Abstract
Members of the cytochrome P450 3A subfamily catalyze the metabolism of endogenous substrates, environmental carcinogens, and clinically important exogenous compounds, such as prescription drugs and therapeutic agents. In particular, the CYP3A4 and CYP3A5 genes play an especially important role in pharmacogenetics, since they metabolize >50% of the drugs on the market. However, known genetic variants at these two loci are not sufficient to account for the observed phenotypic variability in drug response. We used a comparative genomics approach to identify conserved coding and noncoding regions at these genes and resequenced them in three ethnically diverse human populations. We show that remarkable interpopulation differences exist with regard to frequency spectrum and haplotype structure. The non-African samples are characterized by a marked excess of rare variants and the presence of a homogeneous group of long-range haplotypes at high frequency. The CYP3A5*1/*3 polymorphism, which is likely to influence salt and water retention and risk for salt-sensitive hypertension, was genotyped in >1,000 individuals from 52 worldwide population samples. The results reveal an unusual geographic pattern whereby the CYP3A5*3 frequency shows extreme variation across human populations and is significantly correlated with distance from the equator. Furthermore, we show that an unlinked variant, AGT M235T, previously implicated in hypertension and pre-eclampsia, exhibits a similar geographic distribution and is significantly correlated in frequency with CYP3A5*1/*3. Taken together, these results suggest that variants that influence salt homeostasis were the targets of a shared selective pressure that resulted from an environmental variable correlated with latitude.
Introduction
Cytochrome P450 (CYP) genes are abundant in animal, plant, and bacterial genomes and have evolved to metabolize a variety of diverse compounds. Among the most abundant and clinically important of human CYP enzymes are members of the CYP3A subfamily, which catalyze the metabolism of endogenous substrates, ranging from bile acids to steroids such as estrogen and testosterone, as well as environmental carcinogens such as pesticides. CYP3A enzymes also play important roles in the bioavailability and clearance of a wide range of exogenous compounds in the form of prescription drugs and therapeutic agents (Lamba et al. 2002a).
Interindividual variability in clearance of CYP3A substrates can result from the effects of inducers, inhibitors, or genetic or dietary factors that potentially lead to differences in drug toxicity and response. It is estimated that genetic factors are responsible for 70%–90% of interindividual variability in constitutive expression (Ozdemir et al. 2000), but few genetic determinants that underlie this variation are known. Genetic variants that change the amino acid sequence tend to be rare and, although there are a few common noncoding SNPs that affect gene expression and/or activity, the known variants fail to account completely for the observed phenotypic variation (Lamba et al. 2002b).
Members of the CYP3A subfamily are located on chromosome 7q22 and span ∼220 kb. The four genes include CYP3A4 (MIM 124010), which is the dominant adult enzyme and is expressed primarily in liver and small intestine; CYP3A5 (MIM 605325), which is expressed in liver, kidney, intestine, and prostate gland; CYP3A7 (MIM 605340), which is expressed fetally; and CYP3A43 (MIM 606534), which is expressed at low levels in a number of tissues. In many populations, total adult CYP3A protein content consists largely of CYP3A4; however, extreme interpopulation variability exists with regard to CYP3A5 expression. This variation is largely due to a single-base substitution in intron 3 of CYP3A5, which results in an incorrectly spliced mRNA and a nonfunctional protein (Kuehl et al. 2001). This allele, known as “CYP3A5*3,” was reported to have a frequency of ∼27%–50% among African Americans, 85%–95% among whites (Hustert et al. 2001; Kuehl et al. 2001), and 60%–73% among Asians (Hustert et al. 2001). Therefore, the proportion of CYP3A5 in the total liver and intestinal CYP3A pool is lower in carriers of the CYP3A5*3 allele, which creates the potential for differential drug response. In addition, it has been suggested that the expressor allele may be associated with increased systolic blood pressure and mean arterial pressure in African Americans (Givens et al. 2003). The proposed mechanism involves the conversion of cortisol to 6β-hydroxycortisol by CYP3A5 in the kidney, which leads to higher reabsorption of sodium and water retention as well as the clinical phenotype of salt-sensitive hypertension. It was proposed that the expressor allele confers a selective advantage in equatorial populations that may experience water shortages (Kuehl et al. 2001).
Here, we used a comparative genomics approach to pinpoint regions of potential interest for our resequencing study, with the idea that highly conserved sequences between distantly related species may harbor regulatory elements that affect gene expression. We combined comparative genomics with computational predictions of clusters of liver-enriched transcription-factor-binding sites. These sequence elements, together with the coding regions and the known regulatory elements, were surveyed in ethnically diverse samples to uncover additional functional variation, which can be further examined for its possible role in interindividual variability in drug response. In addition, we investigated the role of natural selection in shaping patterns of variation at the CYP3A locus. Our results show a striking haplotype structure, large allele frequency differences between African Americans and non-African populations, and a significant correlation between CYP3A5*1/*3 allele frequency and distance from the equator. We also observed a significant correlation, in the same population samples, between the frequency of an additional unlinked variant that influences salt sensitivity (AGT M235T [MIM 106150]) and distance from the equator. These findings argue for a shared selective pressure on salt-sensitivity variants, with the intensity of selection varying in correlation with latitude.
Material and Methods
Library Screening and BAC-Clone Sequencing
Universal oligonucleotide probes (overgos) were designed on the basis of an alignment of human, mouse, and rat CYP3A coding sequences (Thomas et al. 2002) and were used to screen BAC libraries for the mouse (RPCI-23), rat (RPCI-32), dog (RPCI-81), and olive baboon (RPCI-41). The positive clones were sequenced by the random shotgun approach (Roe 2004).
Comparative Analysis and Resequencing-Study Design
Regions included in the resequencing study were identified by alignment of all nonhuman Cyp3A genes to the human CYP3A4 or CYP3A5 sequences (GenBank accession numbers AC069294 and AC005020, respectively). Mouse and rat sequence information included additional clones from public databases. Because the ancestral relationships between these genes cannot be determined unambiguously, we included multiple genes from each of the nonhuman species that are closely related to CYP3A4 and CYP3A5. Human sequences were analyzed with Cluster-Buster (Frith et al. 2003), to predict clusters of binding sites for the liver-enriched transcription factors HNF1α, HNF4, PXR/CAR, OCT-1, PPAR/RXR, CEBP, HNF3β, and HNF4/COUP-TF. Three regions that contained clusters predicted with high probability and conserved across all five species were included in the resequencing study.
DNA Samples
Human population samples consisted of 24 Europeans, 23 African Americans, and 23 Han from Los Angeles; all individuals are unrelated and are subsets of three Human Variation panels in the Coriell Cell Repositories (Caucasian, African American, and Han People of Los Angeles). Sample information can be obtained from the PharmGKB Web site (accession numbers PS203894 and PS203895). The orthologous regions were sequenced in a western chimpanzee (Pan troglodytes verus). SNP typing was performed on the entire Human Genome Diversity Panel–Centre d’Etude du Polymorphisme Humain (HGDP-CEPH). This study was approved by the institutional review board of the University of Chicago.
PCR Amplification and Sequencing
PCR and sequencing primers (available from the PharmGKB Web site) were designed on the basis of GenBank sequences AC069294 and AC005020 for CYP3A4 and CYP3A5, respectively; all nucleotide positions in this article are numbered according to those sequences. PCR and sequencing were performed as described elsewhere (Wall et al. 2003). The same primers were used to amplify and sequence the human and the chimpanzee samples. In addition to PharmGKB, sample information, primer sequences, polymorphism data, and outgroup sequences are available on the Di Rienzo lab Web site.
Data Analysis
Population genetics summary statistics were calculated using SLIDER and MAXDIP. |D′| and r2 were calculated from diploid data, according to the maximum-likelihood method (Hill 1974). Haplotypes were inferred in each population sample separately, by use of PHASE2 (Stephens et al. 2001). Singleton sites were omitted from the haplotype inference. Fay and Wu’s H test (Fay and Wu 2000) was performed using DnaSP version 4.0 (Rozas and Rozas 1995). Coalescent simulations for the H and haplotype tests were performed assuming values of 4Nr estimated from the data for each population sample.
SNP Genotyping
For the CYP3A5*1/*3 polymorphism, we modified a mismatch PCR-based RFLP protocol by first performing PCR with primers specific to CYP3A5 (Cyp5Ex4F-Cyp5Ex4R) (sequences available from PharmGKB) followed by a nested PCR (Fukuen et al. 2002). The nested PCR product was digested with DdeI and was visualized on 3% agarose gel electrophoresis.
For the AGT M235T polymorphism, we also used a mismatch PCR-based RFLP assay. Genomic DNA was subjected to a first round of amplification with primers 5′-CAACTTCTTGGGCTTCCGTA-3′ and 5′-TGCACATAGTAGGGCAGCAG-3′, under the following conditions: initial denaturation at 94°C for 2 min, followed by 34 cycles of 94°C for 30 s, 64.6°C for 30 s, and 72°C for 1.5 min, and a final extension of 72°C for 1 min. Of this product, 1 μl was diluted 50-fold and was used as template in a nested PCR reaction, by use of primer sequences published elsewhere (Russ et al. 1993) and an annealing temperature of 67°C; 4 μl of product was digested overnight with 5 units of Tth111 I in a 50-μl reaction volume, and the digested products were visualized on a 3% agarose gel.
Results
Sequence Variation and the Frequency Spectrum
The extent of sequence identity across species at the CYP3A4 and CYP3A5 genes is shown in figure A1 (online only). Computational predictions of clusters of liver-enriched transcription-binding sites identified five regions with high probability. Two of them are not conserved and were not included in our survey. The remaining three, two of which fall in intronic sequences and one of which is 10.5 kb upstream of CYP3A4, are conserved across species and were included in our survey. The same segments were sequenced in one chimpanzee.
The surveyed sequence spans ∼150 kb and contains 1.6 kb of coding sequence and 15.2 kb of conserved noncoding sequence (CNS) for CYP3A4 and 1.6 kb of coding sequence and 13.2 kb of CNS for CYP3A5. We identified a total of 92 polymorphic sites (including one multiallelic and six biallelic indels), of which 5 were nonsynonymous, 4 were synonymous, and 83 were noncoding. Twelve noncoding SNPs occurred at highly conserved nucleotide positions—that is, positions where the ancestral allele (inferred by comparison with the chimpanzee sequence) was identical to the base found in the remaining four species. Sequence divergence between human and chimpanzee is relatively low (0.86% and 0.66% for CYP3A4 and CYP3A5, respectively) compared with the genomewide average of 1.24% (Ebersberger et al. 2002).
Summary statistics of the polymorphism data are shown in table 1, and a visual representation of the sequence data, in the form of inferred haplotypes, is shown in figure 1. Polymorphism levels, as summarized by nucleotide diversity (π) and Watterson's estimator of the population mutation rate parameter θ (θW), are low for both genes. Linkage-disequilibrium (LD) levels summarized by ρH01, an estimator of the population-recombination rate parameter (4Nr) (Frisse et al. 2001; Hudson 2001), are also on the low side, especially for CYP3A5 in the non-African samples. The Tajima’s D statistic, which summarizes information about the spectrum of allele frequencies (Tajima 1989), is expected to be near zero, under the neutral equilibrium model. A positive value indicates an excess of intermediate frequency variants consistent with the effects of balancing selection or population subdivision, whereas a negative value indicates a skew toward rare variants and suggests the action of directional selection or population growth. The negative Tajima’s D values at the CYP3A4 and CYP3A5 genes indicate an excess of rare variants; this, together with the low polymorphism levels, raises the possibility of a selective sweep.
Table 1.
Summary Statistics of Polymorphism Data from African American, European, and Han Population Samples
|
Summary Statistics by Population Sample |
|||||||||||||||
| African Americans |
Europeans |
Han |
|||||||||||||
| Gene | Length(bp) | πa | TDb | θWc | θW:divd | ρH01e | πa | TDb | θWc | θW:divd | ρH01e | πa | TDb | θWc | ρH01e |
| CYP3A4 | 16,822 | .46 | −.58 (49) | .55 | 6.7% (6) | .52 | .14 | −1.76 (3) | .31 | 3.7% (11) | .37 | .1 | −.75 | .14 | .62 |
| CYP3A5 | 14,573 | .26 | −1.13 (12) | .39 | 5.5% (2.8) | .36 | .06 | −2.11 (1) | .19 | 2.9% (2.8) | .00 | .06 | −1.21 | .11 | .00 |
Nucleotide diversity per bp (×10-3).
Tajima’s D (Tajima 1989). Numbers in parenthesis indicate the percentile rank of the TD value relative to the distribution of TD values in the SeattleSNP genes.
Watterson’s estimator of the population-mutation-rate parameter θ (=4Nμ) per bp (×10-3) (Watterson 1975).
Ratio of θW to the amount of sequence divergence (div) between human and chimpanzee. Numbers in parenthesis indicate the percentile rank of the θW:div value relative to the distribution of θW:div values in the SeattleSNP genes.
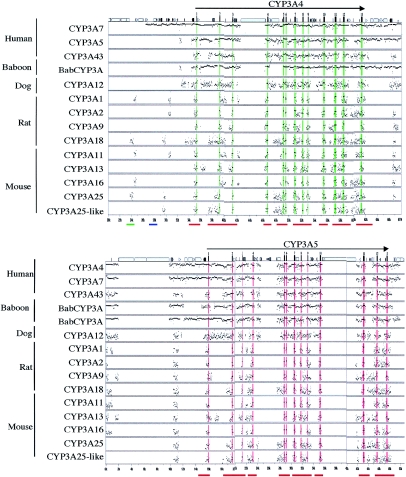
Figure 1.
Inferred haplotypes at CYP3A4 and CYP3A5. Neither singleton sites nor multiallelic indels were included. The chimpanzee sequence was used to infer the ancestral allele at each site. The numbers on the right indicate the number of haplotypes in each population. The asterisk (*) indicates the position of CYP3A5*1/*3. Numbers below the gene names indicate the position of each polymorphic site relative to the reference sequence for each gene (GenBank accession numbers AC069294 and AC005020 for CYP3A4 and CYP3A5, respectively).
An excess of high frequency derived alleles may be expected soon after a selective sweep is completed or at some stage during an ongoing sweep and can be assessed by means of the H test (Fay and Wu 2000). The results of the H test were significant for CYP3A4 in the European and Asian samples (P=.006 and P=.003, respectively). The results for CYP3A5 were not statistically significant for any population sample; however, the power of the test may be low because of the small number of polymorphic sites at this locus. The inferred haplotypes in figure 1 show that the high-frequency-derived alleles tend to be in strong LD (see also fig. A2 [online only]). Two haplotypes, which differ from each other by a single site, account for 60% and 65% of the European and Han haplotypes, respectively. These results suggest that the pattern detected by the H test in the non-African samples is largely due to the increase in frequency of one haplotype class that contains several derived alleles and that spans ∼150 kb. We used the haplotype test (Hudson et al. 1994) to ask if the haplotype structure is indeed inconsistent with the neutral-equilibrium model. We ran coalescent simulations to generate 1,000 samples that contained the same number of polymorphic sites observed in the total surveyed segment (that contained both CYP3A4 and CYP3A5) in the European and Han samples, and we calculated the proportion of simulated samples that contained a subset that accounted for 60% and 65% of the haplotypes, respectively, and contained one or no polymorphic site (Hudson et al. 1994). This proportion is 0.2% and 2% in Europeans and Han, respectively, and it suggests that the haplotype structure is indeed unusual.
To assess whether the low polymorphism levels and the skew toward rare variants is unusual, we compared our results with the data (referred to as “SeattleSNPs”) of the University of Washington–Fred Hutchinson Cancer Research Center Variation Discovery Resource that focuses on genes involved in inflammation. Statistical tests of neutrality, including the H and the haplotype tests, assume a population at equilibrium. However, human populations do not fit the equilibrium expectations. Comparing the CYP3A4 and CYP3A5 genes with an empirical distribution, such as that of the SeattleSNP genes, circumvents this problem. This comparison was facilitated by the fact that the European and African American samples used in our study are the same as those used in the SeattleSNP project; no Asian data is available in the SeattleSNP data set. To take into account the strong evolutionary constraints in the surveyed segments, we normalized the polymorphism levels according to the amount of interspecies sequence divergence by taking the ratio of θW and sequence divergence between human and chimpanzee; the value obtained for the CYP3A4 and CYP3A5 genes was compared with the distribution of the same quantity in the 159 SeattleSNP genes (table 1). The same comparison was made for the Tajima’s D values. A summary of the distribution of the SeattleSNP polymorphism data is given in table 2. Although on the low end of the distribution for the SeattleSNP genes, the polymorphism levels normalized for interspecies divergence observed in CYP3A4 are not unusual; a more marked reduction is seen at CYP3A5, in which the ratio of θW and interspecies divergence is in the 2.8th percentile of the SeattleSNP distribution for both African Americans and Europeans. Tajima’s D values in the European sample were unusually low for both CYP3A4 and CYP3A5 compared with the SeattleSNP data set, whereas the values for the African American sample were well within the observed range. Thus, the comparison of our data with an empirical distribution is in qualitative agreement with the results of statistical tests of neutrality based on the equilibrium model in suggesting a role for directional selection in the CYP3A locus.
Table 2.
Summary of the 159 Genes of the SeattleSNP Data Set
| Tajima’s D |
θW:divb |
||||
| Population | Na | Range | Mean | Range | Mean |
| African Americans | 24 | −1.70 to 1.53 | −.54 | .045 to .336 | .116 |
| Europeans | 23 | −2.80 to 2.46 | .22 | .022 to .206 | .071 |
Number of individuals in the sample.
Ratio of θW to the amount of sequence divergence (div) between human and chimpanzee.
Geographic Distribution of CYP3A5*3 Allele Frequency
One of the variants that defines the haplotype class found at near-fixation frequency in non-Africans is the nonexpressor allele at CYP3A5 (CYP3A5*3). The haplotype structure and marked skew toward rare variants in the non-African samples coupled with the likely phenotypic and fitness effects associated with this allele suggest that it was quickly driven to high frequency by positive natural selection outside Africa. We typed this polymorphism in 1,064 individuals from 52 different world populations in the HGDP-CEPH panel (table A1 [online only]). The frequency of the CYP3A5*3 allele is lowest in sub-Saharan Africa (0.07 in Namibia and the Congo; 0.06 in Nigeria) and highest in European and East Asian populations (0.96 in France; 0.95 in Italy and regions of China). Interestingly, the frequency of the CYP3A5*3 allele increases with distance from the equator (fig. 2A) (Spearman rank correlation score=0.612; P<.0001). A significant rank correlation was also observed for 18 East Asian populations that span 51° of latitude (rank correlation score=0.649; P=.0077), but not for the 7 European populations that span only 21° of latitude.
Figure 2.
Geographic distribution of the CYP3A5*3 and AGT Met235 allele frequencies. A, Plot of CYP3A5*3 and AGT Met235 allele frequencies and distance from the equator (measured in degrees of latitude). B, Plot of CYP3A5*3 versus AGT Met235 allele frequencies, by subpopulation.
Because nearby populations are likely to exchange genes more often than distant populations, it is possible that a correlation between allele frequency and latitude is often observed for this set of populations simply as a result of their geographic structure and history of migration. Ideally, a comparison with a large data set of biallelic markers typed in the same population samples could address this issue. In the absence of biallelic marker data, we compared our findings with the results of a genomewide survey of microsatellite variation at 404 loci typed in exactly the same individuals. These loci were shown to exhibit levels of interpopulation differentiation comparable to those of biallelic markers (Rosenberg et al. 2002). We selected a total of 276 microsatellite alleles, 1 per locus, with global allele frequencies within 5% of that of CYP3A5*3, and we computed the rank correlation score between each microsatellite allele frequency and the distance of each population from the equator. Only two microsatellite alleles had a more extreme correlation score than that observed for CYP3A5*3. We repeated the analysis by matching the allele frequencies in the pool of sub-Saharan African populations instead of matching the global-allele frequencies: we selected 377 alleles, and only 1 of them had a more extreme correlation score than did CYP3A5*3. This indicates that the geographic distribution of the CYP3A5*3 allele frequency is indeed unusual and suggests that it may be due to a selective advantage, conferred by this allele, that increases with increasing latitudes.
Because of the high degree of LD in this region (fig. A2 [online only]), several SNPs in our resequencing survey share the large allele frequency differences between African Americans and non-Africans observed for CYP3A5*3. FST, a statistic that summarizes allele frequency differentiation between samples, was estimated per site between each of the two non-African samples and the African American sample. Whereas average FST values in worldwide human populations are ∼0.123 (Akey et al. 2002), seven and two SNPs in CYP3A4 and CYP3A5, respectively, had an FST >0.5 between African Americans and the non-Africans, reaching a maximum of 0.66. It was previously estimated that only 1.9% of SNPs had a difference in allele frequency between African Americans and Europeans ⩾0.5 (Akey et al. 2002; Rosenberg et al. 2003). In our data set, 8 (21%) of 38 SNPs with minor-allele frequency >5% showed such a large difference in allele frequency. In 5 (11%) of 44 SNPs, the allele frequencies differed by >0.5 between African Americans and Han. This further corroborates the notion that the pattern of geographic differentiation at this locus is not simply the result of human population history.
Geographic Distribution of AGT Met235 Allele Frequency
Variation in the angiotensinogen gene (AGT) was shown to influence risk for human hypertension (Luft 2001) and pregnancy complications (Ward et al. 1993). A promoter A-6G variant exists in strong LD with the M235T variant; the combination of the two is reported to be associated with hypertension (Wu et al. 2004). The CYP3A5*1/*3 polymorphism appears to have similar phenotypic effects as these AGT variants. If the salt-sensitivity phenotypes associated with the CYP3A5 and AGT variants evolved under the same selective pressures, one might expect a similar geographic distribution in these two unlinked genes. To test this hypothesis, we genotyped the AGT M235T variant in our same population samples (i.e., the HGDP-CEPH panel) (table A1 [online only]). As with CYP3A5*3, the frequency of the derived allele, M235 at AGT, increases with distance from the equator (fig. 2A) (rank correlation score 0.712; P<.0001). A significant rank correlation was also observed for 18 East Asian populations (rank correlation score 0.511; P=.0352) but not for the 7 European populations. When compared with 259 microsatellite alleles typed in the same population samples and that have global-allele frequency within 5% of that of AGT M235, no microsatellite allele had a more extreme correlation score than that observed for the M235 allele. Matching the allele frequencies in the pool of sub-Saharan African samples did not change the results; that is, none of 399 alleles with frequencies within the specified range had a more extreme correlation score than did CYP3A5*3. As shown in fig. 2B, the frequencies of the two derived alleles, CYP3A5*3 and AGT M235, are also significantly correlated with each other (rank correlation score 0.680; P<.0001).
Discussion
Our comparative genomics-based survey of sequence variation in three human populations identified a large number of variants in noncoding regions that are conserved across distantly related mammalian species. In addition, it revealed a striking pattern of haplotype structure and an excess of rare variants in the non-African samples, as well as large differences in allele frequencies between samples of African and non-African descent. These results suggest that a relatively homogeneous class of haplotypes that span 150 kb were driven to near-fixation frequency by natural selection in the non-African populations. The observation of a significant rank correlation between the frequency of the CYP3A5*3 allele that defines this haplotype class and distance from the equator further suggests the action of spatially varying selective pressures. The finding of a similar correlation for another variant, AGT M235T, with similar phenotypic effects supports the idea that both variants were the targets of a shared selective pressure. Although we cannot rule out the possibility that other variants in perfect LD with the two SNPs we tested were the true targets of selection, it seems plausible that such hypothetical linked variants must similarly affect salt sensitivity.
Our assessment of the potential signature of natural selection on the CYP3A genes relies on a combination of comparisons of theoretical expectations, based on the neutral equilibrium model, with empirical expectations, based on a large sequence variation data set (SeattleSNP). Because human populations have experienced complex histories, a significant departure in tests based on equilibrium assumptions, such as the H and haplotype tests, may simply be the result of increased variance across loci that is due to past bottlenecks and/or population subdivision. Simulations showed that this is the case specifically for the H test (Przeworski 2002); in addition, human polymorphism data suggest that significant departures in this test may be due to population history rather than selection (Hamblin et al. 2002). If the true history of human populations could be inferred accurately, one could perform the same tests under the assumption of the appropriate null neutral model. In the absence of such information, one can use, for comparison, a distribution obtained from a large set of loci surveyed in the same population samples. If these loci evolved neutrally, such a distribution would simply reflect the stochastic variance associated with the evolutionary process, given the particular history of the populations examined. This is unlikely to be the case for the SeattleSNP data, because of the inclusion of genes that may well have evolved under a variety of selection models (Akey et al. 2004). Hence, the variance in the distribution of SeattleSNP genes is likely to be greater than expected if all the genes were neutral. This makes the comparison of a candidate target of natural selection with the SeattleSNP distribution conservative and our results all the more striking.
A number of characteristic features were observed in the CYP3A4 and CYP3A5 genes in the non-African samples: low polymorphism levels (even after normalization for interspecies divergence), a marked skew toward rare variants, a large fraction of high-frequency-derived alleles, a large subset of inferred haplotypes with low levels of variation, and above-average differences in allele frequency between populations of African and non-African descent. The comparison with the SeattleSNP data for polymorphism levels and the skew toward rare variants allowed us to determine that the results for the European population are indeed unusual. An alternative approach to testing neutrality on the basis of levels of polymorphism and divergence is the HKA test (Hudson et al. 1987), which properly takes the evolutionary variance into account. However, with regard to human populations, this test suffers from the same limitations of other standard tests in that it relies on the theoretical expectations of the standard neutral model. It is not obvious how to perform such an empirical comparison for the aspects of the data used in the H and the haplotype tests; thus, the interpretation of their results is not straightforward. A further caveat for the haplotype test is that we used inferred haplotypes without taking into account the uncertainty in the phase inference, whereas the test assumes that the haplotype phase is known. This may lead to an underestimate of the P value and would not be conservative.
Because of the likely contribution of European genes (discussed below) and the large interethnic differences at this locus, the interpretation of the African American data is not straightforward. Polymorphism levels normalized by interspecies divergence tend to be low in African Americans, even though a number of polymorphic sites may have been introduced through admixture with European genes. Likewise, the occurrence of variants at intermediate frequencies in African Americans, which are nearly fixed in Europeans, suggests that the frequency spectrum in the native African portion of the African American gene pool may be more strongly skewed toward rare variants. Thus, the possibility that natural selection acted on this locus in Africa cannot be excluded on the basis of our data. Resequencing surveys of native African populations will be necessary to address this issue.
Despite these caveats, the functional role of the CYP3A5*1/*3 polymorphism, its likely fitness consequences, and its geographic distribution independently argue for the action of natural selection on this locus. CYP3A enzymes convert circulating cortisol to 6-β-hydroxycortisol, which plays a role in sodium transport in the kidney and is hypothesized to result in defective renal sodium excretion (Ghosh et al. 1995). More specifically, the conversion of cortisol to 6-β-hydroxycortisol by CYP3A5 in the kidney, which results in greater sodium retention, has been proposed to contribute to salt-sensitive hypertension in humans (Schuetz et al. 1992; Givens et al. 2003). Accordingly, 6-β-hydroxycortisol levels were found to be markedly higher in the spontaneously hypertensive rat (SHR) than the normotensive control (Schenkman et al. 1989). In addition, a selective CYP3A inhibitor (TAO) decreases in vivo 6-β-hydroxycortisol and blood pressure in the SHR (Watlington et al. 1992). Together, the correlation between renal CYP3A activity and blood pressure in the SHR is consistent with the idea that increased 6-β-hydroxycortisol production by CYP3A enzymes may play a role in hypertension.
Phenotypic variability in human hypertension susceptibility could be due to genetic variation in genes that underlie salt regulation. The “sodium retention hypothesis” (Gleibermann 1973; Nakajima et al. 2004) proposes that ancient human populations living in hot, humid areas with low sodium availability adapted to their environment by retaining salt, whereas populations in cooler, temperate climates adapted to conditions of greater sodium availability. Since an increased prevalence of hypertension in African American versus white populations is well documented and since blood pressure homeostasis is strongly influenced by sodium regulation and salt sensitivity, population-specific differences in susceptibility to hypertension could be explained, in part, by variation in genes related to salt regulation. This hypothesis provides a general framework for interpreting the patterns of variation observed at genes involved in sodium homeostasis. However, in its original formulation, an ancestral allele that increases salt and water retention, such as CYP3A5*1, is expected to confer a selective advantage that decreases with distance from the equator but is not necessarily predicted to become deleterious. Interestingly, the near-fixation frequency of the derived CYP3A5*3 allele in many non-African populations coupled with the unusual patterns of variation observed in our resequencing survey suggest that CYP3A5*1 does become deleterious at high latitudes. One possible scenario is that the CYP3A5*1 allele has multiple phenotypic effects with opposite fitness consequences; the trade off between the advantage and the disadvantage conferred by this allele changes dependent on an environmental variable correlated with distance from the equator. An example of a possible disadvantage associated with CYP3A5*1 is complications of pregnancy: increased urinary excretion of 6β-hydroxycortisol was observed in women with pre-eclampsia (Frantz et al. 1960). Thus, whereas the selective advantage due to increased salt and water retention decreases and possibly disappears completely with increasing latitude, the selective disadvantage becomes the main influence on the overall fitness, which results in the adaptive rise in frequency of the CYP3A5*3 allele. Alternatively, a different allele in perfect LD with CYP3A5*3 could have conferred a selective advantage at higher latitudes over the CYP3A5*1 haplotypes, which would result in the signature of natural selection suggested by our data.
Our survey of allele frequency strengthens and extends previous findings that the nonexpressor allele (CYP3A5*3) is highly prevalent outside Africa. The unusual geographic distribution of the CYP3A5*3 allele is striking in two respects. First, the correlation between allele frequency and distance from the equator is highly significant and cannot be attributed to patterns of human migration and geographic distance. Second, we observed an enormous range of allele frequencies at this site. Such an extreme degree of interpopulation differentiation suggests the impact of population-specific selective pressures. Interestingly, a similar pattern was observed at AGT, which is involved in risk for hypertension and pregnancy complications (Ward et al. 1993). At the AGT locus, a promoter A-6G variant exists in strong LD with the M235T variant; the combination of the two is reported to be associated with hypertension (Wu et al. 2004). Lifton et al. (1993) suggested that increased salt and water retention associated with the T235 allele may have been advantageous in times of salt scarcity, with the M235 allele rising to higher frequencies following the move to salt-rich areas outside Africa. A survey of the AGT gene in worldwide samples revealed that the A-6 allele is at much higher frequency in African populations compared with non-African ones (Nakajima et al. 2004). A comparison of the CYP3A5*3 and AGT G-6 allele frequencies across populations shared by the two studies reveals a significant positive correlation (Spearman rank correlation P=.0204). In addition, we show that the AGT M235 allele frequency is significantly correlated with distance from the equator and with CYP3A5*3 allele frequency in the HGDP-CEPH panel. The correlation of allele frequency between two unlinked SNPs with similar phenotypic effects is remarkable and suggests a shared selective pressure with regard to sodium regulation and homeostasis. This finding offers a new perspective on the search for candidate variants that increase risk for salt-sensitive hypertension. More generally, one can speculate that a correlation between allele frequencies and environmental variables may identify variants that contribute to common human diseases with different prevalence across ethnic groups.
The low number and frequency of coding variants observed in our sample mirrors the findings of previous efforts to explain phenotypic variation in drug metabolism through coding changes at the CYP3A locus (Lamba et al. 2002b). The most common of these changes was the CYP3A5*6 allele (np 30597), which occurred at a frequency of 11% in the African American sample, the only population in which it was observed. This allele has been reported almost exclusively in African American populations (10%–13% frequency) and results in an alternatively spliced and ultimately truncated protein, which leads to the same effect on function as the CYP3A5*3 allele. The observation that CYP3A5 expression is severely compromised by two common alleles that exist on different haplotypes is clearly unusual and is consistent with the idea that the expression of CYP3A5 may be disadvantageous under some environmental conditions.
The frequency of the CYP3A5*3 allele in African Americans is 33%, whereas its average frequency in the native sub-Saharan African populations is 14%. Given the much higher frequency of this allele in Europeans, this is consistent with previous results on the contribution of European genes to the African American gene pool (Parra et al. 1998). Interestingly, there is a suggestion that CYP3A5*3 haplotypes are more heterogeneous in African Americans than in the non-African populations (fig. 1): most African American haplotypes closely resemble those observed in the non-African samples, but a minority lack the derived alleles at five SNPs (nps 34914, 40104, 51067, 51927 and 55544) that are found in most of CYP3A5*3 haplotypes outside Africa. Two of these SNPs may be functional, since they occur at positions conserved in all five species. This raises the possibility of functional heterogeneity within the CYP3A5*3 haplotypes across native African populations and possibly across different African American populations, depending on the specific origin of their sub-Saharan African gene pool. This may have implications for understanding the contribution of CYP3A variation to interindividual variation in drug response in both African and African American populations.
We used sequence comparisons across five mammalian species and computational predictions to increase the probability of identification of functional variation that may underlie variability in drug response. The utility of comparative genomics has been demonstrated elsewhere, such as the identification of a coordinate regulator of three interleukin genes (Pennacchio and Rubin 2001). We combined this approach with computational prediction of clusters of transcription-factor-binding sites. One conserved predicted cluster was recently confirmed as a functional element important for constitutive activation of CYP3A4 (Matsumura et al. 2004). Four transcription-binding sites—HNF-1, HNF-4, AP-1, and USF-1—are located in this region, and the interaction of these factors with this region was demonstrated through gel shift assays. The HNF-1 and HNF-4 binding sites were correctly predicted in our analysis; AP-1 and USF-1, which are not liver-enriched, were not included in our search. This finding validates the utility of computational predictions and comparative genomics for honing in on putative regulatory elements. However, it is clear that they should be used in combination with other strategies, as evidenced by the well-characterized distal xenobiotic-response-enhancer module (Goodwin et al. 1999), which is neither conserved nor identified by prediction algorithms.
It was previously proposed that drug metabolizing enzyme (DME) genes have diversified through the coevolution of plants and animals. Furthermore, it was hypothesized that differences in DME allele frequencies across human populations reflected differences in diet composition over thousands of years (Nebert 1997). The variety of endogenous and exogenous substrates metabolized by CYP3A4 and CYP3A5 suggests that they may be exposed to multiple selective pressures and that different variants may be the targets of these pressures. Furthermore, variation in other genes within the CYP3A gene cluster, namely CYP3A7 and CYP3A43, could add to the complex evolutionary history of this genomic region. Additional studies of sequence variation in neighboring genes will allow further testing of the hypotheses presented in this article.
Acknowledgments
We are grateful to members of the Pharmacogenetics of Anticancer Agents Research group; especially, to E. Schuetz, for helpful discussions throughout this project; and to E. Schuetz, N. Cox, R. Hudson, and one anonymous reviewer, for insightful comments on the manuscript. We thank D. Nickerson and J. Akey for providing polymorphism and divergence data for the SeattleSNP genes. This work was supported by National Institutes of Health grants GM61393, DK55889, and HG02152. E.E.T. was partially supported by training grant GM07197.
Appendix
Figure A1.
MultiPipMaker plots that illustrate cross-species conservation at CYP3A4 and CYP3A5. The vertical axis ranges between 50% and 100% sequence identity; areas within the plots shaded in light green and pink correspond to the exons of the CYP3A4 and CYP3A5 genes, respectively. Bars below the plots indicate the segments included in the resequencing survey; red bars indicate the sequence included only because of cross-species conservation, green bars indicate the sequence included because of both cross-species conservation and predicted cluster of transcription-factor-binding sites, and blue bars indicate sequences that contain regulatory elements based on prior functional evidence. Numbers on the horizontal axis indicate the position relative to the reference sequence for each gene (GenBank accession numbers AC069294 and AC005020 for CYP3A4 and CYP3A5, respectively).
Figure A2.
Decay of pairwise LD with distance, as measured by |D′| (blue symbols) and r2 (red symbols) across three human population samples. The blackened circles represent the average values of |D′| (blue) and r2 (red) in 10-kb windows.
Table A1.
Frequency of the CYP3A5*3 and AGT Met235 Alleles and Geographic Information from the CEPH-HGDP Panel
|
CYP3A5*3 |
AGT Met235 |
||||
| Population | Frequency | Na | Frequency | Na | Latitude |
| Nigeria (Yoruba) | .06 | 25 | .12 | 20 | 8°N |
| Congo (Mbuti Pygmies) | .07 | 15 | .04 | 13 | 1°N |
| Namibia (San) | .07 | 7 | 0 | 7 | 21°S |
| Central African Republic (Biaka Pygmies) | .11 | 33 | .03 | 31 | 4°N |
| Kenya (Bantu N.E.) | .17 | 12 | .15 | 10 | 3°S |
| S. Africa (Bantu S.E./S.W.) | .19 | 8 | .07 | 7 | 28°S |
| Senegal (Mandenka) | .31 | 24 | .04 | 23 | 12°N |
| Mexico (Pima) | .46 | 25 | .10 | 25 | 29°N |
| China (Dai) | .55 | 10 | .10 | 10 | 21°N |
| China (She) | .55 | 10 | .20 | 10 | 27°N |
| China (Miaozu) | .65 | 10 | .15 | 10 | 28°N |
| China (Tujia) | .65 | 10 | .15 | 10 | 29°N |
| China (Mongolia) | .65 | 10 | .20 | 10 | 48°N |
| Mexico (Maya) | .71 | 24 | .08 | 25 | 19°N |
| China (Naxi) | .72 | 9 | .15 | 10 | 26°N |
| Cambodia | .73 | 11 | .14 | 11 | 12°N |
| Pakistan (Hazara) | .75 | 25 | .27 | 24 | 34°N |
| China (Lahu) | .75 | 10 | .05 | 10 | 22°N |
| China (Han) | .75 | 44 | .16 | 44 | 32°N |
| Pakistan (Kalash) | .76 | 25 | .30 | 25 | 36°N |
| Brazil (Karitiana) | .77 | 24 | 0 | 24 | 10°S |
| Japan | .77 | 31 | .27 | 30 | 38°N |
| China (Xibo) | .78 | 9 | .33 | 9 | 43°N |
| Pakistan (Burusho) | .78 | 25 | .23 | 24 | 36°N |
| New Guinea (Papuan) | .79 | 17 | .09 | 17 | 4°S |
| Pakistan (Balochi) | .80 | 25 | .43 | 21 | 31°N |
| China (Yizu) | .80 | 10 | .25 | 10 | 28°N |
| Melanesian (Bougainville) | .82 | 22 | .05 | 22 | 6°S |
| Pakistan (Sindhi) | .82 | 25 | .16 | 25 | 25°N |
| Italy (Bergamo) | .82 | 14 | .36 | 14 | 46°N |
| Israel (Palestinian) | .82 | 51 | .46 | 50 | 32°N |
| Israel (Bedouin) | .83 | 48 | .44 | 48 | 31°N |
| Brazil (Surui) | .83 | 21 | 0 | 21 | 11°S |
| Orkney Islands (Orcadian) | .84 | 16 | .56 | 16 | 59°N |
| Algeria (Mozabite) | .84 | 29 | .40 | 26 | 32°N |
| Colombia | .85 | 13 | .15 | 13 | 3°N |
| China (Hezhen) | .85 | 10 | .15 | 10 | 47°N |
| China (Daur) | .85 | 10 | .30 | 10 | 48°N |
| Pakistan (Makrani) | .86 | 25 | .26 | 25 | 26°N |
| Pakistan (Brahui) | .88 | 25 | .46 | 25 | 30°N |
| Pakistan (Pathan) | .88 | 25 | .25 | 24 | 33°N |
| Russia Caucasus (Adygei) | .88 | 17 | .59 | 17 | 44°N |
| China (Tu) | .90 | 10 | .20 | 10 | 36°N |
| China (Orogen) | .90 | 10 | .10 | 10 | 50°N |
| Siberia (Yakut) | .90 | 25 | .26 | 25 | 63°N |
| France | .91 | 29 | .66 | 28 | 46°N |
| Israel (Druze) | .92 | 48 | .44 | 48 | 32°N |
| Russian | .92 | 25 | .56 | 24 | 61°N |
| Italy (Tuscan) | .94 | 8 | .28 | 7 | 43°N |
| Italy (Sardinian) | .95 | 28 | .63 | 28 | 40°N |
| China (Uygur) | .95 | 10 | .60 | 10 | 44°N |
| France (Basque) | .96 | 24 | .68 | 20 | 43°N |
Number of genotyped individuals.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Cluster-Buster, http://zlab.bu.edu/cluster-buster/
- Coriell Cell Repositories, http://locus.umdnj.edu/ccr/
- Di Rienzo Lab Web site, http://genapps.uchicago.edu/labweb/pubs.html (for primer sequences, population sample information, and data)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for CYP3A4 [accession number AC069294] and CYP3A5 [accession number AC005020])
- Human Genome Diversity Panel–Centre d’Etude du Polymorphisme Humain (HGDP-CEPH), http://www.cephb.fr/HGDP-CEPH-Panel/
- MAXDIP, http://genapps.uchicago.edu/maxdip/index.html (to generate composite likelihood estimates of recombination rate based on LD data)
- MultiPipMaker, http://bio.cse.psu.edu/pipmaker
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CYP3A4, CYP3A5, CYP3A7, CYP3A43, AGT M235T)
- PharmGKB, http://www.pharmgkb.org/ (for primer sequences and population samples used in resequencing study [accession numbers PS203894 and PS203895])
- SLIDER, http://genapps.uchicago.edu/slider/index.html (for computing summary statistics of population genetic data)
- University of Washington–Fred Hutchinson Cancer Research Center, http://pga.gs.washington.edu/education.html (for SeattleSNPs, the National Heart Lung and Blood Institute's Program for Genomic Applications)
References
- Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD, Nickerson DA, Kruglyak L (2004) Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol 2:E286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD (2002) Interrogating a high-density SNP map for signatures of natural selection. Genome Res 12:1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersberger I, Metzler D, Schwarz C, Pääbo S (2002) Genomewide comparison of DNA sequences between humans and chimpanzees. Am J Hum Genet 70:1490–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI (2000) Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz AG, Katz FH, Jailer JW (1960) 6-β-hydroxy-cortisol: high levels in human urine in pregnancy and toxemia. Proc Soc Exp Biol Med 105:41–43 [DOI] [PubMed] [Google Scholar]
- Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Di Rienzo A (2001) Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet 69:831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Li MC, Weng Z (2003) Cluster-Buster: finding dense clusters of motifs in DNA sequences. Nucleic Acids Res 31:3666–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuen S, Fukuda T, Maune H, Ikenaga Y, Yamamoto I, Inaba T, Azuma J (2002) Novel detection assay by PCR-RFLP and frequency of the CYP3A5 SNPs, CYP3A5*3 and *6, in a Japanese population. Pharmacogenetics 12:331–334 [DOI] [PubMed] [Google Scholar]
- Ghosh SS, Basu AK, Ghosh S, Hagley R, Kramer L, Schuetz J, Grogan WM, Guzelian P, Watlington CO (1995) Renal and hepatic family 3A cytochromes P450 (CYP3A) in spontaneously hypertensive rats. Biochem Pharmacol 50:49–54 [DOI] [PubMed] [Google Scholar]
- Givens RC, Lin YS, Dowling AL, Thummel KE, Lamba JK, Schuetz EG, Stewart PW, Watkins PB (2003) CYP3A5 genotype predicts renal CYP3A activity and blood pressure in healthy adults. J Appl Physiol 95:1297–1300 [DOI] [PubMed] [Google Scholar]
- Gleibermann L (1973) Blood pressure and dietary salt in human populations. Ecol Food Nutr 2:143–156 [Google Scholar]
- Goodwin B, Hodgson E, Liddle C (1999) The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol 56:1329–1339 [DOI] [PubMed] [Google Scholar]
- Hamblin MT, Thompson EE, Di Rienzo A (2002) Complex signatures of natural selection at the Duffy blood group locus. Am J Hum Genet 70:369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG (1974) Estimation of linkage disequilibrium in randomly mating populations. Heredity 33:229–239 [DOI] [PubMed] [Google Scholar]
- Hudson RR (2001) Two-locus sampling distributions and their application. Genetics 159:1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Bailey K, Skarecky D, Kwiatowski J, Ayala FJ (1994) Evidence for positive selection in the superoxide dismutase (SOD) region of Drosophila melanogaster. Genetics 136:1329–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustert E, Haberl M, Burk O, Wolbold R, He YQ, Klein K, Nuessler AC, Neuhaus P, Klattig J, Eiselt R, Koch I, Zibat A, Brockmoller J, Halpert JR, Zanger UM, Wojnowski L (2001) The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 11:773–779 [DOI] [PubMed] [Google Scholar]
- Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E (2001) Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 27:383–391 [DOI] [PubMed] [Google Scholar]
- Lamba JK, Lin YS, Schuetz EG, Thummel KE (2002a) Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev 54:1271–1294 [DOI] [PubMed] [Google Scholar]
- Lamba JK, Lin YS, Thummel K, Daly A, Watkins PB, Strom S, Zhang J, Schuetz EG (2002b) Common allelic variants of cytochrome P4503A4 and their prevalence in different populations. Pharmacogenetics 12:121–132 [DOI] [PubMed] [Google Scholar]
- Lifton RP, Warnock D, Acton RT, Harman L, Lalouel JM (1993) High prevalence of hypertension-associated angiotensinogen variant T235 in African Americans. Clin Res 260A [Google Scholar]
- Luft FC (2001) Molecular genetics of salt-sensitivity and hypertension. Drug Metab Dispos 29:500–504 [PubMed] [Google Scholar]
- Matsumura K, Saito T, Takahashi Y, Ozeki T, Kiyotani K, Fujieda M, Yamazaki H, Kunitoh H, Kamataki T (2004) Identification of a novel polymorphic enhancer of the human CYP3A4 gene. Mol Pharmacol 65:326–334 [DOI] [PubMed] [Google Scholar]
- Nakajima T, Wooding S, Sakagami T, Emi M, Tokunaga K, Tamiya G, Ishigami T, Umemura S, Munkhbat B, Jin F, Guan-jun J, Hayasaka I, Ishida T, Saitou N, Pavelka K, Lalouel J-M, Jorde LB, Inoue I (2004) Natural selection and population history in the human angiotensinogen gene (AGT): 736 complete AGT sequences in chromosomes from around the world. Am J Hum Genet 74:898–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW (1997) Polymorphisms in drug-metabolizing enzymes: what is their clinical relevance and why do they exist? Am J Hum Genet 60:265–271 [PMC free article] [PubMed] [Google Scholar]
- Ozdemir V, Kalowa W, Tang BK, Paterson AD, Walker SE, Endrenyi L, Kashuba AD (2000) Evaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration method. Pharmacogenetics 10:373–388 [DOI] [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD (1998) Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 63:1839–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Rubin EM (2001) Genomic strategies to identify mammalian regulatory sequences. Nat Rev Genet 2:100–109 [DOI] [PubMed] [Google Scholar]
- Przeworski M (2002) The signature of positive selection at randomly chosen loci. Genetics 160:1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe B (2004) Shotgun library construction for DNA sequencing in methods in molecular biology. In: Zhao S, Stodolski M (eds) Bacterial artificial chromosomes, volume 1: library construction, physical mapping, and sequencing, Vol 255: Methods in molecular biology. Humana Press, Totowa, NJ, pp 171–187 [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Li LM, Ward R, Pritchard JK (2003) Informativeness of genetic markers for inference of ancestry. Am J Hum Genet 73:1402–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science 298:2381–2385 [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R (1995) DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput Appl Biosci 11:621–625 [DOI] [PubMed] [Google Scholar]
- Russ AP, Maerz W, Ruzicka V, Stein U, Gross W (1993) Rapid detection of the hypertension-associated Met235→Thr allele of the human angiotensinogen gene. Hum Mol Genet 2:609–610 [DOI] [PubMed] [Google Scholar]
- Schenkman JB, Thummel KE, Favreau LV (1989) Physiological and pathophysiological alterations in rat hepatic cytochrome P-450. Drug Metab Rev 20:557–584 [DOI] [PubMed] [Google Scholar]
- Schuetz EG, Schuetz JD, Grogan WM, Naray-Fejes-Toth A, Fejes-Toth G, Raucy J, Guzelian P, Gionela K, Watlington CO (1992) Expression of cytochrome P450 3A in amphibian, rat, and human kidney. Arch Biochem Biophys 294:206–214 [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, Prasad AB, Summers TJ, Lee-Lin SQ, Maduro VV, Idol JR, Ryan JF, Thomas PJ, McDowell JC, Green ED (2002) Parallel construction of orthologous sequence-ready clone contig maps in multiple species. Genome Res 12:1277–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Frisse LA, Hudson RR, Di Rienzo A (2003) Comparative linkage-disequilibrium analysis of the β-globin hotspot in primates. Am J Hum Genet 73:1330–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward K, Hata A, Jeunemaitre X, Helin C, Nelson L, Namikawa C, Farrington PF, Ogasawara M, Suzumori K, Tomoda S, Berrebi S, Sasaki M, Corvol P, Lifton RP, Lalouel JM (1993) A molecular variant of angiotensinogen associated with preeclampsia. Nat Genet 4:59–61 [DOI] [PubMed] [Google Scholar]
- Watlington CO, Kramer LB, Schuetz EG, Zilai J, Grogan WM, Guzelian P, Gizek F, Schoolwerth AC (1992) Corticosterone 6 β-hydroxylation correlates with blood pressure in spontaneously hypertensive rats. Am J Physiol 262:F927–931 [DOI] [PubMed] [Google Scholar]
- Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276 [DOI] [PubMed] [Google Scholar]
- Wu SJ, Chiang FT, Chen WJ, Liu PH, Hsu KL, Hwang JJ, Lai LP, Lin JL, Tseng CD, Tseng YZ (2004) Three single-nucleotide polymorphisms of the angiotensinogen gene and susceptibility to hypertension: single locus genotype vs. haplotype analysis. Physiol Genomics 17:79–86 [DOI] [PubMed] [Google Scholar]