Abstract
We previously reported that the genomes of gonadal germ cells at 11.5–19.5 days postcoitum (dpc) are incompetent to support full-term development of cloned mouse embryos. In this study, we performed nuclear transfer using primordial germ cells (PGCs) from earlier stages at 8.5–10.5 dpc. When PGC nuclei at 8.5, 9.5, and 10.5 dpc were transferred into enucleated oocytes, seven cloned embryos developed into full-term offspring. Of these, five, all derived from 8.5- or 9.5-dpc PGCs, developed into healthy adults with normal fertility. Of the remaining two offspring derived from 10.5-dpc PGCs, one died shortly after birth, and the other showed slight growth retardation but subsequently developed into a fertile adult. We examined allele-specific methylation at the imprinted H19 and Snrpn loci in 9.5- to 11.5-dpc PGCs. Although the beginning of methylation erasure was evident on the H19 paternal allele at 9.5 dpc, most PGCs did not demonstrate significant erasure of paternal allele-specific methylation until 10.5 dpc. Maternal allele-specific methylation was largely erased from Snrpn by 10.5 dpc. By 11.5 dpc, the majority of PGCs showed nearly complete or complete erasure of allele-specific methylation in both H19 and Snrpn. These results demonstrate that at least some genomic imprints remain largely intact in 8.5- to 9.5-dpc PGCs and then undergo erasure at ≈10.5 dpc as the PGCs enter the genital ridges. Thus, migrating PGCs at 8.5–9.5 dpc can be successfully used as donors for nuclear transfer, whereas gonadal PGCs at 11.5 dpc and later are incompetent to support full-term development.
Keywords: developmental totipotency, DNA methylation, imprinted genes, nuclear transfer
In the mouse, primordial germ cells (PGCs) develop from the pluripotent epiblast and are first detected as alkaline phosphatase-positive cells in the embryo at ≈7.25 days postcoitum (dpc) (1, 2). PGCs rapidly proliferate although migrating along the hindgut and dorsal mesentery (8.5–9.5 dpc), reaching the genital ridges by 10.5–11.5 dpc. After colonizing the genital ridges, they continue proliferating until 13.5 dpc. Female germ cells then enter meiotic prophase and undergo meiotic arrest, whereas male germ cells remain mitotic until ≈15.5 dpc, when they enter a state of mitotic arrest.
During development and differentiation of germ cells in the mouse, the germline genome undergoes dramatic epigenetic reprogramming. This includes erasure and resetting of epigenetic modifications that normally distinguish maternal and paternal alleles of imprinted genes (3–5). Previous studies have shown that differential DNA methylation patterns are established in imprinted genes during gametogenesis in each sex and are required for monoallelic expression of these genes in somatic cells of developing embryos (6–9). These differential methylation patterns are preserved during somatic development but must be erased and reset during gametogenesis. In germ cells, the inherited methylation imprints are completely erased by 13.5 dpc, and new imprints are reestablished on both alleles according to the individual's sex (10, 11). Imprinting erasure is associated with a global loss of methylation in PGCs at about the time when these cells colonize the genital ridges (7, 12, 13). The Igf2r differentially methylated region-2 begins to lose methylation as early as 9.5 dpc (14), whereas other imprinted genes initiate demethylation 1–2 days later at 10.5–11.5 dpc (13, 15). Sex-specific imprints are then reestablished to a similar extent on both alleles during germ cell development, although this occurs at different times on the two alleles, indicating that some form of parent-of-origin-specific epigenetic distinction persists after erasure of differential DNA methylation (4, 16, 17).
Although the available data indicate that imprints are erased soon after arrival of PGCs in the genital ridge (7, 12, 13), these studies were based on examination of a small number of imprinted genes. Because DNA methylation is not the only modification that distinguishes parental alleles of imprinted genes, we sought an alternative approach to assessing the developmental potential of PGCs at different stages. The nuclear transfer (cloning) procedure (18) provides a means of evaluating the developmental potential of the entire genome in any individual donor cell, without bias related to the selection of specific genes for analysis or reliance upon a single parameter (e.g., DNA methylation) for this assessment. Because the development to term of cloned embryos requires normal allele-specific imprinting in the donor cell genome, nuclear transfer can be used to provide a definitive functional assay of exactly when during PGC development erasure of imprints makes these cells incompetent to support normal development. Additionally, nuclear transfer provides a way of determining whether imprint erasure during PGC development differs between males and females. Our previous studies revealed that PGCs at 11.5–19.5 dpc were not able to support full-term development after nuclear transfer (19). Here, we show that nuclei from PGCs at earlier stages are able to direct normal embryonic development but subsequently lose this ability at a stage that correlates with a global erasure of imprints during PGC development.
Materials and Methods
Animals. PGC donors were collected from transgenic mice (Tg OG2) expressing GFP driven by the promoter/enhancer region of the germ-cell specific Oct4 gene (a generous gift from J. R. Mann, Beckman Research Institute of the City of Hope, Duarte, CA) (20). Female mice homozygous for the Tg OG2 transgene were mated with DBA/2 male mice to produce (OG2 × DBA/2) F1 hybrids. Female CD-1 mice were mated with male Tg OG2 mice to produce (CD-1 × OG2) F1 hybrids. PGCs were isolated from both types of F1 hybrids as described (19). In each case, an aliquot of isolated PGCs was stained for alkaline phosphatase activity to confirm that these were indeed germ cells. Oocytes to be enucleated and used as recipients for nuclear transfer were collected from B6D2F1 (C57BL/6 × DBA/2) female mice, as described (18). Adult female CD-1 mice, rendered pseudopregnant by mating with vasectomized CD-1 males, were used as surrogate mothers.
The B6(CAST 7) (B6/CAST) substrain of mice has a M. musculus castaneus (CAST) chromosome 7 on a C57BL/6J (B6) background (21). For analysis of allele-specific methylation patterns of the imprinted H19 and Snrpn genes, PGCs were obtained from F1 hybrid fetuses produced by crosses between B6(CAST 7) females and Tg OG2 males (B6/CAST × OG2). This combination facilitated the selection of PGCs and distinction of maternal alleles from paternal alleles. Several polymorphisms have been described that distinguish alleles of imprinted genes on chromosome 7 from B6 and CAST mice (21, 22). Protocols for the handling and treatment of all animals were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Hawaii.
Media. PGCs from dissected fetuses were collected into DMEM (GIBCO/BRL) supplemented with 20% FBS. Oocytes and two-cell embryos were cultured in CZB medium (23) at 37°C under 5% CO2 in air. Oocyte manipulation was carried out in Hepes-CZB (24) at room temperature.
Identification of PGCs. Staging of fetuses followed the system whereby the day of vaginal plug was designated as 0.5 dpc. General morphology and the number of paired somites were recorded in each case. Fetuses were individually dissected, then pooled in Hepes-MEM (GIBCO/BRL) with 20% FBS. Specific regions containing PGCs, including the hindgut at 8.5 dpc, the dorsal mesentery at 9.5 dpc, and the genital ridges plus immediate surrounding tissues at 10.5 dpc, were dissected from the fetuses. These tissue fragments were then incubated individually in 0.02% EDTA/phosphate solution for 10 min at room temperature followed by gentle pipettings to dissociate into individual cells. GFP-positive cells were visualized under epiillumination and collected randomly from the cell suspension by using a micropipette fitted to an inverted fluorescence microscope (Olympus, Melville, NY; IX70) equipped with a FITC filter.
Alkaline Phosphatase Staining of Isolated PGCs. GFP-positive cells were histochemically stained for alkaline phosphatase (ALP) activity to assess the purity of the recovered PGC population. Approximately 100 GFP-positive cells collected from fetuses at each stage (8.5, 9.5, and 10.5 dpc) were placed on glass slides. After fixation with 4% paraformaldehyde, the cells were washed with Ca2+- and Mg2+-free PBS and histochemically stained by using the ALP detection kit (Chemicon), according to the manufacturer's instructions.
Nuclear Transfer, Oocyte Activation, and Embryo Transfer. PGCs to be used for cloning were placed in Hepes-CZB medium containing 12% (wt/vol) polyvinylpyrrolidone (average Mr, 360,000; ICN). The plasma membrane of each PGC was disrupted within a micropipette and the nucleus, and some associated cytoplasm were injected into a mouse oocyte from which metaphase II chromosomes had been previously removed (18). Reconstructed oocytes were incubated in CZB medium for 2 h at 37°C followed by culture for 5 h in Ca2+-free CZB containing 10 mM Sr2+ and 5 μg/ml cytochalasin B to induce oocyte activation without emission of chromosomes of PGCs. Activated oocytes with two distinct pronuclei were cultured in CZB medium for 1 day, and the embryos reaching the two-cell stage were transferred into the oviducts of pseudopregnant surrogate females. The day of transfer was considered day 0.5 of pregnancy (0.5 dpc). Full-term cloned offspring on day 19.5 dpc were delivered by cesarean section and raised by lactating foster mothers.
Bisulfite Genomic Sequencing. To examine allele-specific methylation patterns at imprinted loci, bisulfite genomic sequencing (25) was used. DNA was prepared and subjected to bisulfite modification as described (26), with modifications. Genomic DNA was extracted from 190–230 PGCs of (B6/CAST × OG2) F1 fetuses. As a control, we collected DNA from tail tissue of a (B6/CAST × OG)F1 pup at 3 days postpartum. DNA was subjected to bisulfite modification by using a DNA methylation kit (Zymo Research, Orange, CA). Genomic DNA was also prepared from tails or skin of cloned mice of (OG2 × DBA/2) F1 donor PGCs. Bisulfite-converted DNA was subjected to PCR amplification of the H19- and Snrpn-differentially methylated domains (DMDs). Nested primer sets of the H19 DMD (BMsp2t1/BHha1t3 followed by BMsp2t2/BHha1t4) were used as described (26). In some cases, a different set of nested primers was used [H19CT-R4/H19CT-F4 followed by H19CT-R3/H19CT-F3 based on GenBank sequence data for the H19 gene (GenBank accession no. U19619)]. The primer sequences were as follows: H19CT-R4, 5′-TTTTCACACAATAACRCTAATAACCCCA-3′; H19CT-F4, 5′-TAGAGATTTTATTTTTATGTTYGGGGGA-3′; H19CT-R3, 5′-CAAAACCCTATAAATCAAATACCTAAAA-3′; H19CT-F3, 5′-GAGYGTGTAGGGTATTTATATTTAGGAT-3′.
Nested PCR primer sets specific to the Snrpn DMD were used as described (17). The first PCR reaction included two cycles of 2 min at 94°C, 1 min at 55°C, 1 min at 72°C, followed by 28 cycles of 30 seconds at 94°C, 1 min at 55°C, 1 min at 72°C followed by a 10-min extension at 72°C. The second PCR reaction was similar, except that the first two cycles were omitted. PCR products were subcloned into the pGEM-T Easy vector (Promega) and sequenced. Maternal and paternal alleles were distinguished on the basis of a DNA polymorphism unique to CAST and not present in M. musculus, as described (21, 22).
Results
Mice Cloned from PGCs. To assess directly the developmental potential of migrating PGCs at stages before or coincident with their arrival at the genital ridges, we performed nuclear transfer using PGCs at 8.5, 9.5, and 10.5 dpc as nuclear donors. Analysis of alkaline phosphatase activity in GFP-expressing cells collected from 8.5-, 9.5-, and 10.5-dpc fetuses showed that the purity of the collected PGCs was 100% (Fig. 1). When the nuclei of 8.5-dpc PGCs were transferred into enucleated oocytes, ≈40% of activated oocytes developed into two-cell embryos (Table 1). Two (1.7%) of 117 embryos transferred developed to term (Table 1). When the nuclei of 9.5-dpc PGC were transferred into enucleated oocytes, 37% of activated oocytes developed into two-cell embryos (Table 1). Three (4.5%) fertile offspring were obtained after transfer of 66 cloned embryos (Table 1). The average body and placental weights at birth of five pups (four males and one female) cloned from 8.5- and 9.5-dpc PGCs were 1.90 ± 0.23 and 0.38 ± 0.10 g, respectively (Table 2). All five of these pups developed into healthy adults and produced offspring with normal litter size after mating with B6D2F1 mice (Table 2). When the nuclei of 10.5-dpc PGCs were used for cloning, 44% of activated oocytes developed into two-cell embryos. Two (1.3%) of 149 embryos developed to full term (Table 1). However, unlike the clones produced from 8.5- or 9.5-dpc PGCs, both of these cloned pups showed developmental abnormalities. One male pup (OG77-1F) was extremely heavy at birth (2.88 g) and died 2 days later (Table 2). The other female pup (OG74-1F) was born with a body weight within the normal range (1.69 g), but the placenta was extremely large (0.52 g) (Table 2). This female survived to weaning (26 days postpartum) but displayed growth retardation. Its body weight at weaning was 7.08 g. The average body weight of normal mice at weaning was 15.5 g. Nevertheless, this mouse was bred and became pregnant three times, each time killing all her pups shortly after delivery (Table 2).
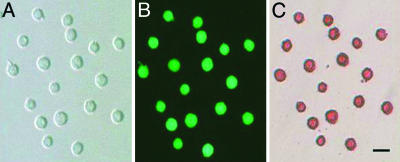
Fig. 1.
GFP-positive cells of 10.5-dpc fetuses examined under a phase-contrast (A) or epiillumination using a fluorescence-inverted microscope with a FITC filter (B) or after staining for alkaline phophatase activity (C). (Bar, 20 μm.)
Table 1. Mice cloned from early primordial germ cells.
| PGC donors (♀ × ♂) F1 | Age of donor PGCs, dpc | No. of nucleus-transferred oocytes | No. of activated oocytes | No. of two-cell embryos (%)* | No. of embryos transferred | No. of full-term cloned pups (%)† | Cloned pups that survived |
|---|---|---|---|---|---|---|---|
| (OG2 × DBA) F1 | 8.5 | 144 | 137 | 50 (36.5) | 50 | 1 (2.0) | 1 |
| (CD-1 × OG2) F1 | 8.5 | 162 | 153 | 67 (43.8) | 67 | 1 (1.5) | 1 |
| (OG2 × DBA) F1 | 9.5 | 191 | 178 | 66 (37.1) | 66 | 3 (4.5) | 3 |
| (OG2 × DBA) F1 | 10.5 | 351 | 338 | 149 (44.1) | 149 | 2 (1.3) | 1‡ |
Percent of activated oocytes.
Percent of two-cell embryos.
One pup died 2 days after birth.
Table 2. Growth and reproductive performance of cloned mice.
| Clones
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Identity of individual clones | Age of donor PGCs, dpc | Sex | Body weight at birth, g | Placenta weight, g | Development to adult | No. of test mating* | No. of pregnancy | Average litter size |
| OG55-1F | 8.5 | ♂ | 1.89 | 0.46 | Yes | 2 | 2 | 10 |
| OG58-1F | 8.5† | ♂ | 2.17 | 0.38 | Yes | 2 | 2 | 8 |
| OG67-1F | 9.5 | ♂ | 2.07 | 0.43 | Yes | 2 | 2 | 10.5 |
| OG67-2F | 9.5 | ♀ | 1.64 | 0.2 | Yes | 2 | 2 | 9.5 |
| OG70-1F | 9.5 | ♂ | 1.71 | 0.41 | Yes | 2 | 2 | 8.5 |
| OG74-1F | 10.5 | ♀ | 1.69 | 0.52 | Yes | 3 | 3 | 0§ |
| OG77-1F | 10.5 | ♂ | 2.88 | 0.37 | No‡ | - | - | - |
Mated with B6D2F1 female or male mice.
Collected from (CD-1 × OG2) F1 fetuses. The other PGCs were collected from (OG2 × DBA) F1 fetuses.
Cloned pup died 2 days after birth.
Mother killed all newborn pups.
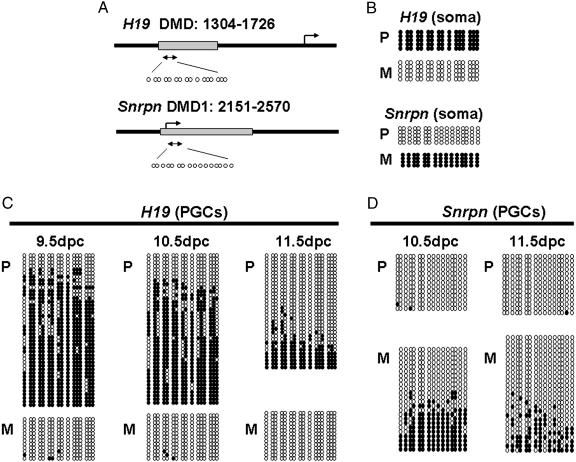
Allele-Specific Methylation of the H19 and Snrpn DMDs in PGCs of Normal Mice. To examine the demethylation process at imprinted loci in PGCs, we first analyzed the methylation profile of the DMD in the imprinted H19 and Snrpn genes in PGCs from normal fetuses at 9.5, 10.5, and 11.5 dpc. These fetuses were produced by normal mating of B6/CAST females with Tg OG2 males (B6/CAST × OG2). The H19 gene is normally paternally silenced, with paternally inherited alleles showing hypermethylation in the DMD leading to transcriptional repression, whereas maternally inherited alleles are hypomethylated in the DMD and actively expressed (Fig. 2B). This differential allele-specific methylation is completely erased in fetal PGCs by 13.5 dpc (13, 16). In PGCs at 9.5 dpc, the paternal allele of the H19 gene was predominantly methylated (81% of alleles were >50% methylated in the DMD), whereas the maternal allele was almost completely unmethylated (98% of all CpG in the DMD were unmethylated) (Fig. 2C). Thus, at this migrating stage, a small proportion of the PGCs have already initiated demethylation of the paternal H19 allele, but most of the paternal alleles remain hypermethylated (Fig. 2C). In PGCs at 10.5 dpc, the H19 DMD was further demethylated on the paternal allele (Fig. 2C). About 24% of paternal H19 alleles were unmethylated in the DMD at this stage (Fig. 2C). By 11.5 dpc, when all germ cells had reached the genital ridges, >75% of the paternal alleles of H19 were demethylated in the DMD (Fig. 2C). This indicates that the erasure of imprints in the DMD of the paternal allele of H19 occurs progressively as PGCs colonize the genital ridge. The maternal allele remained almost completely unmethylated throughout this period (99% of CpGs in the DMD were unmethylated, Fig. 2C).
Fig. 2.
Allele-specific methylation of the H19- and Snrpn-DMDs in 9.5- to 11.5-dpc PGCs. (A) Schematic representation of the H19 upstream DMD and Snrpn promoter DMD1. Boxes, DMDs of the genes examined; arrows, transcription start sites of the genes; double arrows, the regions where methylation was analyzed; small circles, individual CpG residues within the areas amplified. Sixteen cytosines of H19 DMD are located at the following positions: 1330, 1360, 1362, 1372, 1374, 1391, 1397, 1538, 1546, 1568, 1617, 1621, 1624, 1638, 1645, and 1668 (numbers are in accordance with GenBank accession no. U19619). Sixteen cytosines of Snrpn DMD1are located at the following positions: 2267, 2279, 2334, 2363, 2376, 2400, 2406, 2416, 2418, 2420, 2439, 2451, 2470, 2483, 2510, and 2520 (GenBank accession no. AF081460). (B) Allele-specific methylation of the H19 and Snrpn DMDs in somatic cells. DNA from tail of a (B6/CAST × OG) F1 pup at 3 days postpartum was used. Each line corresponds to a single strand of DNA, and each circle represents a CpG dinucleotide on the strand. Each black circle designates a methylated cytosine. Each white circle corresponds to an unmethylated cytosine. Paternal (P) and maternal (M) alleles were distinguished during sequence analysis by DNA polymorphisms between M. musculus castaneus (B6/CAST) and M. musculus musculus (Tg OG2) in the DMD sequence (21). (C) Allele-specific methylation of the H19 DMD in 9.5-, 10.5-, and 11.5-dpc PGCs. PGCs were collected from (B6/CAST × OG) F1 fetuses. (D) Allele-specific methylation of the Snrpn DMD1 in 10.5- and 11.5-dpc PGCs.
In contrast to the H19 gene, maternal alleles of the Snrpn gene were normally hypermethylated in the DMD, with hypomethylated paternal alleles (Fig. 2B). Analysis of allele-specific methylation of this locus in PGCs at 10.5 and 11.5 dpc showed a similar pattern of methylation erasure as was seen at the H19 locus, coincident with the entry of PGCs into the genital ridges (Fig. 2D). Thus, in PGCs at 10.5 dpc, only 37.5% of the maternal alleles retained >50% methylation in the Snrpn DMD and by 11.5 dpc, this was reduced to 18.5% (Fig. 2D). The paternal alleles of Snrpn were completely hypomethylated in the DMD in PGCs at 10.5 ∼ 11.5 dpc (Fig. 2D).
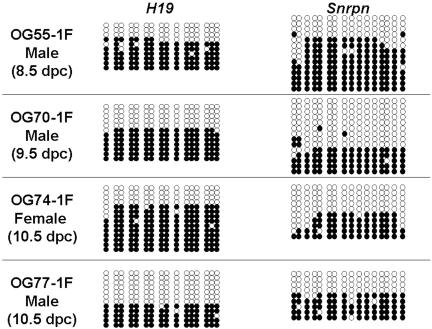
Methylation Status of the H19 and Snrpn Genes in Mice Cloned from PGC Nuclei. We examined the methylation status of the H19- and Snrpn-DMDs in somatic cells of four mice cloned from 8.5-, 9.5-, or 10.5-dpc PGCs (Fig. 3). DNA was collected from tails of two fertile adult mice cloned from 8.5- and 9.5-dpc PGC nuclei (OG55-1F, OG70-1F) and from one fertile adult mouse cloned from a 10.5-dpc PGC (OG74-1F). DNA was also collected from skin of one full-term fetus cloned from a 10.5-dpc PGC that died 2 days after birth (OG77-1F). Because all cloned mice were produced by using PGC of (OG2 × DBA) F1 hybrids, it was not possible to distinguish parental alleles. Nevertheless, the DMD methylation patterns detected in each case showed two distinct classes of alleles, hypomethylated and hypermethylated (Fig. 3). This supports the notion that cloned embryos that develop to term or beyond are typically derived from nuclei bearing properly imprinted genes (at least to the extent that the status of the H19 and Snrpn genes are indicative).
Fig. 3.
Methylation status of the H19 and Snrpn DMDs in mice cloned from 8.5- ∼10.5-dpc PGCs. Genomic DNA was collected from tails of three adults cloned from 8.5-, 9.5-, 10.5-dpc PGCs and from skin of a dead pup cloned from a 10.5-dpc PGC (OG77-1F). PGCs were collected from (OG2 × DBA) F1 hybrid fetuses; the parental alleles could not be distinguished.
Discussion
We (19) and others (15, 27) reported that attempts to clone mice from fetal germ cells at 11.5 dpc and later stages were consistently unsuccessful. This was attributed to the absence of allele-specific epigenetic modifications at imprinted loci in gonadal germ cells due to erasure of differential methylation (15, 19, 27), although this explanation has not been formally tested. Here we report the successful cloning of mice from early PGCs. This success was specific to the use of PGCs at 8.5 or 9.5 dpc and, in one case, at 10.5 dpc. This confirms that PGCs at 11.5 dpc and thereafter lack proper epigenetic programming to support full development of embryos. Thus it appears that PGCs are initially totipotent but then lose their developmental potential at a stage roughly coincident with their entry into the developing genital ridges. Because the timing of this loss of potential coincides with the loss of differential methylation at imprinted loci, this appears to be a basis for the loss of developmental totipotency in fetal germ cells.
Previous studies have indicated that nuclear transfer often results in improper epigenetic programming in the majority of cloned embryos produced, resulting in their failure to develop past implantation (21, 28). However, in the small proportion of cloned embryos that successfully develop to term, epigenetic programming of imprinted loci appears to be predominantly retained from the donor nucleus (29). Genomic imprinting is of critical importance for proper development of eutherian mammalian embryos and fetuses through the imposition of parent-of-origin-specific monoallelic gene expression (5–9, 30). Aberrant (biallelic or null) expression of imprinted genes, including Igf2, Igf2r, Mash2, and H19, is known to result in deleterious effects on fetal development (31–34). Given that successful development of cloned embryos requires maintenance of the epigenetic status of genomic imprints present in the donor nucleus, it appears to be critical that a donor nucleus used for cloning is properly imprinted at the time of nuclear transfer to support subsequent development to term and beyond. Thus, our results indicate that most migrating PGCs possess a properly imprinted genome, whereas postmigratory germ cells do not (19).
In this study, we examined the erasure of genomic imprinting in PGCs in two ways. First, we directly examined the allele-specific methylation status of imprinted genes in migratory and postmigratory PGCs. Our data (Fig. 2) agree with those from previous studies in that erasure of parental allele-specific methylation begins in migratory PGCs (14), but the majority of differential methylation is erased at or immediately after the time PGCs enter the genital ridges (12, 13). However, only a small proportion of imprinted genes have been examined in this direct manner. Furthermore, differential methylation is not the only allele-specific epigenetic distinction at imprinted loci. It is known that, after the erasure of allele-specific differential methylation, biallelic methylation patterns are established in an asynchronous manner in germ cells, with remethylation occurring earlier on the allele that was previously methylated than on the allele that was inherited in a hypomethylated state (4, 16).
A more global assessment of proper epigenetic programming and, hence, genomic imprinting in any particular cell type are afforded by the second method we used to assess developmental potential of PGCs, the nuclear transfer (cloning) process. By definition, successful development to term and beyond of cloned embryos requires that a totipotent genome is contributed by the donor nucleus. Thus, we were able to examine the developmental potential of PGC genomes by using cloning as a functional bioassay. Our observation that a very large proportion (99.3%) of two-cell embryos cloned from PGCs at 10.5 dpc fail to give rise to viable offspring suggests that significant erasure of imprints required for normal development has already occurred by this stage. However, our finding that one embryo cloned from a 10.5-dpc PGC [=1/149 two-cell embryos (0.7%)] was able to develop into a fertile adult, albeit with growth retardation at weaning, and another term clone died with overgrowth suggests this erasure process is not completely synchronous in all PGCs, and that a very small proportion of these cells retain a properly imprinted genome competent for use in cloning. Similar results were reported by Miki et al. (35) in that two of four term offspring cloned from 10.5-dpc PGCs [=2/1,200 two-cell embryos (0.2%)] grew into healthy adults, although the other two were stillborn. These results support the contention that 10.5 dpc marks a stage of transition in PGCs from a state in which allele-specific epigenetic distinctions are stabilized by differential DNA methylation in migratory PGCs to a state in which this distinction is erased in gonadal germ cells. The inability to generate any cloned embryos from gonadal germ cells at 11.5 dpc or later confirms this functional loss of allele-specific imprinting (15, 19, 27, 35). More importantly, however, our results demonstrate that healthy fertile offspring can develop from cloned embryos derived from nuclei of early PGCs before 10.5 dpc, at 8.5 or 9.5 dpc, with an efficiency (1.5–4.5% of two-cell embryos) similar to that of cloned embryos derived from properly imprinted somatic cells. This confirms that migrating PGCs possess sufficient allele-specific distinctions at key imprinted loci to facilitate normal development and are thus developmentally totipotent. This contention is further confirmed by our finding of differentially methylated alleles at the H19 and Snrpn loci in somatic cells of cloned offspring derived from PGC nuclei (Fig. 3).
For successful development of a cloned embryo, proper genomic imprinting of the genome of a donor nucleus is clearly required. However, this is not the only determinant of developmental potency. Regardless of the source of a donor nucleus, significant reprogramming of gene expression is required after nuclear transfer to facilitate proper embryonic development. Bortvin et al. (36) suggested that ES cells are more successful than somatic cells as nuclear donors for cloning, because they initially express a set of at least 11 “Oct4-related” genes that are molecular markers for developmental pluripotency in early embryonic cells. Interestingly, 10.5-dpc PGCs also express the same set of genes (36). More recently, it was suggested that ES cells may be of germ cell origin (37). Thus it might be assumed that PGCs should be as effective as ES cells when used as donors for nuclear transfer. A distinction was previously noted in the developmental kinetics of embryos cloned from somatic cells compared to those cloned from ES cells in that somatic clones show a relatively higher rate of successful development to the blastocyst stage than do ES clones, whereas ES clones show better development from the blastocyst stage to term than do somatic clones (36). Our data suggest that embryos cloned from early-stage PGCs develop with kinetics similar to those of embryos cloned from differentiated somatic cells (data from ref. 19 and this study). This may reflect the fact that, whereas PGCs at 8.5–9.5 dpc continue to express genes characteristic of pluripotent embryonic cells, they have also begun to express genes unique to the germ cell lineage (38) and so are distinct from early embryonic cells. Nevertheless, the results of this study and that of Miki et al. (35) demonstrate that the genomes of early PGCs are developmentally totipotent, and that these cells can be used as nuclear donors for the cloning process.
Acknowledgments
We thank Dr. Jeff R. Mann (Beckman Research Institute of the City of Hope, Duarte, CA) for generously providing the OG2 transgenic mice. Also we acknowledge Mr. Christopher B. Geyer and Ms. Susan S. Lee for technical support and advice on the bisulfite genomic sequencing. This work was supported by National Institutes of Health Grant HD042772.
Author contributions: Y.Y. designed research; Y.Y., E.W.L., and K.I. performed research; Y.Y. and Y.M. contributed new reagents/analytic tools; Y.Y., M.S.B., J.R.M., and R.Y. analyzed data; and Y.Y., M.S.B., J.R.M., and R.Y. wrote the paper.
Abbreviations: PGC, primordial germ cell; dpc, days postcoitum; DMD, differentially methylated domain.
References
- 1.Ginsburg, M., Snow, M. H. & McLaren, A. (1990) Development (Cambridge, U.K.) 110, 521–528. [DOI] [PubMed] [Google Scholar]
- 2.Lawson, K. A. & Hage, W. J. (1994) Ciba Found. Symp. 182, 68–84, and discussion (1994) 182, 84–91. [DOI] [PubMed] [Google Scholar]
- 3.Kafri, T., Ariel, M., Brandeis, M., Shemer, R., Urven, L., McCarrey, J., Cedar, H. & Razin, A. (1992) Genes Dev. 6, 705–714. [DOI] [PubMed] [Google Scholar]
- 4.Lucifero, D., Mann, M. R., Bartolomei, M. S. & Trasler, J. M. (2004) Hum. Mol. Genet. 13, 839–849. [DOI] [PubMed] [Google Scholar]
- 5.Monk, M., Boubelik, M. & Lehnert, S. (1987) Development (Cambridge, U.K.) 99, 371–382. [DOI] [PubMed] [Google Scholar]
- 6.Li, E., Beard, C. & Jaenisch, R. (1993) Nature 366, 362–365. [DOI] [PubMed] [Google Scholar]
- 7.Szabo, P. E. & Mann, J. R. (1995) Genes Dev. 9, 3097–3108. [DOI] [PubMed] [Google Scholar]
- 8.Tucker, K. L., Beard, C., Dausmann, J., Jackson-Grusby, L., Laird, P. W., Lei, H., Li, E. & Jaenisch, R. (1996) Genes Dev. 10, 1008–1020. [DOI] [PubMed] [Google Scholar]
- 9.Bartolomei, M. S. & Tilghman, S. M. (1997) Annu. Rev. Genet. 31, 493–525. [DOI] [PubMed] [Google Scholar]
- 10.Brandeis, M., Kafri, T., Ariel, M., Chaillet, J. R., McCarrey, J., Razin, A. & Cedar, H. (1993) EMBO J. 12, 3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shemer, R., Birger, Y., Dean, W. L., Reik, W., Riggs, A. D. & Razin, A. (1996) Proc. Natl. Acad. Sci. USA 93, 6371–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surani, M. A. (2001) Nature 414, 122–128. [DOI] [PubMed] [Google Scholar]
- 13.Hajkova, P., Erhardt, S., Lane, N., Haaf, T., El-Maarri, O., Reik, W., Walter, J. & Surani, M. A. (2002) Mech. Dev. 117, 15–23. [DOI] [PubMed] [Google Scholar]
- 14.Sato, S., Yoshimizu, T., Sato, E. & Matsui, Y. (2003) Mol. Reprod. Dev. 65, 41–50. [DOI] [PubMed] [Google Scholar]
- 15.Lee, J., Inoue, K., Ono, R., Ogonuki, N., Kohda, T., Kaneko-Ishino, T., Ogura, A. & Ishino, F. (2002) Development (Cambridge, U.K.) 129, 1807–1817. [DOI] [PubMed] [Google Scholar]
- 16.Davis, T. L., Yang, G. J., McCarrey, J. R. & Bartolomei, M. S. (2000) Hum. Mol. Genet. 9, 2885–2894. [DOI] [PubMed] [Google Scholar]
- 17.Lucifero, D., Mertineit, C., Clarke, H. J., Bestor, T. H. & Trasler, J. M. (2002) Genomics 79, 530–538. [DOI] [PubMed] [Google Scholar]
- 18.Wakayama, T., Perry, A. C., Zuccotti, M., Johnson, K. R. & Yanagimachi, R. (1998) Nature 394, 369–374. [DOI] [PubMed] [Google Scholar]
- 19.Yamazaki, Y., Mann, M. R., Lee, S. S., Marh, J., McCarrey, J. R., Yanagimachi, R. & Bartolomei, M. S. (2003) Proc. Natl. Acad. Sci. USA 100, 12207–12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabo, P. E., Hubner, K., Scholer, H. & Mann, J. R. (2002) Mech. Dev. 115, 157–160. [DOI] [PubMed] [Google Scholar]
- 21.Mann, M. R., Chung, Y. G., Nolen, L. D., Verona, R. I., Latham, K. E. & Bartolomei, M. S. (2003) Biol. Reprod. 69, 902–914. [DOI] [PubMed] [Google Scholar]
- 22.Tremblay, K. D., Duran, K. L. & Bartolomei, M. S. (1997) Mol. Cell. Biol. 17, 4322–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatot, C. L., Lewis, J. L., Torres, I. & Ziomek, C. A. (1990) Biol. Reprod. 42, 432–440. [DOI] [PubMed] [Google Scholar]
- 24.Kimura, Y. & Yanagimachi, R. (1995) Biol. Reprod. 52, 709–720. [DOI] [PubMed] [Google Scholar]
- 25.Clark, S. J., Harrison, J., Paul, C. L. & Frommer, M. (1994) Nucleic Acids Res. 22, 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis, T. L., Trasler, J. M., Moss, S. B., Yang, G. J. & Bartolomei, M. S. (1999) Genomics 58, 18–28. [DOI] [PubMed] [Google Scholar]
- 27.Kato, Y., Rideout, W. M., 3rd, Hilton, K., Barton, S. C., Tsunoda, Y. & Surani, M. A. (1999) Development (Cambridge, U.K.) 126, 1823–1832. [DOI] [PubMed] [Google Scholar]
- 28.Rideout, W. M., 3rd, Eggan, K. & Jaenisch, R. (2001) Science 293, 1093–1098. [DOI] [PubMed] [Google Scholar]
- 29.Inoue, K., Kohda, T., Lee, J., Ogonuki, N., Mochida, K., Noguchi, Y., Tanemura, K., Kaneko-Ishino, T., Ishino, F. & Ogura, A. (2002) Science 295, 297. [DOI] [PubMed] [Google Scholar]
- 30.Ueda, T., Abe, K., Miura, A., Yuzuriha, M., Zubair, M., Noguchi, M., Niwa, K., Kawase, Y., Kono, T., Matsuda, Y., et al. (2000) Genes Cells 5, 649–659. [DOI] [PubMed] [Google Scholar]
- 31.DeChiara, T. M., Robertson, E. J. & Efstratiadis, A. (1991) Cell 64, 849–859. [DOI] [PubMed] [Google Scholar]
- 32.Guillemot, F., Nagy, A., Auerbach, A., Rossant, J. & Joyner, A. L. (1994) Nature 371, 333–336. [DOI] [PubMed] [Google Scholar]
- 33.Lau, M. M., Stewart, C. E., Liu, Z., Bhatt, H., Rotwein, P. & Stewart, C. L. (1994) Genes Dev. 8, 2953–2963. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Z. Q., Fung, M. R., Barlow, D. P. & Wagner, E. F. (1994) Nature 372, 464–467. [DOI] [PubMed] [Google Scholar]
- 35.Miki, H., Inoue, K., Kohda, T., Honda, A., Ogonuki, N., Yuzuriha, M., Mise, N., Matsui, Y., Baba, T., Abe, K., et al. (2005) Genesis 41, 81–86. [DOI] [PubMed] [Google Scholar]
- 36.Bortvin, A., Eggan, K., Skaletsky, H., Akutsu, H., Berry, D. L., Yanagimachi, R., Page, D. C. & Jaenisch, R. (2003) Development (Cambridge, U.K.) 130, 1673–1680. [DOI] [PubMed] [Google Scholar]
- 37.Zwaka, T. P. & Thomson, J. A. (2005) Development (Cambridge, U.K.) 132, 227–233. [DOI] [PubMed] [Google Scholar]
- 38.Saitou, M., Barton, S. C. & Surani, M. A. (2002) Nature 418, 293–300. [DOI] [PubMed] [Google Scholar]