Abstract
We have inactivated transcription factor TFIID subunit TBP-associated factor 4 (TAF4) in mouse embryonic fibroblasts. Mutant taf4−/− cells are viable and contain intact TFIID comprising the related TAF4b showing that TAF4 is not an essential protein. TAF4 inactivation deregulates more than 1000 genes indicating that TFIID complexes containing TAF4 and TAF4b have distinct target gene specificities. However, taf4−/− cell lines have altered morphology and exhibit serum-independent autocrine growth correlated with the induced expression of several secreted mitotic factors and activators of the transforming growth factor β signalling pathway. In addition to TAF4 inactivation, many of these genes can also be induced by overexpression of TAF4b. A competitive equilibrium between TAF4 and TAF4b therefore regulates expression of genes controlling cell proliferation. We have further identified a set of genes that are regulated both by TAF4 and upon adaptation to serum starvation and which may be important downstream mediators of serum-independent growth. Our study also shows that TAF4 is an essential cofactor for activation by the retinoic acid receptor and CREB, but not for Sp1 and the vitamin D3 receptor.
Keywords: Affymetrix, apoptosis, fibrosis, homologous recombination, TGFβ
Introduction
General transcription factor TFIID is composed of the TATA-binding protein (TBP) and 13–14 evolutionarily conserved TBP-associated factors (TAFs) (Tora, 2002). Study of temperature-sensitive (TS) TAF mutations indicates that up to 80% of yeast genes require one or several TAFs for their expression (Shen et al, 2003). In vertebrate cells, targeted inactivation of TAF9 in chicken DT40 cells or of TAF10 in mouse embryonal carcinoma cells leads to cell cycle arrest and apoptosis (Metzger et al, 1999; Chen and Manley, 2000). Similarly, inactivation of TAF10 or TAF8 (Taube nuss) in mice leads to early embryonic lethality characterised by apoptosis of the blastocyst inner cell mass (Voss et al, 2000; Mohan et al, 2003). Collectively, these results indicate that TAFs are required for viability and cell cycle progression.
TAF4 is an evolutionarily conserved, ubiquitously expressed TFIID subunit that is also present in TFTC (Tanese et al, 1996; Mengus et al, 1997; Brand et al, 1999; Sanders and Weil, 2000; Thuault et al, 2002). TAF4 is essential in yeast and plays a general role in transcription in Caenorhabditis elegans embryos (Sanders and Weil, 2000; Walker et al, 2001). The TAF4 family also comprises TAF4b, preferentially expressed in the testis, ovary and lymphoid B cells (Dikstein et al, 1996; Freiman et al, 2001). Targeted inactivation of the taf4b gene in mice leads to female sterility due to a block in oocyte maturation and to male sterility due to defective spermatogenesis (Falender et al, 2005).
In vitro studies and transient expression assays have suggested that TAF4 is a coactivator for the Sp1, CREB and RAR transcription factors (Gill et al, 1994; Mengus et al, 1997; Saluja et al, 1998; Gangloff et al, 2000; Asahara et al, 2001); however, the in vivo function of TAF4 is presently unknown. We have inactivated TAF4 in murine cells and show that it has a major and unexpected function in negatively regulating cell proliferation in addition to signalling by the retinoid and cAMP pathways.
Results
Inactivation of the taf4 gene
The TAF4 CR-II region contains a histone fold domain (HFD), required for heterodimerisation with TAF12, where the α3 helix is separated from the α2 helix by an extended conserved linker region (Gangloff et al, 2000; Werten et al, 2002; Figure 1A).
Figure 1.
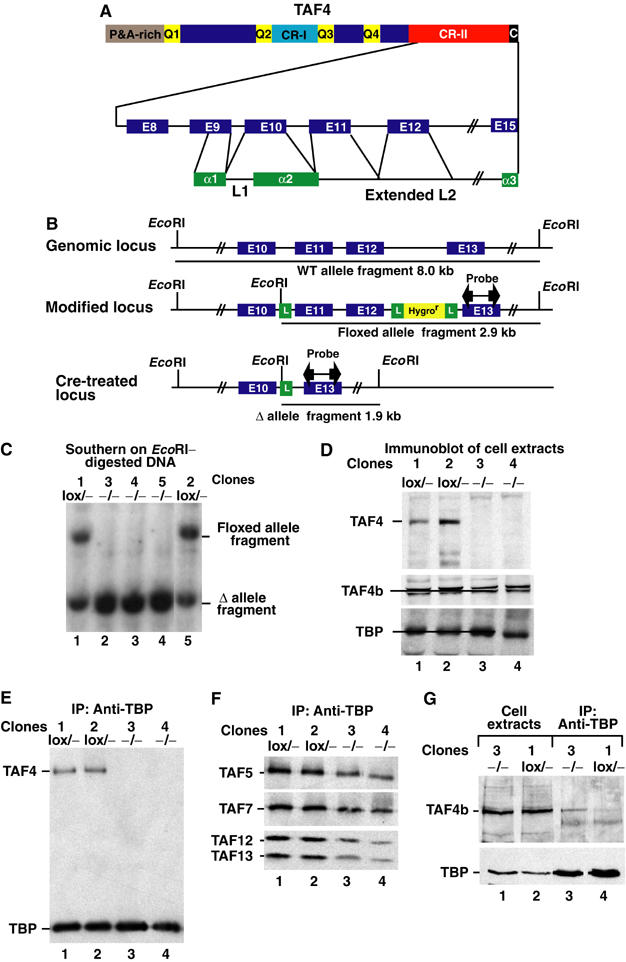
Structure of TAF4. (A) The TAF4 protein is schematised showing the proline–alanine-rich (P&A) and glutamine-rich (Q1–Q4) regions and conserved regions I and II (CR-I and CR-II). Lower part: The intron/exon (E) structure of the CR-II region is schematised in relation to the HFD. (B) The native and modified taf4 alleles are schematised. The exons are indicated along with EcoRI sites, loxP sites (L) and the hygromycin resistance cassette. The location of the probe used in Southern blots is shown along with the EcoRI fragments generated from each allele. (C) Southern blots used to genotype fibroblast clones. The genotype of each clone is shown above the lane. (D) Immunoblots of cell extracts from the clones indicated above each lane were probed to reveal TAF4, TAF4b and TBP. A 20 μg portion of total cell extract was loaded in each lane. On some gels, TAF4b resolves into two closely migrating species. (E, F) Immunoblots of anti-TBP immunoprecipitations to reveal the indicated TAFs. The filter was first probed to reveal TBP and TAF4 and then reprobed sequentially to reveal the indicated TAFs. The clones and their genotypes are shown above each lane. (G) The indicated cell extracts were precipitated with antibody against TBP and probed for the presence of TBP and TAF4b.
A loxP site was inserted upstream of exon 11 and a hygromycin resistance cassette flanked by loxP sites downstream of exon 12 (Figure 1B). Cre recombinase-mediated deletion of exons 11 and 12 would remove amino acids within the conserved linker region putting the downstream exons (13–15) out of frame. After homologous recombination in embryonic stem (ES) cells, mice were generated carrying this allele (taf4lox/+). The ES cells were also transfected with a vector expressing Cre recombinase and ES cell clones with a deleted allele were used to create mice (taf4−/+).
Breeding taf4−/+ mice failed to yield any taf4−/− offspring, but taf4−/− embryos could be detected until E9.5 (data not shown). This suggests that TAF4 is not essential for cell viability. To test this possibility, taf4lox/− mice were bred and used to generate an embryonic fibroblast cell line. These cells were transfected with a vector expressing Cre recombinase and we isolated taf4lox/− lines where no recombination had taken place (Figure 1C, clones 1 and 2) and taf4−/− lines where the floxed allele had been deleted (Figure 1C, clones 3–5).
Immunoblotting with an antibody directed against an N-terminal epitope indicated that no full-length or truncated TAF4 was seen in the taf4−/− cells (Figure 1D). In contrast, equivalent amounts of TBP were present in both extracts. These results indicate that no stable protein is made from the deleted taf4 alleles, but the resultant taf4−/− cells are viable.
Extracts were then precipitated with the TBP antibody. TAF4 was co-precipitated with TBP from the taf4lox/− cell extracts, but no TAF4 was seen in the immunoprecipitates from taf4−/− extracts (Figure 1E). To investigate TFIID integrity in taf4−/− cells, the immunoprecipitates were probed for other TFIID subunits. TAF5, TAF6, TAF7, TAF13, and TAF12, the TAF4 heterodimerisation partner, were all co-precipitated with TBP in extracts from both cell types (Figure 1F and data not shown). Only a small reduction in the amount of the TAFs relative to TBP could be observed in the immunoprecipitates from the taf4−/− cells. Loss of TAF4 does not therefore disrupt the TFIID complex.
One possible explanation for the integrity of TFIID and taf4−/− cell viability would be the presence of TAF4b, which also heterodimerises with TAF12 (Gangloff et al, 2000). Immunoblotting indicates that TAF4b is expressed at comparable levels in both taf4−/− and taf4lox/− cells (Figure 1D and G). TAF4b immunoprecipitates with TBP from extracts of the taf4−/− cells, while lower levels are observed in the immunoprecipitates from the taf4lox/− cells (Figure 1G, lanes 3 and 4). Hence, TAF4b may compensate the loss of TAF4 to maintain TFIID integrity and viability in taf4−/− cells.
Loss of TAF4 leads to serum-independent autocrine growth
Compared to the rounder and flatter taf4lox/− cells, taf4−/− cells are more elongated (Supplementary Figure 1). This is clearly seen at high density where the taf4lox/− cells form a monolayer of regular round cells, whereas the taf4−/− cells are much more irregular, elongated and densely packed (Figure 2A). Although each cell type grows at comparable rates in serum-rich media, the taf4−/− cells grow to a higher density (Figure 2B). In low serum, the taf4lox/− cells adopt a flattened morphology typical of quiescent cells, whereas the taf4−/− cells remain elongated (Figure 2C and Supplementary Figure 2) and grow more rapidly (Figure 2C and D).
Figure 2.
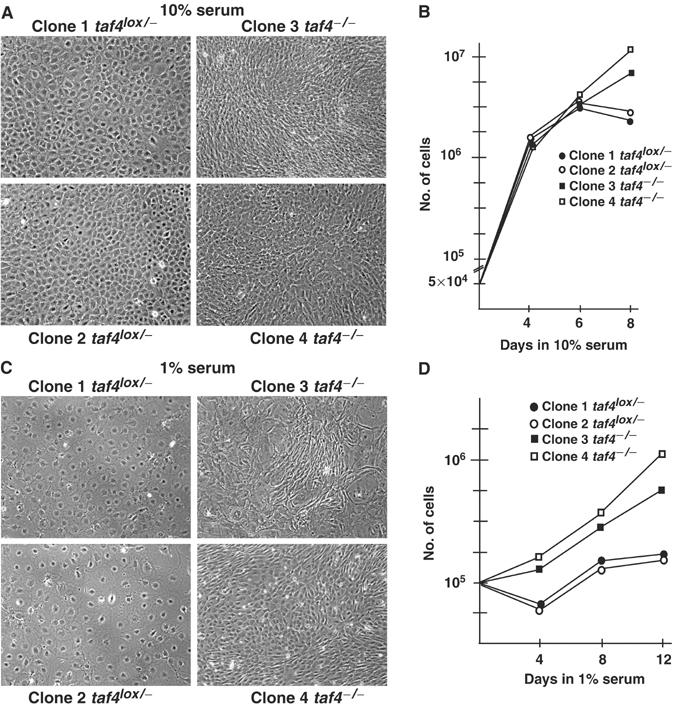
Growth of taf4−/− fibroblasts. (A) Phase-contrast images of taf4−/− and taf4lox/− monolayers grown in 10% serum. (B) Comparison of the growth of taf4−/− and taf4lox/− cells in 10% serum. A total of 5 × 104 cells were seeded in 10 cm plates on day 0. At each indicated day, a plate was trypsinised and the cells counted. The average values of three experiments are shown. (C) Phase-contrast images of the indicated clones grown for 4 days in 1% serum. (D) Growth of taf4−/− and taf4lox/− cells in 1% serum. The experiment was performed as described in panel B. In this case, 105 cells were seeded on day 0 and grown in 1% serum.
In the absence of serum, taf4lox/− cells stop proliferating and rapidly undergo apoptotic death (Figure 3A and B). In contrast, taf4−/− cells survive almost indefinitely in the absence of serum. Although some apoptotic cells are observed (visible as refractory cells in Figure 3A), this is offset by the presence of proliferating cells expressing the characteristic Ki67 marker, whereas no proliferating taf4lox/− cells are observed (Figure 3D).
Figure 3.
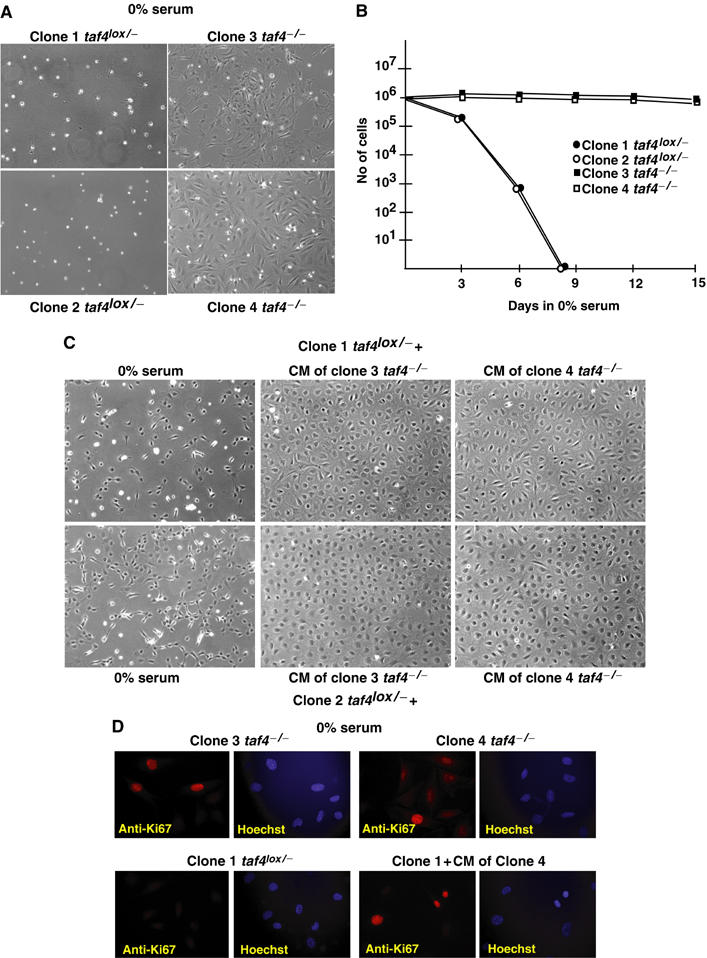
Growth of taf4−/− cells in the absence of serum. (A) Phase-contrast images of the clones after 4 days in 0% serum. (B) Comparison of the growth of taf4−/− and taf4lox/− cells in the absence of serum. A total of 106 cells were seeded in 10 cm plates on day 0, and counted at the indicated days. (C) A total of 106 taf4lox/− cells were seeded in either serum-free medium (upper and lower left panels) or serum-free conditioned media (CM) in which the indicated taf4−/− cells had been grown for 2 days (centre and right panels). Images were taken 2 days after seeding. (D) Immunofluorescence to detect proliferation marker Ki67. In each case, the immunostaining and Hoechst-stained DNA are shown side by side.
The above results suggest that the taf4−/− cells secrete a growth factor(s) to assure their survival. To test this, taf4−/− cells were grown at high density for 2 days in serum-free medium. In comparison to the serum-free medium, the conditioned media clearly supported survival (Figure 3C) and proliferation of the taf4lox/− cells as evidenced by the induction of Ki67 expression (Figure 3D). Hence, loss of TAF4 leads to secretion of mitotic factors that support autocrine growth and also the survival and growth of taf4lox/− cells.
The conserved TAF4 CR-II domain is necessary and sufficient to regulate cell growth
To demonstrate that loss of TAF4 is responsible for the phenotypic changes, the taf4−/− cells were stably transfected with vectors expressing either the full-length human TAF4 or deletion mutants containing the CR-II and flanking region or the CR-II alone (Figure 4A and B, lanes 1–9). Extracts from these clones were precipitated with the anti-TBP antibody showing that each TAF4 derivative was co-precipitated with TBP (Figure 4B, lanes 10–15, and data not shown). This confirms that the CR-II region suffices for integration into TFIID in vivo.
Figure 4.
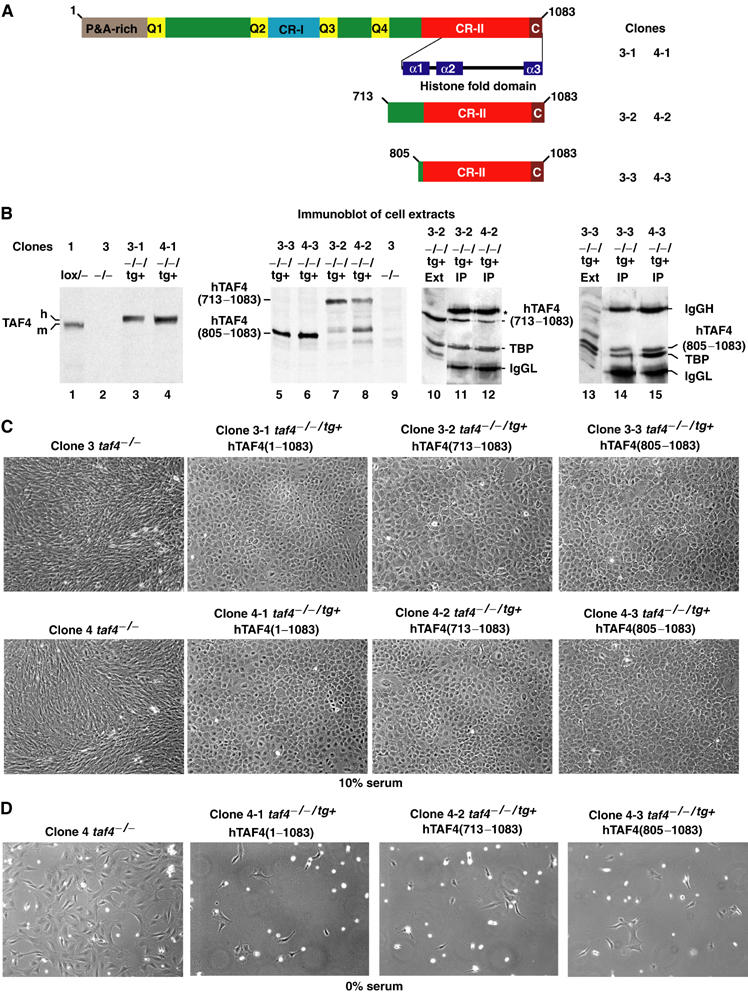
Re-expression of TAF4 suppresses serum-independent growth. (A) Schematic structure of TAF4 showing the domains described in Figure 1. The deletion mutants are indicated along with the nomenclature of the resulting clones. (B) Immunoblots (lanes 1–9) of the cell extracts indicated above each lane. In lanes 10–15, the indicated cell extracts were precipitated with antibody against TBP and the precipitated proteins revealed with antibody against TBP and TAF4 (20TA). The first lane in each panel shows one of the starting extracts as reference. The immunoglobulin heavy (*) and light chains of the precipitating antibodies are also indicated. (C) Morphology of revertant clones. Phase-contrast images of confluent monolayers of the indicated cell types grown in 10% serum. (D) Re-expression of TAF4 abrogates serum independence. A total of 106 cells from the different clones were seeded in serum-free media. Images were acquired after 4 days incubation.
Strikingly, taf4−/− cells re-expressing full-length TAF4 revert to a morphology resembling that of taf4lox/− cells and distinct from the parental taf4−/− cells (see Figures 4C and 2A). In addition, these cells lose their serum independence (Figure 4D). Similar observations were made with the clones expressing each of the deletion mutants (see relevant panels in Figure 4C and D). Loss of endogenous TAF4 can hence be compensated by exogenous TAF4 expression, which represses autocrine growth. Moreover, this repression requires only the presence of the CR-II domain in the TFIID complex.
TAF4 inactivation affects a large and diverse set of genes
To make a comprehensive evaluation of TAF4 inactivation on gene expression, RNA was prepared from exponentially growing taf4lox/− and taf4−/− clones in 10% serum and used for Affymetrix gene profiling. We searched for genes that were reproducibly up- or downregulated in duplicate samples of each taf4lox/− and taf4−/− clone. The expression of several of these genes was also verified by reverse transcription (RT)–PCR. Analysis of the expression of more than 14 500 genes indicated that loss of TAF4 resulted in a greater than two-fold upregulation of 479 genes (Supplementary Figure 3; full data can be found at http://igbmc.fr/recherche/Dcp_Trans/Eq_IDavi/Publi/Paper1.html). Of these, over a 100 were upregulated more than four-fold. In contrast, 775 genes were downregulated, 280 of which showed more than a four-fold repression (Supplementary Figure 3).
The affected genes did not belong to a specific pathway, but to many different aspects of cell metabolism (Supplementary Figure 4). Among the most affected are genes that modulate cell–matrix interactions and cell–cell adhesion and communication (for example, integrin α6 (ITGα6), cadherin 11 (CDH11), fibromodulin (FMOD), lumicam (LUM) and decorin (DCN); Figure 5A and B) or cytoskeleton dynamics and cell shape (the oncogene VAV 3). Their differential regulation likely contributes to the changes in cell morphology seen upon loss of TAF4.
Figure 5.
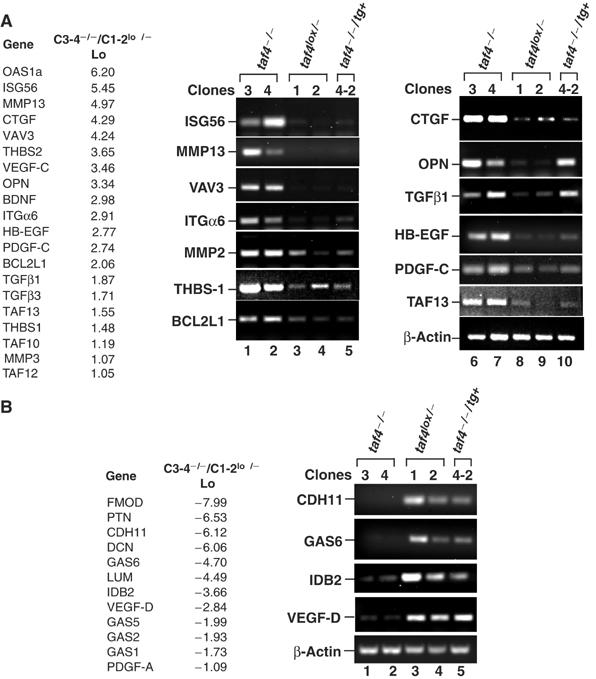
Changes in gene expression in taf4−/− cells. (A, B) A short list of some relevant up- and downregulated genes. The full list is in Supplementary Figure 3. The log 2 values of induction and repression are shown. RT–PCR was performed on the indicated genes using RNA from the cells indicated above each lane. These RNA preparations are independent of those used in the profiling analysis and therefore provide a further confirmation of the results.
Among the most strongly upregulated are a series of 17 interferon-inducible genes, for example 2′–5′ oligoadenylate synthetase 1A (OAS1a) and ISG56 (Ifit1) (Figure 5A and B, and Supplementary Figure 3). However, the interferons themselves are not expressed. The mechanism by which these genes are activated therefore remains to be determined. Loss of TAF4 also upregulates the expression of other TFIID subunits, TAF10, TAF12 and TAF13, and the antiapoptotic gene BCL2L1. In contrast, several growth arrest specific (Gas) genes are downregulated upon loss of TAF4 (Figure 5A and B, and Supplementary Figure 3).
Loss of TAF4 activates components of the TGFβ signalling pathway
Several secreted mitotic factors are among the affected genes. Connective tissue growth factor (CTGF) (Figure 5A), a well-characterised secreted growth factor capable of supporting autocrine growth (Brigstock, 2003), is upregulated. CTGF is a major mitogen in fibroblasts induced by transforming growth factor β (TGFβ) that acts as a secondary growth factor in TGFβ-stimulated cell proliferation (Leask and Abraham, 2004). Loss of TAF4 additionally upregulated both TGFβ1 and TGFβ3 (Figure 5A and Supplementary Figure 3). We also observed upregulation of MMP13, MMP3 and MMP2 (Figure 5A and Supplementary Figure 3), matrix serine proteases of which MMP2 is known to cleave and activate the latent form of TGFβ1 (Yu and Stamenkovic, 2000; Annes et al, 2003). Similarly, thrombospondin 1 and 2 (THBS), two further activators of latent TGFβ1 (Murphy-Ullrich and Poczatek, 2000), were also strongly induced. TAF4 inactivation thus induces TGFβ1 and TGFβ3, as well as MMP2, THBS1, THBS2 and CTGF all of which potentiate their function.
This observation suggests that the taf4−/− cells secrete active TGFβ. To test this, 3T3 cells harbouring an integrated luciferase gene under the control of SMAD-binding sites (Inman et al, 2002) were used. In serum-free medium, low luciferase levels were observed (Figure 6A, lanes 1 and 5), which were strongly stimulated by addition of recombinant TGFβ1 (lanes 2 and 6) or conditioned taf4−/− cell media (lanes 3, 4 and 7, 8) showing that they contain high levels of active TGFβ. This result was confirmed by the observation that the conditioned media induced phosphorylation of SMAD2/3 in taf4−/− and taf4lox/− cells as well as in 3T3 and HeLa cells (Figure 6B). Constitutive SMAD2/3 phosphorylation was observed in taf4−/− cells grown in the absence of serum, but not in the other cell types (Figure 6B, lane 1). It is also noteworthy that SMAD2/3 phosphorylation could be induced by a short exposure to serum and is constitutive in 10% serum in the taf4lox/− and taf4−/− cells, but not in 3T3 or HeLa cells where it was induced only by recombinant TGFβ or the conditioned media. Moreover, serum-independent growth of the taf4−/− cells could be blocked using SB431542, an inhibitor of the TGFβ-activated ALK5 protein kinase (Laping et al, 2002) (Figure 6E). Similarly, the addition of TGFβ1 and/or TGFβ3 to serum-free medium was sufficient to induce survival of taf4lox/− cells (Figure 6F). Together, these results indicate that the taf4−/− cells secrete active TGFβ required for their serum-independent growth.
Figure 6.
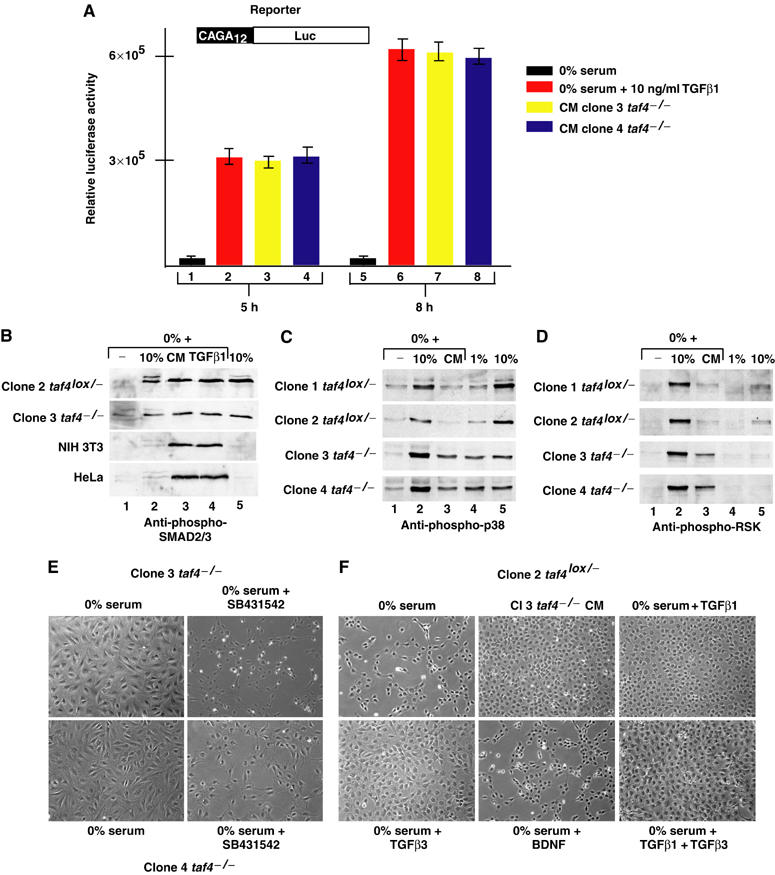
Activation of SMAD and p38 signalling pathways. (A) 3T3 cells harbouring the schematised integrated reporter were incubated in serum-free media with or without recombinant TGFβ or in conditioned media from the taf4−/− clones for 5 or 8 h as indicated before performing luciferase assays. Relative luciferase activity (arbitrary units) is shown on the Y-axis. (B) Conditioned media induce phosphorylation of SMAD3/3. In lanes 2–4, each cell line was grown overnight in serum-free medium before a 60 min exposure to either 10% serum, conditioned media from the clone 3 taf4−/− cells or recombinant TGFβ (10 ng/ml). In lanes 1 and 5, cells were grown for 24 h in 0 or 10% serum, respectively. After treatment, cells were immediately lysed in Laemmli loading buffer and analysed by SDS–PAGE and staining with Coomassie brilliant blue. Equivalent amounts of cell extract were then separated by SDS–PAGE and probed with antibody recognising SMAD2/3 phosphorylated at S465/467. (C, D) Cells were grown in the absence of serum and stimulated for 15 min prior to lysis with serum or conditioned media as indicated. Immunoblots were performed using antibodies recognising p38 phosphorylated at Thr180/Tyr182 or the RSK1–3 family phosphorylated at Thr577. (E) Phase-contrast images (× 20 magnification) of taf4−/− cells after 4 days growth in the absence of serum with or without the ALK5 inhibitor SB431542 (20 μM). (F) Phase-contrast images of taf4lox/− cells after 3 days growth in the absence of serum with conditioned media, TGFβ1, β3 (10 ng/ml) or BDNF (100 ng/ml) as indicated.
TAF4 inactivation additionally induces expression of osteopontin (OPN, or secreted phosphoprotein 1, SPP1), a growth and antiapoptotic factor found upregulated in many transformed cells, heparin-binding epidermal growth factor (HB-EGF, or DTR), platelet-derived growth factor C (PDGF-C) and vascular endothelial growth factor C (VEGF-C) (Figure 5A and Supplementary Figure 3). Paradoxically however, expression of pleiotrophin (PTN), another heparin-binding growth factor, PDGF-A and VEGF-D (Fos-induced growth factor, FIGF) were strongly repressed (Figure 5B and Supplementary Figure 3). Thus, loss of TAF4 affects both the nature and the balance of growth factor expression, suggesting that autocrine growth may involve cooperation between TGFβ and other mitogens.
While the conditioned media can activate the SMAD pathway, we verified if the presence of other mitogens could activate Map kinase pathways. Strong induction of phopho-p38 can be observed in taf4−/− cells after exposure to either 10% serum or conditioned medium from taf4−/− cells (Figure 6C, lanes 2 and 3). Induction of p38 phosphorylation by both serum and conditioned media was less efficient in taf4lox/− cells, perhaps because their viability is compromised even by short serum starvation. Similarly, comparable high levels of p38 phosphorylation are observed in taf4−/− cells grown in either 10 or 1% serum (lanes 4 and 5), whereas phosphorylation is serum dependent in taf4lox/− cells. Analogous results were obtained with antibodies against phospho-RSK1–3 (Merienne et al, 2000; Figure 6D, lanes 1–3), although little constitutive phosphorylation was seen (lanes 4 and 5).
The CR-II domain of TAF4 restores normal gene expression
We examined gene expression in the taf4−/−/tg+ cells expressing the CR-II domain that is minimally required for phenotypic reversal. The presence of the CR-II domain restored normal expression of the majority of the affected genes. For example, CTGF, MMP2 and THBS1 were repressed, while CDH11 and GAS 6 were induced (Figure 5A and B). Thus, the CR-II domain corrects the expression of most of the deregulated genes in agreement with the observed phenotypic reversion. Similar results were seen with the full-length TAF4 (data not shown). In contrast, neither the CR-II domain nor the full-length TAF4 fully repressed TGFβ1, TGFβ3 or OPN expression (Figure 5A and Figure 7B, lanes 7 and 14, and data not shown). Upregulation of TGFβ by itself is not therefore sufficient for autocrine growth, but expression of potentiating factors, such as CTGF, MMP2 and THBS1, and/or HB-EGF are critical.
Figure 7.
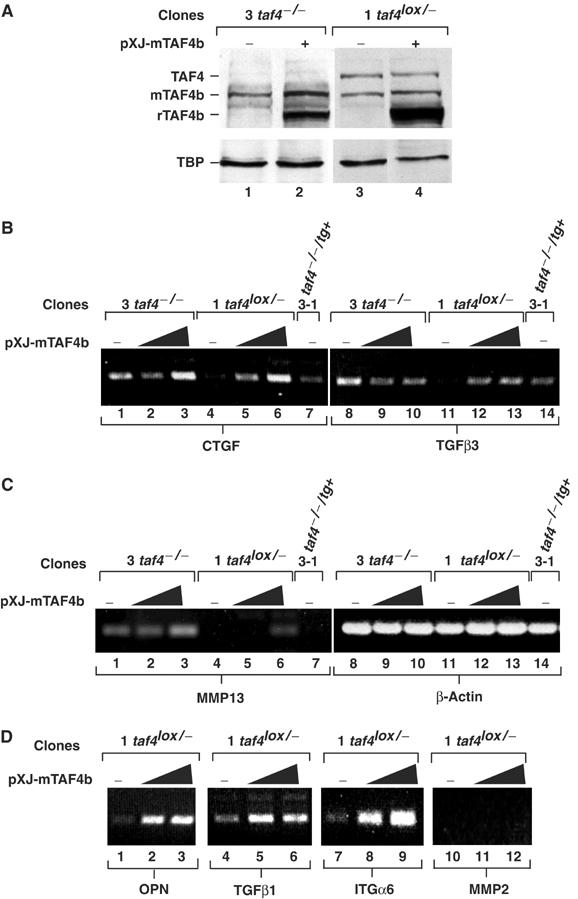
Overexpression of TAF4b induces gene expression. (A) Indicated clones were transfected with 4 μg of the pXJ-TAF4b expression vector or with empty vector (+ and −, respectively). After 48 h, protein extracts were probed with antibodies against TAF4b, TAF4 and TBP. Endogenous TBP, TAF4 and TAF4b are shown along with the transfected recombinant (r)TAF4b. (B–D) The indicated clones were transfected with 2 or 4 μg of the pXJ-TAF4b expression vector or with empty vector (−). RT–PCR was performed for the indicated genes 48 h after transfection.
TAF4b induces growth factor expression
The changes in gene expression reflect the loss of TAF4 and hence the activity of the residual TFIID complex containing TAF4b showing that it has a different target gene specificity. To test whether this situation may be mimicked by overexpression of TAF4b even in the presence of TAF4, we transfected TAF4b in the taf4lox/− cells to ask if genes that were upregulated in the absence of TAF4 could also be activated by TAF4b overexpression (Figure 7A).
Transfection of TAF4b in taf4−/− cells has little effect on CTGF or TGFβ3 expression (Figure 7B, lanes 1–3 and 8–10). In contrast, overexpression of TAF4b in taf4lox/− cells results in a potent induction of TGFβ1, TGFβ3, CTGF, ITGα6, OPN or HB-EGF (Figure 7B, lanes 4–6 and 11–13, and Figure 7D, and data not shown). The expression of these genes is therefore controlled by the relative levels of TAF4 and TAF4b. These results are consistent with the idea that TAF4 controls their expression by limiting the amount of TFIID containing TAF4b. In contrast, little induction of MMP13 and MMP2 expression was observed (Figure 7C, lanes 4–6, and Figure 7D, lanes 10–12). It is possible that full induction of these genes is a secondary event regulated by the induced growth factors and is not seen during the shorter course of the transfection experiments.
Genes involved in fibroblast survival under low-serum conditions
TAF4 inactivation induces several secreted mitogens to support autrocrine growth, but many other factors that may act as downstream mediators are also deregulated. To highlight potential candidates, we used Affymetrix gene profiling to examine gene expression in taf4−/− cells grown in 0% serum and in taf4lox/− cells grown in 1% serum. This will identify genes that may have a particular relevance for growth and survival under limiting serum conditions that can then be compared with those deregulated upon loss of TAF4.
After 3 days in 0% serum, a greater than two-fold upregulation of 182 genes and downregulation of 117 genes is seen in the taf4−/− cells (Figure 8A and Supplementary Figure 5). In an analogous experiment in 1% serum, upregulation of 144 genes and repression of 213 genes was observed in the taf4lox/− cells (Figure 8B and Supplementary Figure 5b). By comparison with the TAF4-regulated genes, we identified eight genes that were upregulated upon loss of TAF4 and further induced in 0% serum. Three of these were also upregulated in the taf4lox/− cells in 1% serum, further strengthening the notion that they are involved in adaptation to low serum. Among, these eight genes are the ETS1 oncogene and THBS1. Moreover, the three genes that are upregulated under all conditions comprise BCL6 and Kruppel-like factor 5 (KLF5). The third gene is brain-derived neurotrophic factor (BDNF), characterised for its role in neuron survival and synapse plasticity. However, despite the observed upregulation, addition of recombinant BDNF to serum-free medium did not mediate survival of the taf4lox/− cells (Figure 6E).
Figure 8.
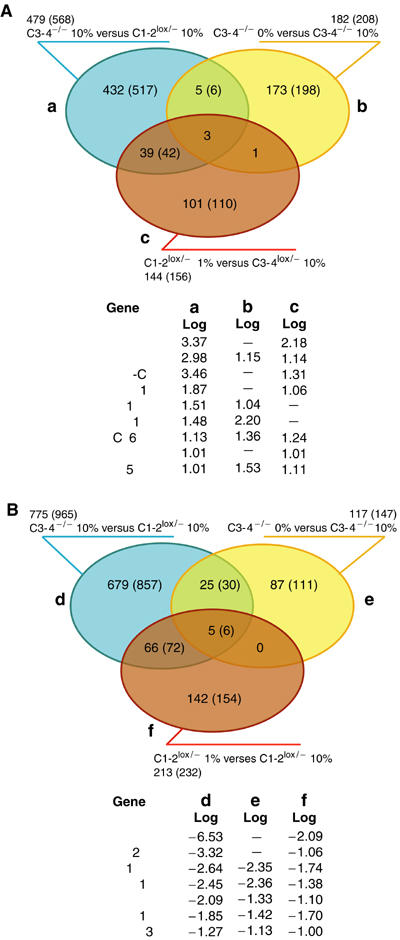
Comparison of TAF4- and serum-regulated genes in fibroblasts. (A, B) The genes are divided into groups, (a–c) upregulated, (d–f) downregulated as defined, and represented as a coloured ellipse. The number of individual genes (probe sets) in each group is shown outside the ellipse. Coregulated genes are indicated in the overlapping areas. The identities of the genes regulated under all conditions are shown below along with the signal log value in each group. Full lists are in Supplementary Figures 3, 5 and 6.
In addition to the above, 39 genes upregulated in taf4−/− cells were also induced in the taf4lox/− cells in low serum including TGFβ1, PDGF-C, and MAFF and MAFK, two oncogenic leucine zipper-containing transcription factors (Figure 8A and Supplementary Figure 5). Again the coregulation of these genes suggests a role in adaptation to low-serum conditions.
In an analogous comparison, we identified 66 genes downregulated in the taf4−/− cells in 10% serum and in taf4lox/− cells in low serum. Furthermore, 25 genes downregulated upon loss of TAF4 were further repressed in 0% serum (Figure 8B and Supplementary Figure 6). In this case, five genes were downregulated under all conditions.
TAF4 is a critical cofactor for activation by CREB and the retinoic acid receptor
TAF4 has been reported to interact with the transcriptional activator Sp1 and function as a potential coactivator. We tested the requirement for TAF4 as a cofactor for activation by Sp1 by transfecting each cell line with a GAL4-Sp1B expression vector and a GAL4-responsive reporter gene. GAL4-Sp1B activates transcription to comparable extents in all cell lines tested, indicating that TAF4 is not an essential Sp1 cofactor in this context (Figure 9A).
Figure 9.
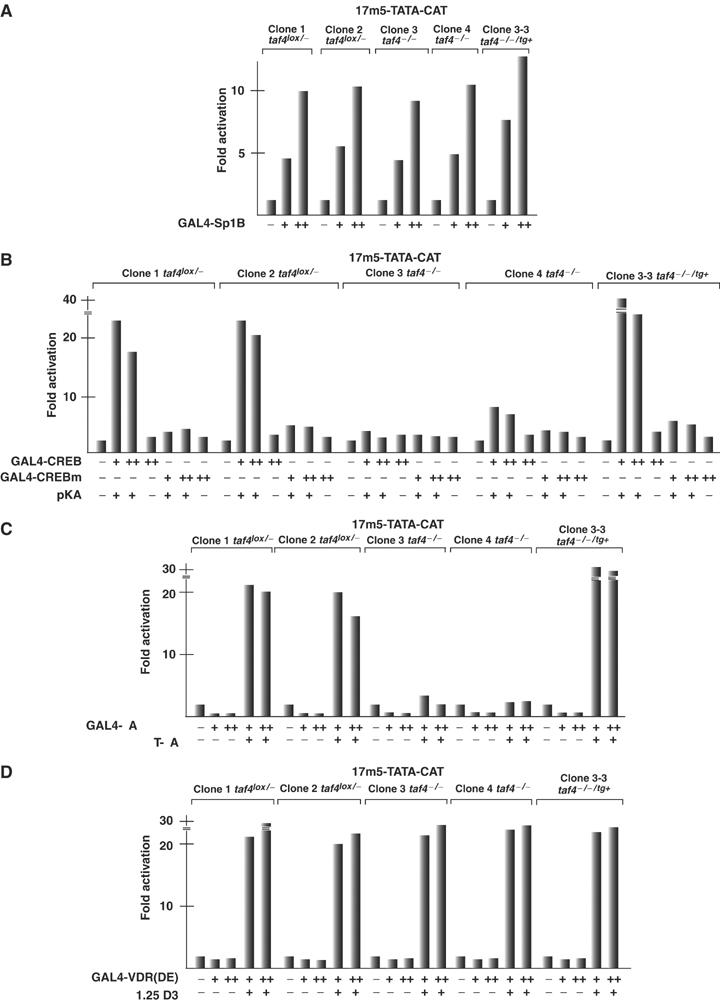
TAF4 is a critical cofactor for CREB and RAR. (A–D) The indicated cell lines were transfected with the 17m5-TATA-CAT reporter (2.5 μg), the CMV-based β-galactosidase expression vector (2.5 μg) and the activator plasmids shown below each lane. CREBm contains the S133A mutation. In panel B, cotransfection of the catalytic subunit of protein kinase A is indicated, and in panels C and D, the presence or absence of 10−6 M all-trans-retinoic acid or 1alpha,25(OH)2-vitamin D3 is indicated. + represents 100 ng and ++ 250 ng of expression vector. The value of the 17m5-TATA-CAT reporter plasmid alone is taken as 1 and the fold activation is expressed relative to this value.
TAF4 also interacts with the CREB/CREM proteins and has been proposed to act as a cofactor for their function (Saluja et al, 1998; Asahara et al, 2001). Transfection of GAL4-CREB together with the protein kinase A catalytic subunit leads to transcriptional activation in the taf4lox/− cells (Figure 9B). In contrast, in taf4−/− clones, activation is strongly diminished. However, GAL4-CREB strongly activated transcription in 3-3 taf4−/−/tg+ cells showing that the CR-II region of TAF4 is necessary and sufficient to act as a CREB cofactor. In all cells, strong activation is dependent on expression of the PKA catalytic subunit and is abolished by mutation of the PKA acceptor serine within CREB (S133A). Similar results were observed with CREM (data not shown).
We further examined transcriptional activation by the ligand-dependent activation function (AF)-2 of the retinoic acid receptor RARα. Efficient ligand-dependent transactivation by GAL4-RARα(DE) was observed in the taf4lox/− cells, but not in the taf4−/− cells (Figure 9C). Expression of only the CR-II domain in the clone 3-3 taf4−/−/tg+ cells restored a strong activation by GAL4-RARα(DE), consistent with our previous overexpression experiments (Gangloff et al, 2000). In contrast, inactivation of TAF4 did not affect ligand-dependent activation by the vitamin D3 receptor, which showed comparable activity in all cell lines (Figure 9D).
Discussion
TAF4 is not essential for cell viability
We describe mouse embryonic fibroblasts lacking TAF4. Compared to other ubiquitously expressed TAFs whose loss leads to cell cycle arrest and apoptosis, taf4−/− cells are viable, showing that mouse TAF4 is not essential for viability and cell cycle progression. Although biochemical studies suggest that TAF4 is essential for TFIID integrity (Gangloff et al, 2000), taf4−/− cells contain intact TFIID. This may be explained by the presence of TAF4b, which associates with TBP in the absence of TAF4 to preserve TFIID integrity and the viability of taf4−/− cells. However, even in taf4−/− cells, a large proportion of the TAF4b is not associated with TFIID. It remains to be determined whether this TAF4b is present in another complex acting independently of TFIID.
The changes in gene expression and altered growth of the taf4−/− cells indicate that the TFIID complex comprising TAF4b exhibits distinct functional properties and target gene specificity. Overexpression of TAF4b mimics loss of TAF4 in that it induces several genes upregulated in taf4−/− cells. This suggests that competition between TAF4 and TAF4b for integration into TFIID regulates the relative amounts of the complex containing each TAF. If this equilibrium is disturbed by loss of TAF4 or overexpression of TAF4b, the expression of a set of target genes is deregulated. This model helps explain why expression of only the TAF4 CR-II can restore normal levels of expression of many genes and suppress autocrine growth, as this domain is necessary and sufficient for integration into the TFIID complex and thus competes with TAF4b.
TAF4 regulates genes involved in the control of cell proliferation
Although study of TS mutants in yeast has allowed the identification of genes dependent on different TAFs (Shen et al, 2003), little is known about TAF-dependent target genes and signalling pathways in mammalian cells since loss of TAF function often results in cell cycle arrest and apoptosis. The taf4−/− cells offer a unique system in which to study the functional properties of one of the major TFIID subunits. Here, we show that loss of TAF4 affects positively or negatively the expression of more than 1000 genes involved in multiple biological processes.
The major phenotype of the taf4−/− cells is, however, serum-independent autocrine growth due to deregulated production of mitotic factors. Several lines of evidence suggest that enhanced TGFβ signalling is one of the principal pathways involved. Loss of TAF4 leads to increased expression of TGFβ1 and TGFβ3 as well as of the secondary TGFβ-induced mitogen CTGF (Brigstock, 2003; Leask and Abraham, 2003). CTGF has intrinsic growth factor properties and acts as a secondary mediator of the proliferative response of fibroblasts to TGFβ, in particular during the development of fibrosis (Leask and Abraham, 2004). CTGF also potentiates TGFβ signalling, by interacting directly with TGFβ1 and facilitating binding to its surface receptor (Abreu et al, 2002). Thus, the requirement for CTGF for autocrine growth may be explained both by its intrinsic mitogenic activity and its ability to potentiate TGFβ1 signalling.
In addition to CTGF, MMP2, THBS1 and THBS2, factors known to activate latent TGFβ (Yu and Stamenkovic, 2000; Annes et al, 2003) are also induced. As a consequence, taf4−/− cells secrete high levels of active TGFβ that activate the SMAD pathway. Loss of TAF4 therefore leads to induction of a positive autoregulatory feedback loop of TGFβ signalling. The taf4−/− conditioned media also activate the p38 (but not p42–p44, data not shown) Map kinase pathway. The p38 pathway may be activated by TGFβ via the TAK-1 kinase or alternatively by another of the growth factors in the conditioned media.
Indeed, TAF4 inactivation induces OPN as well as HB-EGF. HB-EGF can remain associated with the cell membrane acting as a juxtacrine factor or be released through cleavage by several proteases, one of which, MMP3, is also upregulated by loss of TAF4 (Suzuki et al, 1997). Thus, both OPN and HB-EGF may cooperate with TGFβ signalling to allow autocrine growth.
We additionally observed upregulation of PDGF-C and VEGF-C. Paradoxically however, PDGF-A and VEGF-D are repressed. This suggests that these growth factor families may not be directly implicated in regulating the proliferation of the taf4lox/− and taf4−/− cells, or that these two members of each family have distinct functional properties in fibroblasts. Together, our results show that TAF4 inactivation induces multiple growth factors that may cooperate with TGFβ to promote the serum independence of the taf4−/− cells.
Re-expression of the TAF4 CR-II domain suppresses serum independence. However, expression of TGFβ1/3 and OPN are not repressed either by the CR-II domain or full-length TAF4. In contrast, expression of CTGF, THBS1, THBS2 and MMP2 are all downregulated along with HB-EGF and PDGF-C. The inability of the taf4−/−/tg+ cells to survive in the absence of serum despite elevated TGFβ and OPN expression highlights the critical role played by the proteins that activate and potentiate TGFβ signalling and/or the requirement for cooperation with other induced mitogens.
As discussed above, enhanced TGFβ signalling reflects the activity of the TFIID complex containing TAF4b. This finding is reminiscent of previous results showing TAF4b involvement in signalling by inhibin and activin, members of the TGFβ superfamily. TAF4b expression in granulosa cells of the ovarian follicle is required for the inhibin–activin-induced expression of genes required for synthesis and release of follicle-stimulating hormone (Freiman et al, 2001). Our results suggest that TAF4b may play a broader role in TGFβ signalling. In addition, TAF4b has been suggested to be involved in the activation of antiapoptotic genes by the NF-κB transcription factor in response to TNFα signalling (Yamit-Hezi and Dikstein, 1998). Our results show that enhancing TAF4b function through loss of TAF4 also results in the activation of antiapoptotic pathways.
Downstream factors involved in fibroblast survival in low serum
TAF4 inactivation modulates the expression of a diverse set of genes. In addition to the secreted mitogens, we have tried to highlight potential downstream mediators important for serum independence by identifying genes whose expression is further deregulated when the taf4−/− cells are grown in 0% serum or genes whose expression is normally modulated in taf4lox/− cells in low-serum conditions. These comparisons showed that a significant proportion of genes whose expression was modified in taf4lox/− cells in low serum were also deregulated in the taf4−/− cells in 10% serum. Furthermore, the deregulated expression of a small number of genes in the taf4−/− cells was exacerbated upon growth in 0% serum. Collectively, the regulation of these genes suggests they may be critical in promoting survival in low serum and hence may contribute to serum independence.
Among the genes upregulated under all conditions are BCL6, a transcriptional repressor and an oncogene normally involved in B-cell lymphomas (Polo et al, 2004), and KLF5, a transcriptional activator associated with cell proliferation (Nandan et al, 2004). The function of these factors in promoting survival and proliferation shows an obvious correlation with adaptation to low serum, suggesting that they are potentially important regulators of fibroblast proliferation.
In contrast, it is surprising to find BDNF in this class. BDNF is a secreted mitogen that normally functions as a survival factor in neurons and is involved in synapse plasticity (Binder and Scharfman, 2004). Nevertheless, no significant expression of the high-affinity BDNF receptor is seen in either the taf4lox/− or taf4−/− cells and recombinant BDNF does not mediate their serum-independent growth. It is also interesting to note that sequestration of TAF4 in intranuclear aggregates has been previously suggested to contribute to development of neurodegenerative diseases (Shimohata et al, 2000) where decreased BDNF expression and secretion may be a major contributing factor (Gauthier et al, 2004). The fact that loss of TAF4 induces BDNF and a general stimulation of antiapoptotic factors argues against the current model for TAF4 function in neurodegenerative disease. It will be interesting to determine whether loss of TAF4 in neurons has similar consequences as in fibroblasts.
Among the genes downregulated under all conditions are OLFL3, a cell surface protein of unknown function, preproenkephalin (PENK), a neural expressed opioid peptide, and the interleukin 1 receptor antagonist (IL1RN). The potential functional relevance of these proteins in this context remains to be determined. On the other hand, GREM-1 is an antagonist of the BMP subfamily of TGFβ factors that binds BMPs and prevents them from interacting with their receptors, but has no known activity towards the TGFβ1 and TGFβ3 family members. BMP1 and BMP4 are expressed in both taf4lox/− and taf4−/− cells. Their activity is perhaps enhanced by the downregulation of GREM-1. HOD (HOP1) contains an atypical homeodomain that does not bind to DNA, but interacts with the serum response factor to block mitogenic signals (Chen et al, 2002; Shin et al, 2002). Repression of this gene may therefore favour the proliferative response in fibroblasts, as has been previously shown in other cell types.
TAF4 is a coactivator for CREB and RAR
Despite previous observations of TAF4 interactions with Sp1 (Gill et al, 1994; Saluja et al, 1998), activation by this factor is not affected in the taf4−/− cells. In contrast, TAF4 is an essential cofactor for activation by CREB and the RAR both of which exhibit severely reduced activity in the taf4−/− cells. CREB interacts directly with the CR-II of TAF4 (Asahara et al, 2001) and the CR-II alone is sufficient to mediate CREB activity. Similarly, the CR-II is sufficient for RAR activity, in agreement with our overexpression experiments (Gangloff et al, 2000). One important difference with previous observations is that TAF4 is not required for activation by the VDR, at least on the transfected reporter. There is therefore a difference between these receptors in this loss-of-function assay that was not observed in our previous overexpression experiments (Mengus et al, 1997).
Taken together, our results indicate that an equilibrium between TAF4 and TAF4b acts to regulate a large set of genes involved in the control of cell proliferation. The TFIID containing TAF4b more efficiently mediates TGFβ signalling, whereas the presence of TAF4 in TFIID is required for the activity of CREB and RAR. These observations highlight the distinct functional properties of TAF4 and TAF4b.
Materials and methods
Construction of targeting vector and inactivation of taf4
The murine taf4 locus was isolated by screening an ES genomic DNA library with the TAF4 cDNA. Genomic DNA was subcloned into the NotI site of plasmid pZERO (InvitroGen) and sequenced. Using the cloned locus as template, PCR was used to generate a targeting vector in which a PGK promoter-driven hygromycin resistance gene flanked by LoxP sites was inserted downstream of exon 12 and a second loxP site inserted upstream of exon 11 along with an additional EcoRI site. All constructions were verified by restriction enzyme digests and automated DNA sequencing.
Genotyping of ES cell clones and mice was performed by Southern blot using the probe and digests outlined in Figure 1. The taf4lox/+ ES cell clone was electroporated with a vector expressing the Cre recombinase and 40 subclones were analysed by Southern blot for the presence of the deleted allele. Several taf4+/− ES clones were thus identified. Blastocyst injection and generation of the mutant mice from the taf4lox/+ and taf4−/+ ES cell clones was performed as described previously (Martianov et al, 2001).
Isolation of embryonic fibroblasts
taf4lox/− embryos were isolated at E13.5 and a primary fibroblast culture was initiated. Fibroblasts were passed at high dilution until a population of immortalised cells was obtained. These cells were transfected with vectors expressing the Cre recombinase and the green fluorescent protein. After 48 h, cells were sorted by FACS and individual GFP-expressing cells were seeded in 96-well plates. Clones were amplified and genotyped to yield more than 5–10 clones of each taf4lox/− and taf4−/− genotype. Two representative clones of each genotype are shown. Cells were grown in Dulbecco's minimal essential media supplemented with glutamax and 10 or 1% fetal calf serum as indicated.
To re-express TAF4, pXJ40-based vectors expressing full-length human TAF4 or deletion mutants as described (Mengus et al, 1997; Perletti et al, 2001) were transfected into the taf4−/− clone 3 and clone 4 cells along with a vector encoding resistance to puromycin. Resistant clones were amplified and TAF4 expression verified by immunoblot. More than 10 revertant clones for each construct were isolated and the results for two representative clones are shown.
Monoclonal antibodies and immunoblots
Antibodies against TAF4, TBP, TAF5, TAF6, TAF7, TAF12 and TAF13 were as described previously (Mengus et al, 1995, 1997). A rabbit polyclonal antibody was generated against the TAF4b peptide KSRSNKEDPEQLRLKQKAK. Positively reacting antisera were identified by immunoblot analysis on extracts of Cos cells transfected with a TAF4b expression vector and affinity purified against the immobilised peptide. The TAF4b expression vector comprises the C-terminal 784 amino acids of mTAF4b, corresponding to those described for the human TAF4b (Dikstein et al, 1996). Databases predict that mTAF4b should comprise 850 amino acids; however, the N-terminus of the endogenous protein has not been characterised. TBP immunoprecipitations were performed as previously described using monoclonal antibodies 3G3 or 2C1 (Mengus et al, 1997). Phosphorylated SMAD2/3 was detected using antibody (3101) from Cell Signaling.
Affymetrix gene profiling and data analysis
Duplicate independent preparations of total RNA from both taf4lox/− and taf4−/− clones exponentially growing in 10% serum (eight in total) were analysed on the Mouse Genome GeneChip 430A 2.0 comprising 22 600 probe sets representing over 14 500 mouse genes. To analyse gene expression in low-serum conditions, duplicate samples of cells from the taf4−/− clones or the taf4lox/− clones were transferred in 0 or 1% serum, respectively, for 3 days before preparation of RNA. Profiling and statistical analyses were performed as described in Supplementary data.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Data
Acknowledgments
We thank Dr K Merienne and L Tora for gift of antibodies, Dr C Hill for the TGFβ-responsive 3T3 cells, D Dembelé for help with statistical analysis, A Dierich, E Metzger and the staff of the IGBMC ES cell and animal facilities, G Duval for the rabbit polyclonal antibodies, M Oulad-Abdelghani and the monoclonal antibody facility, the DNA sequencing, the oligonucleotide and peptide synthesis facilities. This work was supported by grants from the CNRS, the INSERM, the Hôpital Universitaire de Strasbourg, the Ministère de la Recherche et de la Technologie, the Association pour la Recherche contre le Cancer, the Ligue Nationale et Départementale Région Alsace contre le Cancer and the European Union.
References
- Abreu JG, Ketpura NI, Reversade B, De Robertis EM (2002) Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol 4: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annes JP, Munger JS, Rifkin DB (2003) Making sense of latent TGFbeta activation. J Cell Sci 116: 217–224 [DOI] [PubMed] [Google Scholar]
- Asahara H, Santoso B, Guzman E, Du K, Cole PA, Davidson I, Montminy M (2001) Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB. Mol Cell Biol 21: 7892–7900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder DK, Scharfman HE (2004) Brain-derived neurotrophic factor. Growth Factors 22: 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Yamamoto K, Staub A, Tora L (1999) Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem 274: 18285–18289 [DOI] [PubMed] [Google Scholar]
- Brigstock DR (2003) The CCN family: a new stimulus package. J Endocrinol 178: 169–175 [DOI] [PubMed] [Google Scholar]
- Chen F, Kook H, Milewski R, Gitler AD, Lu MM, Li J, Nazarian R, Schnepp R, Jen K, Biben C, Runke G, Mackay JP, Novotny J, Schwartz RJ, Harvey RP, Mullins MC, Epstein JA (2002) Hop is an unusual homeobox gene that modulates cardiac development. Cell 110: 713–723 [DOI] [PubMed] [Google Scholar]
- Chen Z, Manley JL (2000) Robust mRNA transcription in chicken DT40 cells depleted of TAF(II)31 suggests both functional degeneracy and evolutionary divergence. Mol Cell Biol 20: 5064–5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikstein R, Zhou S, Tjian R (1996) Human TAFII 105 is a cell type-specific TFIID subunit related to hTAFII130. Cell 87: 137–146 [DOI] [PubMed] [Google Scholar]
- Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, Morris PL, Tjian R, Richards JS (2005) Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev 19: 794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R (2001) Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 293: 2084–2087 [DOI] [PubMed] [Google Scholar]
- Gangloff YG, Werten S, Romier C, Carre L, Poch O, Moras D, Davidson I (2000) The human TFIID components TAF(II)135 and TAF(II)20 and the yeast SAGA components ADA1 and TAF(II)68 heterodimerize to form histone-like pairs. Mol Cell Biol 20: 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, Rangone H, Cordelieres FP, De Mey J, MacDonald ME, Lessmann V, Humbert S, Saudou F (2004) Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 118: 127–138 [DOI] [PubMed] [Google Scholar]
- Gill G, Pascal E, Tseng ZH, Tjian R (1994) A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA 91: 192–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Hill CS (2002) Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol Cell 10: 283–294 [DOI] [PubMed] [Google Scholar]
- Laping NJ, Grygielko E, Mathur A, Butter S, Bomberger J, Tweed C, Martin W, Fornwald J, Lehr R, Harling J, Gaster L, Callahan JF, Olson BA (2002) Inhibition of transforming growth factor (TGF)-beta1-induced extracellular matrix with a novel inhibitor of the TGF-beta type I receptor kinase activity: SB-431542. Mol Pharmacol 62: 58–64 [DOI] [PubMed] [Google Scholar]
- Leask A, Abraham DJ (2003) The role of connective tissue growth factor, a multifunctional matricellular protein, in fibroblast biology. Biochem Cell Biol 81: 355–363 [DOI] [PubMed] [Google Scholar]
- Leask A, Abraham DJ (2004) TGF-beta signaling and the fibrotic response. FASEB J 18: 816–827 [DOI] [PubMed] [Google Scholar]
- Martianov I, Fimia GM, Dierich A, Parvinen M, Sassone-Corsi P, Davidson I (2001) Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol Cell 7: 509–515 [DOI] [PubMed] [Google Scholar]
- Mengus G, May M, Carre L, Chambon P, Davidson I (1997) Human TAF(II)135 potentiates transcriptional activation by the AF-2s of the retinoic acid, vitamin D3, and thyroid hormone receptors in mammalian cells. Genes Dev 11: 1381–1395 [DOI] [PubMed] [Google Scholar]
- Mengus G, May M, Jacq X, Staub A, Tora L, Chambon P, Davidson I (1995) Cloning and characterization of hTAFII18, hTAFII20 and hTAFII28: three subunits of the human transcription factor TFIID. EMBO J 14: 1520–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merienne K, Jacquot S, Zeniou M, Pannetier S, Sassone-Corsi P, Hanauer A (2000) Activation of RSK by UV-light: phosphorylation dynamics and involvement of the MAPK pathway. Oncogene 19: 4221–4229 [DOI] [PubMed] [Google Scholar]
- Metzger D, Scheer E, Soldatov A, Tora L (1999) Mammalian TAF(II)30 is required for cell cycle progression and specific cellular differentiation programmes. EMBO J 18: 4823–4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan WS Jr, Scheer E, Wendling O, Metzger D, Tora L (2003) TAF10 (TAF(II)30) is necessary for TFIID stability and early embryogenesis in mice. Mol Cell Biol 23: 4307–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy-Ullrich JE, Poczatek M (2000) Activation of latent TGF-beta by thrombospondin-1: mechanisms and physiology. Cytokine Growth Factor Rev 11: 59–69 [DOI] [PubMed] [Google Scholar]
- Nandan MO, Yoon HS, Zhao W, Ouko LA, Chanchevalap S, Yang VW (2004) Kruppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene 23: 3404–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perletti L, Kopf E, Carre L, Davidson I (2001) Coordinate regulation of RARgamma2, TBP, and TAFII135 by targeted proteolysis during retinoic acid-induced differentiation of F9 embryonal carcinoma cells. BMC Mol Biol 2: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Dell'Oso T, Ranuncolo SM, Cerchietti L, Beck D, Da Silva GF, Prive GG, Licht JD, Melnick A (2004) Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med 10: 1329–1335 [DOI] [PubMed] [Google Scholar]
- Saluja D, Vassallo MF, Tanese N (1998) Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol Cell Biol 18: 5734–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Weil PA (2000) Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J Biol Chem 275: 13895–13900 [DOI] [PubMed] [Google Scholar]
- Shen WC, Bhaumik SR, Causton HC, Simon I, Zhu X, Jennings EG, Wang TH, Young RA, Green MR (2003) Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J 22: 3395–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, Naruse S, Kimura T, Koide R, Nozaki K, Sano Y, Ishiguro H, Sakoe K, Ooshima T, Sato A, Ikeuchi T, Oyake M, Sato T, Aoyagi Y, Hozumi I, Nagatsu T, Takiyama Y, Nishizawa M, Goto J, Kanazawa I, Davidson I, Tanese N (2000) Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat Genet 26: 29–36 [DOI] [PubMed] [Google Scholar]
- Shin CH, Liu ZP, Passier R, Zhang CL, Wang DZ, Harris TM, Yamagishi H, Richardson JA, Childs G, Olson EN (2002) Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell 110: 725–735 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Raab G, Moses MA, Fernandez CA, Klagsbrun M (1997) Matrix metalloproteinase-3 releases active heparin-binding EGF-like growth factor by cleavage at a specific juxtamembrane site. J Biol Chem 272: 31730–31737 [DOI] [PubMed] [Google Scholar]
- Tanese N, Saluja D, Vassallo MF, Chen JL, Admon A (1996) Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc Natl Acad Sci USA 93: 13611–13616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuault S, Gangloff YG, Kirchner J, Sanders S, Werten S, Romier C, Weil PA, Davidson I (2002) Functional analysis of the TFIID-specific yeast TAF4 (yTAF(II)48) reveals an unexpected organization of its histone-fold domain. J Biol Chem 277: 45510–45517 [DOI] [PubMed] [Google Scholar]
- Tora L (2002) A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev 16: 673–675 [DOI] [PubMed] [Google Scholar]
- Voss AK, Thomas T, Petrou P, Anastassiadis K, Scholer H, Gruss P (2000) Taube nuss is a novel gene essential for the survival of pluripotent cells of early mouse embryos. Development 127: 5449–5461 [DOI] [PubMed] [Google Scholar]
- Walker AK, Rothman JH, Shi Y, Blackwell TK (2001) Distinct requirements for C. elegans TAF(II)s in early embryonic transcription. EMBO J 20: 5269–5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werten S, Mitschler A, Romier C, Gangloff YG, Thuault S, Davidson I, Moras D (2002) Crystal structure of a subcomplex of human transcription factor TFIID formed by TATA binding protein-associated factors hTAF4 (hTAF(II)135) and hTAF12 (hTAF(II)20). J Biol Chem 277: 45502–45509 [DOI] [PubMed] [Google Scholar]
- Yamit-Hezi A, Dikstein R (1998) TAFII105 mediates activation of anti-apoptotic genes by NF-kappaB. EMBO J 17: 5161–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Stamenkovic I (2000) Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev 14: 163–176 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Data


