarbamide (PTC [MIM 171200]) was first recognized in the early 1930s, when A. J. Fox discovered the polymorphism in himself and a coworker, organic chemist C. R. Noller (Anonymous 1931; Fox 1932). The inability of some individuals to taste PTC was found to segregate in a nearly Mendelian recessive manner (Blakeslee 1932), and subsequent studies went on to estimate the frequency of presumed “taster” and “nontaster” alleles in hundreds of populations worldwide (Guo and Reed 2001). These studies showed that the frequency of the nontaster allele in human populations (estimated under the assumption that the inability to taste PTC is attributable to the recessive allele in a one-locus, two-allele system) varies around a mean of ∼50%, as shown in figure 1.
Figure 1.
Frequency of PTC “nontaster” allele, estimated from 348 populations described by Guo and Reed (2001) under the assumption that the inability to taste PTC is attributable to the recessive allele in a one-locus, two-allele system.
Although PTC itself has not been found in nature, the ability to taste PTC is correlated strongly with the ability to taste other naturally occurring bitter substances, many of which are toxic (Harris and Kalmus 1949; Barnicot et al. 1951; Tepper 1998). Furthermore, variation in PTC taste sensitivity has been correlated with dietary preferences that may have significant health effects (Bartoshuk et al. 1994). For example, PTC is similar in structure to isothiocyanates and goitrin, both of which are bitter substances found in cruciferous vegetables like cabbage and broccoli (Tepper 1998). Variable aversions to these compounds have been implicated in the variable rates of thyroid-deficiency disease in PTC tasters and nontasters, with nontasters being more susceptible (Drewnowski and Rock 1995).
The correlation of PTC perception with the perception of other toxic bitter substances and the association of the inability to perceive PTC with disease susceptibility combine to suggest that natural selection may have been an important factor in the evolution of this trait. R. A. Fisher hypothesized that the pervasive phenotypic variation in PTC perception is due to balancing natural selection, which may have favored heterozygotes (Fisher et al. 1939). However, such hypotheses have been difficult to test without knowledge of the genetic underpinnings of the trait. The recent discovery of a gene that accounts for up to 85% of the phenotypic variance in PTC perception allowed us to address this problem (Drayna et al. 2003; Kim et al. 2003).
Here, we describe an investigation of selective effects on the PTC gene by use of analyses of molecular genetic data. By examining patterns of DNA sequence variation, we were able to test the PTC gene for evidence of long-term selective pressures. As predicted by Fisher >60 years ago (Fisher et al. 1939), we found support for the hypothesis that balancing natural selection has acted to maintain taster and nontaster alleles in human populations.
Methods
DNA sequences from the entire coding region of PTC (1,002 bp, 333 amino acids) were obtained by use of methods described in the work of Kim et al. (2003) and Drayna et al. (2003) from 165 individuals of the following descents: African (9 sub-Saharan Africans from Coriell Human Variation panel HD12, 22 Cameroonians), Asian (17 Chinese, 13 Japanese, 12 Koreans, 7 Middle Easterners, 10 Pakistanis, 10 other Southeast Asians), European (10 Hungarians, 45 Utah samples from Centre d'Etude du Polymorphisme Humain), and North American (10 Southwest Native Americans). For comparison, sequences were also obtained from one chimpanzee (Pan troglodytes) and one gorilla (Gorilla gorilla).
Ambiguous haplotypes were resolved using molecular techniques. In such individuals, the two allelic versions of the gene were cloned as single PCR products and individual clones were sequenced to reveal both haplotypes. Phylogenetic relationships among haplotypes were inferred using the neighbor-joining algorithm of the PHYLIP software package (Felsenstein 1993). This tree, which was rooted using the gorilla sequence, was then used to determine the polarity of character states. Evolutionary relationships among haplotypes were visualized using a minimum spanning tree generated by the ARLEQUIN computer program (Schneider et al. 2000).
Tajima’s D (1989) and Fu and Li’s D and F statistics (1993) were used to test the hypothesis that patterns of diversity in humans are consistent with the hypothesis of neutrality. To avoid confusion, we refer to Tajima’s D as “DT” and Fu and Li’s D as “DF.” These tests were performed by simulating 10,000 gene genealogies and comparing statistics obtained from the simulations with the observed statistic, as described by Tajima (1989) and Fu and Li (1993). To incorporate varying assumptions about population size change in human populations, these simulations were performed using the algorithm of Rogers (1995). This algorithm assumes that human population sizes increased suddenly from an ancient population size (N0) to a larger population size (N1), t generations ago, with infinite-sites mutation rate μ. Patterns of genetic diversity produced under these conditions approximate those produced under more complicated conditions, such as exponential and logistic growth (Wooding and Rogers 2002).
Tests for excesses of synonymous and nonsynonymous nucleotide substitutions were performed using the methods of McDonald and Kreitman (1991) and Li et al. (1985). The McDonald-Kreitman test (McDonald and Kreitman 1991) uses a Fisher’s exact test to determine whether the ratio of synonymous and nonsynonymous substitutions differs between two categories: polymorphisms that are variable within species and polymorphisms that distinguish species (i.e., fixed differences). We used the McDonald-Kreitman test to examine polymorphisms found in humans and chimpanzees. The KA/KS test determines whether there is an overall excess of synonymous or nonsynonymous nucleotide substitutions (Li et al. 1985).
Tests for genetic differentiation between populations were performed using Slatkin’s linearized FST statistic (Slatkin 1991). The statistical significance of these values was assessed using the bootstrap method of Excoffier et al. (1992), in which observed values are compared with FST values simulated by randomly allocating chromosomes to different populations. These tests used 10,000 bootstrap replications.
Results and Discussion
DNA sequencing revealed five variable nucleotides in humans. These variants were partitioned into seven haplotypes, as shown in figure 2. The chimpanzee and gorilla were both homozygous at all nucleotide positions and thus carried one haplotype each. Human and chimpanzee sequences differed by an average of 8.3 nucleotides, as did human and gorilla sequences. The chimpanzee and gorilla sequences differed by six nucleotides.
Figure 2.
Variable nucleotide positions in PTC haplotypes. Each haplotype is summarized in two rows. The top row summarizes nucleotide variation in the haplotype, and the bottom row summarizes amino acid variation in the haplotype. Each column represents a codon containing a variable nucleotide position, indicated at the top of the column. The number of occurrences of each haplotype is indicated to the right for the African (Af), Asian (As), European (Eu), and North American (NA) samples. Haplotype counts are not given for the chimpanzee and gorilla haplotypes (ptA and ggA, respectively), which were each observed twice. Shaded columns indicate the three variable amino acid positions used for haplotype designation by Kim et al. (2003).
All five nucleotide substitutions observed in humans caused amino acid substitutions, as did three of the six substitutions distinguishing human and chimpanzee. The observed rate of nonsynonymous substitution was substantially higher than is usually observed in human genes, suggesting that positive natural selection may have acted to preserve new nonsynonymous variants (Makalowski and Boguski 1998; Yang and Bielawski 2000; Nekrushenko et al. 2001; Bamshad and Wooding 2003). To test the hypothesis that an excess of nonsynonymous substitutions was present, we first analyzed the human and chimpanzee sequences by use of a McDonald-Kreitman test (1991), as described in the “Methods” section. This test showed that the excess of nonsynonymous substitutions observed in humans was not statistically significant (P>.10). A KA/KS test showed that the overall ratio of synonymous to nonsynonymous substitutions did not differ significantly from expectation under neutrality (P>.10). These nonsignificant results may be attributable to the low number of polymorphisms observed, which weakens the tests. Thus, although notable for being higher than in most genes, the bias toward nonsynonymous variants in our sample was not sufficient to reject the hypothesis of neutrality.
The minimum spanning tree revealed that the human sample was dominated by two major haplotypes, hsA and hsG, differing by three amino acid substitutions, as shown in figure 3A. These two haplotypes, which account for >90% of sampled chromosomes (fig. 3), are strongly associated with taster (hsA) and nontaster (hsG) status, respectively (Drayna et al. 2003; Kim et al. 2003). In addition, the hsA and hsG haplotypes were both found at intermediate frequencies: 0.55 and 0.38. A variety of factors, including population subdivision and balancing natural selection, can lead to the presence of two or more intermediate-frequency haplotypes in gene genealogies (Marjoram and Donnelly 1994; Bamshad and Wooding 2003). The evolution of two or more intermediate-frequency clusters is also surprisingly common under selectively neutral conditions (Slatkin and Hudson 1991).
Figure 3.
Minimum spanning tree of relationships between PTC gene haplotypes and haplotype frequencies. A, Each circle represents a haplotype, and the size of each circle represents the haplotype’s relative frequency. Within each circle, shading indicates the fraction of observations in the population indicated. Each connection between haplotypes corresponds to one nucleotide substitution, except where indicated by parentheses. For example, haplotype A differs from haplotype E by one nucleotide, but it differs from the chimpanzee haplotype by six nucleotides. Differences between haplotypes can be determined from figure 2. The three-letter codes above the network diagram indicate the haplotype designation used by Kim et al. (2003). B, Haplotype frequencies in each sample.
To test whether patterns of DNA sequence variation in PTC fit expectations under the hypothesis of evolutionary neutrality, we analyzed the sequences by use of the DT, DF, and F statistics. These statistics are functions of the number of variable nucleotide positions in a sample of sequences, the mean pairwise difference between sequences, and the number of derived variants that are only observed once in the sample, all of which are affected by natural selection (Tajima 1989; Fu and Li 1993). For example, positive natural selection leading to the rapid fixation of a single, advantageous haplotype will result in a decrease in the expected number of polymorphic sites, a decrease in the mean pairwise difference between sequences, and an increase in the number of variants observed only once in the sample (Fu and Li 1993). In contrast, balancing natural selection can lead to an increase in all three of these values (Fu and Li 1993).
Tests of these statistics performed under the standard assumption of constant population size failed to reject the hypothesis of neutrality in PTC (DT = 1.55, P>.05; DF = −1.46, P>.90; F=-0.50, P>.60). However, several lines of evidence based on archaeology, genetics, and linguistics suggest that human populations have grown dramatically (>100-fold) over the past 100,000 years (Ruhlen 1994; Klein 1999; Stiner et al. 1999; Excoffier 2002). Such growth is known to have strong effects on genetic diversity (Rogers and Harpending 1992). For example, diversity patterns in populations that have grown are often characterized by an excess of low-frequency genetic variants and a low mean pairwise difference between sequences, both of which lead to reductions in the expected values of all three of the statistics we tested (Wooding and Rogers 2002). Given evidence for population increase in the Upper Pleistocene and the possible effects of this growth on patterns of genetic diversity, the assumption of constant population size is likely inappropriate.
To investigate the possibility that incorrect assumptions about population history were causing a type II statistical error (i.e., a failure to reject the null hypothesis of neutrality when it is false) in our initial tests, we devised new tests of the DT, DF, and F statistics that take population growth into account. These tests were performed as described in the “Methods” section, under the assumption that human populations increased suddenly from an ancient effective population size of 10,000 to a larger effective population size, N1, t years before present, with a nucleotide-substitution rate of 10-9/site/year. These values are representative of those inferred for nuclear genes in humans (Tishkoff and Verrelli 2003). Because there is some disagreement about the timing and magnitude of this expansion (Hey 1997; Fay and Wu 1999; Harris and Hey 1999; Hey and Harris 1999; Harpending and Rogers 2000; Wall and Przeworski 2000; Excoffier 2002; Ptak and Przeworski 2002), we iteratively tested the DT, DF, and F statistics for population histories with magnitudes of population growth from 1-fold to 1,000-fold and dates of population expansion from 0 to 200,000 years ago.
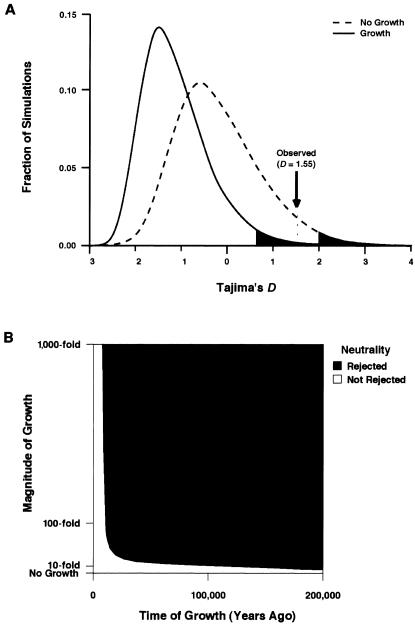
This procedure revealed that tests of all three statistics are highly sensitive to assumptions about population growth. For example, the assumption of 100-fold growth 100,000 years ago resulted in a change of DT’s P value from .07 to .01, as shown in figure 4A. CIs generated for the DT statistic showed that the hypothesis of neutrality was rejected (at a two-tailed P value cutoff of .025) under all population histories in which the human population expanded between 15-fold and 1,000-fold between 10,000 and 200,000 years ago, as shown in figure 4B. In addition, under the population history parameters for which observed DT values differed significantly from expectation, the values were greater than expected. Thus, our data departed from expectation in a direction consistent with the hypothesis of balancing natural selection (Tajima 1989; Fu and Li 1993). Results were similar for CIs generated using the DF and F statistics, which rejected the hypothesis of neutrality for all population histories in which the human population expanded between 15-fold and 1,000-fold between 30,000 and 200,000 years ago. Thus, the hypothesis of neutrality in PTC was rejected by these tests under all but the most conservative assumptions about population growth in humans.
Figure 4.
Tests of the DT statistic. A, Comparison of the probability distribution of the DT statistic under the assumption that human population sizes have been constant and under the assumption that, 100,000 years ago, human population sizes increased 100-fold. Shaded areas are upper 2.5% confidence limits. B, Tests under varying population history parameters. Blue areas indicate parameters under which the hypothesis of neutrality was rejected (P<.025). Yellow areas indicate parameters under which the hypothesis of neutrality was not rejected.
The sensitivity of the DT, DF, and F statistics to population growth has implications beyond the detection of natural selection in PTC. All three of these statistics are widely used in tests for natural selection in humans, usually under the assumption that human population sizes have remained constant (Tishkoff and Verrelli 2003). As we have shown, this assumption is highly conservative in the detection of balancing natural selection. However, the assumption of constant population size is anticonservative in the detection of positive natural selection, which leads to reductions in diversity nearly identical to those caused by population growth (Tajima 1989; Fu and Li 1993). For this reason, tests for positive natural selection that use the DT, DF, and F statistics are vulnerable to type I statistical errors (i.e., the rejection of the null hypothesis when it is true) if human population increases are not taken into account. With this in mind, the significance of many earlier tests of the DT, DF, and F statistics in humans, including our own (e.g., Wooding and Rogers 2000; Wooding et al. 2002), may need to be reconsidered.
Balancing selection is not the only force that can lead to significantly high DT, DF, and F values. Such patterns can also be caused by population subdivision, which allows the persistence of divergent haplotypes in different geographical regions (Kaplan et al. 1991; Hudson et al. 1992; Wakeley 2001; Laporte and Charlesworth 2002). In our analyses, two sources of population subdivision were potentially important: subdivision between continents and subdivision within Africa.
Population subdivision between continents is not large (Tishkoff and Verrelli 2003), but it could be sufficient to confound statistics like DT, DF, and F. To test the hypothesis that the presence of the two intermediate-frequency haplotypes in our data is the result of subdivision between continents, we analyzed patterns of genetic differentiation among continental populations by use of the FST statistic, which compares the level of genetic diversity within subpopulations to levels of diversity in the population as a whole (Hartl and Clark 1997). In our data, diversity patterns were driven largely by the frequencies of the hsA and hsG haplotypes, which were present at similar frequencies in most populations, as shown in figure 3. The FST value observed among all four continental samples was 0.056. This value is significantly different from zero (P<.025) but is lower than is typically observed in nuclear genes, which generally have values of ∼0.15 (Przeworski et al. 2001; Schneider et al. 2003; Tishkoff and Verrelli 2003; Watkins et al. 2003). This FST value is lower than 80% of those reported for 25,549 SNPs by Akey et al. (2002), for instance, and is also lower than 45% of those reported for 1,627 genes by Schneider et al. (2003). The latter sample would be expected to have exceptionally low FST values because it included a large number of individuals from admixed populations, such as African-Americans and Hispanic-Latinos. The FST observed in our sample suggests that continental populations are less different with respect to variation in PTC than they are with respect to most other genes, not more different as would be expected if population subdivision or local adaptation had occurred.
Between-continent FST values were strongly affected by the inclusion of the North American sample, owing to the very high frequency of the hsA haplotype (0.95) in that group. The FST value excluding the North American sample was substantially lower than for the sample as a whole: 0.025. This value is significantly greater than zero (P<.025) but is lower than 75% of FST values reported by Schneider et al. (2003) and lower than 90% of values reported by Akey et al. (2002). The strong effect of the North American sample could be due to a variety of factors. First, our North American sample is small and may not provide an accurate representation of genetic diversity in North Americans. Estimates of the frequency of nontaster alleles in larger North American samples vary widely (Cavalli-Sforza et al. 1994). Second, archaeological and linguistic evidence suggest that North America was not inhabited by humans until recently (∼15,000 years ago) (Goebel 1999; Nettle 1999), and genetic evidence suggests that North and South American populations descended from a relatively small number of founders that entered the Americas via the Bering Strait (Torroni et al. 1993a, 1993b). Both of these factors can have strong effects on FST values (Urbanek et al. 1996).
Evidence for extensive subdivision has also been found within Africa (Schneider and Excoffier 1999; Tishkoff and Williams 2002; Yu et al. 2002). As with continental subdivision, subdivision within Africa could inflate DT, DF, and F statistics, yielding a false signature of balancing natural selection. To test the hypothesis that subdivision within the African sample in our study was responsible for the high observed values of these statistics in our sample as a whole, we performed separate DT, DF, and F tests for each continent under the assumptions of (1) no growth and (2) 100-fold growth 100,000 years ago. As shown in table 1, these statistics were significantly higher than expected in Asia and Europe, even when population growth was not taken into account. Furthermore, the P values of the DT, DF, and F statistics in the African population alone were greater than for the sample as a whole, not lower as would be expected if subdivision within Africa were causing the presence of high overall D values. Thus, substructure in African populations cannot solely explain the high DT values observed in PTC.
Table 1.
Results of Statistical Tests[Note]
|
P Value for |
|||||||||
|
Observed Value for Statistic |
No Growth |
Growth |
|||||||
| Sample | DT | DF | F | DT | DF | F | DT | DF | F |
| Africa | .46 | −.89 | −.56 | .18 | .80 | .56 | .01 | .03 | .01 |
| Asia | 2.94 | −.67 | .57 | .01 | .76 | .28 | .01 | .01 | .01 |
| Europe | 2.91 | −.62 | .59 | .01 | .81 | .28 | .01 | .01 | .01 |
| North America | −2.66 | −2.58 | −3.05 | .99 | .99 | .68 | .99 | .91 | .87 |
| All | 1.55 | −1.46 | −.50 | .08 | .90 | .64 | .01 | .01 | .01 |
Note.— P values given are the fraction of simulations that yielded a greater value than observed. The “Growth” columns show P values calculated under the assumption that, 100,000 years ago, the human population expanded 100-fold.
Taken together, three lines of evidence suggest that balancing natural selection has acted to maintain high levels of diversity in human populations. First, two haplotypes strongly associated with functionally divergent phenotypes dominate the sample. Second, under reasonable assumptions about human population history, the distribution of polymorphism frequencies in our sample has significantly more intermediate-frequency variants than expected under neutrality (P<.01). Third, the geographical distribution of the taster and nontaster alleles is not consistent with the hypothesis that they have arisen through population subdivision within or between continents. Thus, R. A. Fisher’s hypothesis that balancing natural selection has maintained taster and nontaster alleles appears to hold true in humans (Fisher et al. 1939).
It is of some interest that one of the original motivations for R. A. Fisher’s balancing selection hypothesis was his finding that variation in PTC perception exists in chimpanzees and orangutans, as well as in humans (Fisher et al. 1939). Fisher’s observation raises a basic question: does the divergence of the PTC taster and nontaster alleles pre-date the divergence of humans and chimpanzees? This question cannot be answered with accuracy by use of our data because such a small number of mutations (five) are present in humans and also because our data from chimpanzees are limited. However, under the assumption that the nucleotide substitution rate in PTC is 10−9/site/year, the estimated divergence date of the hsA and hsG haplotypes is ∼1.5 million years. This estimate has a variance that is larger than the estimate itself. However, if correct, the date implies that the human taster and nontaster alleles diverged after humans and chimpanzees did, ∼5 million years ago. Thus, given that the taster allele is ancestral in humans it seems unlikely that humans and apes share commonly derived nontaster alleles. We propose an alternative hypothesis—that PTC alleles conferring nontaster status evolved independently in humans and apes.
Evidence for balancing selection at the PTC locus does not imply that other selective pressures have been absent. For instance, it is possible that positive natural selection led to the rapid evolution of the nontaster allele, which was then maintained by balancing selection. This possibility might explain the unusually large number of nonsynonymous nucleotide substitutions found in this gene. It is also possible that specific PTC alleles have been favored by positive natural selection in particular environments, resulting in local adaptation. Such effects might account for the high frequency of PTC taster alleles in New World populations and the significant low DT, DF, and F in our North American sample.
The mechanism through which balancing natural selection has maintained divergent PTC alleles in human populations remains unclear. No stop codons or deletions, which might yield nonfunctional alleles, have yet been found at the PTC locus. In addition, although several haplotypes are present in our sample, two account for >90% of observations: hsA and hsG. If nontaster alleles were simply “broken” taster alleles, it seems likely that a greater diversity of nontaster alleles would be expected (Harding et al. 2000). One possibility is that PTC heterozygotes gain a fitness advantage through the perception and avoidance of a larger repertoire of bitter toxins than homozygotes. We hypothesize that PTC nontaster alleles encode functional receptor molecules that bind to toxic bitter substances other than PTC. Definitive tests of this hypothesis will require additional information about the functional properties of the PTC protein.
Acknowledgments
Mark Leppert, Jon Seger, and David Witherspoon provided helpful discussion. Tom Friedman and Robert Morell made useful comments on the manuscript. This project was supported by grants NIH ES12125 (to S.W.), NIH GM59290 and NSF BCS0218370 (to L.B.J.), and NIH/NIDCD Z01-000046-04 (to D.D.).
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PTC) [PubMed] [Google Scholar]
References
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD (2002) Interrogating a high-density SNP map for signatures of natural selection. Genome Res 12:1805–1814 10.1101/gr.631202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous (1931) Tasteblindness. Science Suppl 73:14 [Google Scholar]
- Bamshad M, Wooding S (2003) Signatures of natural selection in the human genome. Nat Rev Genet 4:99–111 10.1038/nrg999 [DOI] [PubMed] [Google Scholar]
- Barnicot NA, Harris H, Kalmus H (1951) Taste thresholds of further eighteen compounds and their correlation with PTC thresholds. Ann Eugen 16:119–128 [DOI] [PubMed] [Google Scholar]
- Bartoshuk LM, Duffy VB, Miller IJ (1994) PTC/PROP tasting: anatomy, psychophysics, and sex effects. Physiol Behav 56:1165–1171 10.1016/0031-9384(94)90361-1 [DOI] [PubMed] [Google Scholar]
- Blakeslee AF (1932) Genetics of sensory thresholds: taste for phenylthiocarbamide. Proc Natl Acad Sci USA 18:120–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Drayna D, Coon H, Kim UK, Elsner T, Cromer K, Otterud B, Baird L, Peiffer AP, Leppert M (2003) Genetic analysis of a complex trait in the Utah Genetic Reference Project: a major locus for PTC taste ability on chromosome 7q and a secondary locus on chromosome 16p. Hum Genet 112:567–572 [DOI] [PubMed] [Google Scholar]
- Drewnowski A, Rock CL (1995) The influence of genetic taste markers on food acceptance. Am J Clin Nutr 62:506–511 [DOI] [PubMed] [Google Scholar]
- Excoffier L (2002) Human demographic history: refining the recent African origin model. Curr Opin Genet Dev 12:675–682 10.1016/S0959-437X(02)00350-7 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI (1999) A human population bottleneck can account for the discordance between patterns of mitochondrial versus nuclear DNA variation. Mol Biol Evol 16:1003–1005 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1993) PHYLIP: (phylogenetic inference package) version 3.5d. Computer program distributed by the author, Department of Genetics, University of Washington, Seattle [Google Scholar]
- Fisher RA, Ford EB, Huxley J (1939) Taste-testing the anthropoid apes. Nature 144:750 [Google Scholar]
- Fox AL (1932) The relationship between chemical constitution and taste. Proc Natl Acad Sci USA 18:115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel T (1999) Pleistocene human colonization of Siberia and peopling of the Americas: an ecological approach. Evol Anthropol 8:208–227 [DOI] [Google Scholar]
- Guo SW, Reed DR (2001) The genetics of phenylthiocarbamide perception. Ann Hum Biol 28:111–142 10.1080/03014460151056310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding RM, Healy E, Ray AJ, Ellis NS, Flanagan N, Todd C, Dixon C, Sajantila A, Jackson IJ, Birch-Machin MA, Rees JL (2000) Evidence for variable selective pressures at MC1R. Am J Hum Genet 66:1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpending H, Rogers A (2000) Genetic perspectives on human origins and differentiation. Annu Rev Genomics Hum Gen 1:361–385 10.1146/annurev.genom.1.1.361 [DOI] [PubMed] [Google Scholar]
- Harris EE, Hey J (1999) Human demography in the Pleistocene: do mitochondrial and nuclear genes tell the same story? Evol Anthropol 8:81–86 [DOI] [Google Scholar]
- Harris H, Kalmus H (1949) The measurement of taste sensitivity to phenylthiourea (PTC). Ann Eugen 15:32–45 [DOI] [PubMed] [Google Scholar]
- Hartl DL, Clark AG (1997) Principles of population genetics, 3rd ed. Sinauer Associates, Sunderland, MA [Google Scholar]
- Hey J (1997) Mitochondrial and nuclear gene trees present conflicting portraits of human origins. Mol Biol Evol 14:166–172 [DOI] [PubMed] [Google Scholar]
- Hey J, Harris E (1999) Population bottlenecks and patterns of human polymorphism. Mol Biol Evol 16:1423–1426 [DOI] [PubMed] [Google Scholar]
- Hudson RR, Boos DD, Kaplan NL (1992) A statistical test for detecting geographic subdivision. Mol Biol Evol 9:138–151 [DOI] [PubMed] [Google Scholar]
- Kaplan N, Hudson RR, Iizuka M (1991) The coalescent process in models with selection, recombination and geographic subdivision. Genet Res 57:83–91 [DOI] [PubMed] [Google Scholar]
- Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D (2003) Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science 299:1221–1225 10.1126/science.1080190 [DOI] [PubMed] [Google Scholar]
- Klein RG (1999) The human career: human biological and cultural origins. University of Chicago Press, Chicago [Google Scholar]
- Laporte V, Charlesworth B (2002) Effective population size and population subdivision in demographically structured populations. Genetics 162:501–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Wu CI, Luo CC (1985) A new method for estimating synonymous and nonsynonmous rates of nucleotide substitution considering the relative likelihood of nucleotide codon changes. Mol Biol Evol 2:150–174 [DOI] [PubMed] [Google Scholar]
- Makalowski W, Boguski MS (1998) Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences. Proc Natl Acad Sci USA 95:9407–9412 10.1073/pnas.95.16.9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjoram P, Donnelly P (1994) Pairwise comparisons of mitochondrial DNA sequences in subdivided populations and implications for early human evolution. Genetics 136:673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654 10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- Nekrushenko A, Makova KD, Li WH (2001) The KA/KS ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res 12:198–202 10.1101/gr.200901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettle D (1999) Linguistic diversity of the Americas can be reconciled with a recent colonization. Proc Natl Acad Sci USA 96:3325–3329 10.1073/pnas.96.6.3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Hudson RR, Di Rienzo A (2001) Adjusting the focus on human variation. Trends Genet 16:296–302 10.1016/S0168-9525(00)02030-8 [DOI] [PubMed] [Google Scholar]
- Ptak SE, Przeworski M (2002) Evidence for population growth in humans is confounded by fine-scale population structure. Trends Genet 18:559–563 10.1016/S0168-9525(02)02781-6 [DOI] [PubMed] [Google Scholar]
- Rogers AR (1995) Genetic evidence for a Pleistocene population explosion. Evolution 49:608–615 [DOI] [PubMed] [Google Scholar]
- Rogers AR, Harpending HC (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569 [DOI] [PubMed] [Google Scholar]
- Ruhlen M (1994) The origin of language. John Wiley and Sons, New York [Google Scholar]
- Schneider JA, Pungliya MS, Choi JY, Jiang R, Sun XJ, Salisbury BA, Stephens JC (2003) DNA variability of human genes. Mech Ageing Dev 124:17–25 10.1016/S0047-6374(02)00165-3 [DOI] [PubMed] [Google Scholar]
- Schneider S, Excoffier L (1999) Why hunter–gatherer populations do not show signs of Pleistocene demographic expansions. Proc Natl Acad Sci USA 96:10597–10602 10.1073/pnas.96.19.10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN version 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva, Geneva, Switzerland [Google Scholar]
- Slatkin M (1991) Inbreeding coefficients and coalescence times. Genet Res 58:167–175 [DOI] [PubMed] [Google Scholar]
- Slatkin M, Hudson RR (1991) Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129:555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiner MC, Munro ND, Surovell TA, Tchernov E, Bar-Yosef O (1999) Paleolithic population growth pulses evidenced by small animal exploitation. Science 283:190–194 10.1126/science.283.5399.190 [DOI] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper BJ (1998) 6-n-propylthiouracil: a genetic marker for taste, with implications for food preference and dietary habits. Am J Hum Genet 63:1271–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Verrelli BC (2003) Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet 4:293–340 10.1146/annurev.genom.4.070802.110226 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM (2002) Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet 3:611–621 [DOI] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo CM, Wallace DC (1993a) Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet 53:563–590 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Sukernik RI, Schurr TG, Starikorskaya YB, Cabell MF, Crawford MH, Comuzzie AG, Wallace DC (1993b) MtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. Am J Hum Genet 53:591–608 [PMC free article] [PubMed] [Google Scholar]
- Urbanek MG, Goldman D, Long JC (1996) The apportionment of dinucleotide repeat diversity in Native Americans and Europeans: a new approach to measuring gene identity reveals asymmetric patterns of divergence. Mol Biol Evol 13:943–953 [DOI] [PubMed] [Google Scholar]
- Wakeley J (2001) The coalescent in an island model of population subdivision with variation among demes. Theor Popul Biol 59:133–144 10.1006/tpbi.2000.1495 [DOI] [PubMed] [Google Scholar]
- Wall JD, Przeworski M (2000) When did the human population size start increasing? Genetics 155:1865–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins WS, Rogers AR, Ostler CT, Wooding S, Bamshad MJ, Brassington AM, Carroll ML, Nguyen SV, Walker JA, Batzer MA, Prasad BVR, Reddy G, Das PK, Jorde LB (2003) Genetic variation among world populations: inferences from 100 Alu insertion polymorphisms. Genome Res 13:1607–1618 10.1101/gr.894603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S, Rogers A (2000) A Pleistocene population X-plosion? Hum Biol 72:693–695 [PubMed] [Google Scholar]
- Wooding S, Rogers A (2002) The matrix coalescent and an application to human single-nucleotide polymorphisms. Genetics 161:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding SP, Watkins WS, Bamshad MJ, Dunn DM, Weiss RB, Jorde LB (2002) DNA sequence variation in a 3.7 kb noncoding sequence 5′ of the CYP1A2 gene: implications for human population history and natural selection. Am J Hum Genet 71:528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Bielawski B (2000) Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15:496–503 10.1016/S0169-5347(00)01994-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Chen FC, Ota S, Jorde LB, Pamilo P, Patthy L, Ramsay M, Jenkins T, Shyue SK, Li WH (2002) Larger genetic differences within Africans than between Africans and Eurasians. Genetics 161:269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]