Abstract
Centrins, small calcium binding EF-hand proteins, function in the duplication of a variety of microtubule organizing centers. These include centrioles in humans, basal bodies in green algae, and spindle pole bodies in yeast. The ciliate Tetrahymena thermophila contains at least four centrin genes as determined by sequence homology, and these have distinct localization and expression patterns. CEN1's role at the basal body was examined more closely. The Cen1 protein localizes primarily to two locations: one is the site at the base of the basal body where duplication is initiated. The other is the transition zone between the basal body and axoneme. CEN1 is an essential gene, the deletion of which results in the loss of basal bodies, which is likely due to defects in both basal body duplication and basal body maintenance. Analysis of the three other centrins indicates that two of them function at microtubule-rich structures unique to ciliates, whereas the fourth is not expressed under conditions examined in this study, although when artificially expressed it localizes to basal bodies. This study provides evidence that in addition to its previously known function in the duplication of basal bodies, centrin is also important for the integrity of these organelles.
INTRODUCTION
Centrins, also called caltractins, are widely conserved members of the EF-hand superfamily of calcium-binding proteins (Salisbury, 1995). Although sharing ∼50% identity with the ubiquitous calcium sensor calmodulin, centrins perform distinct functions within the eukaryotic cell. In particular, centrins have a variety of roles at microtubule organizing centers (MTOCs) and microtubule-based structures (Salisbury, 1995; Vaughn and Harper, 1998; Chapman et al., 2000). Additionally, centrins are constituents of calcium-responsive contractile fibers found in many protists (Salisbury et al., 1984; Wright et al., 1985).
One of the widely recognized roles for centrin is in the duplication of microtubule organizing centers, including centrosomes, basal bodies, and the yeast spindle pole body (Baum et al., 1986; Salisbury et al., 2002; Koblenz et al., 2003). Centrosomes consist of two orthogonally oriented centrioles surrounded by amorphous pericentriolar material from which spindle and astral microtubules are organized. Centrioles are distinguished by 9-fold symmetry of microtubules in a “flower” arrangement, with the majority of metazoans displaying triplet microtubules at these positions. Basal bodies are structurally similar to centrioles and differ primarily in the manner in which they organize microtubules. Cilia and flagella, which consist of nine doublet microtubules surrounding a central microtubule pair, are nucleated from the distal end of the basal body (reviewed in Beisson and Wright, 2003). In some situations, it is the identical organelle that acts as a centriole and then is converted to a basal body, or vice versa, at particular points in the life cycle of the cell. This is the case in many vertebrate cells where the oldest centriole is capable of forming a single “9 + 0” cilium (known as the primary cilium) that is assembled during the G1 phase of the cell cycle and disassembled during mitosis (Jensen et al., 1987). The budding yeast spindle pole body is structurally distinct from the centriole and basal body. This trilaminar structure remains embedded within the nuclear envelope throughout the cell cycle. Spindle and astral microtubules emanate from the inner and outer plaques, respectively. Although dissimilar in appearance to the centrosome, the spindle pole body performs the same task of organizing mitotic and meiotic spindles, and some components of these organelles, including centrin, are conserved between these divergent organelles (Adams and Kilmartin, 2000).
Temperature-sensitive mutations in the yeast centrin gene CDC31 result in failure in an early step of spindle pole body duplication and a cell cycle arrest in G2/M phase, such that cells arrest with a single, enlarged spindle pole body (SPB) (Byers, 1981; Baum et al., 1986). Like SPB duplication, basal body and centriole duplication is dependent on centrin function, as demonstrated by RNA interference (RNAi) experiments in both human cells and Chlamydomonas. Thus far, three centrin genes have been cloned from human cells (Lee and Huang, 1993; Errabolu et al., 1994; Middendorp et al., 1997). When human centrin2 levels are reduced by RNAi, cells exhibit defects in centriole duplication (Salisbury et al., 2002). The same is true when levels of the Chlamydomonas centrin (Vfl2) are decreased to 5% of the normal level. In these cells, the number of basal bodies is reduced to ∼0.3 per cell from the normal condition of two per cell, indicating a failure in basal body duplication (Koblenz et al., 2003). Defects also were observed in the structure of remaining basal bodies, indicating a possible role for centrin in basal body maturation or stability.
In addition to its essential role in MTOC duplication, centrin is involved in other cellular processes. In yeast, Cdc31 is required for Kic1 kinase activity that is important for proper cell wall development (Sullivan et al., 1998). Cdc31 also is involved in mRNA export at the nuclear pore complex (Fischer et al., 2004). The contractile properties of centrin are important in a number of structures, including the nucleus-basal body connector and stellate fibers associated with Chlamydomonas basal bodies (Sanders and Salisbury, 1989; Taillon et al., 1992; Sanders and Salisbury, 1994), and the infraciliary lattice, a contractile cortical network in Paramecium (Madeddu et al., 1996; Klotz et al., 1997). Centrin also has been associated with the inner dynein arm important for axonemal beating (LeDizet and Piperno, 1995; Guerra et al., 2003).
In some organisms, such as yeast and Chlamydomonas, the product of a single centrin gene carries out multiple functions. However, other species have multiple centrin genes that may encode products that are specialized to carry out different tasks or to carry out similar tasks at different stages of the life cycle. In humans, centrin1 expression is confined to the testes and retinal cells, whereas centrins 2 and 3 are more ubiquitously expressed (Hart et al., 1999; Laoukili et al., 2000). Strikingly, when a Leishmania centrin gene was deleted, one of the two lifestyle forms of this parasite was unable to duplicate its basal bodies and was unable to progress through mitosis, whereas the other was unaffected, suggesting that a different centrin was active during this stage of its life cycle (Selvapandiyan et al., 2004).
Because the ciliate Tetrahymena thermophila has ∼750 basal bodies per cell (Frankel, 2000; Frankel and Nelsen, 2001) and sophisticated genetic and molecular biological techniques are available for its study (Bruns and Cassidy-Hanley, 2000a,b; Gaertig and Kapler, 2000; Hai et al., 2000), we turned to Tetrahymena to gain insights into basal body/centriole duplication. Centrin has been localized to basal bodies and cilia in Tetrahymena, and a centrin gene has been cloned (Guerra et al., 2003). We independently cloned this centrin gene, called CEN1, and have begun characterizing it and three other centrins we identified through sequence similarity in the recently sequenced Tetrahymena genome. By using fusions with the green fluorescent protein (GFP) (Chalfie et al., 1994), we localized Cen1 and the second of the centrins (CEN2) to basal bodies. We created a null mutation in CEN1 and have shown it is required for basal body duplication and stability. Furthermore, we found that the third centrin (CEN3) localizes to specialized structures in the oral apparatus, a complex network of cilia, and accessory structures that sweeps food into the cell. The fourth centrin (CEN4) also localizes to these structures in the oral apparatus as well as to contractile vacuole pores (the openings for the contractile vacuoles that maintain the proper osmolarity of the cells) and to basal bodies (Frankel, 2000).
MATERIALS AND METHODS
Strains, Culture Conditions, and Matings
T. thermophila strains B2086, CU427, and CU428 (all generous gifts from Dr. Peter Bruns, Cornell University, Ithaca, NY) were the starting point for new strain development. Strains with constructs encoding GFP-Centrin fusion proteins were created as described below. UCB8 and UCB9 are CEN1 knockout heterokaryons (described below). B*VI (gift from Dr. Aaron Turkewitz, University of Chicago, Chicago, IL) was used for star crosses in strain construction (Hai et al., 2000). All were grown in SSP (2% proteose peptone, 0.1% yeast extract, 0.2% glucose, 0.003% FeEDTA) at 30°C. Starvation media consisted of 10 mM Tris, pH 7.5 (Orias et al., 2000). For matings, an equal number of cells of different mating types, previously starved for 18 h, were mixed (Gaertig and Kapler, 2000).
Gene Identification and Sequence Analysis
To search for centrin genes in Tetrahymena, PCR using degenerate primers EIK-F and VAK-R, designed to represent regions of high conservation between centrins, was conducted (Supplemental Table 1 lists all primers used in this study). Standard molecular biology techniques were used to isolate a centrin gene, which was subsequently reported by Guerra et al. (2003). To search for additional centrin genes, preliminary sequence data were obtained from The Institute for Genomic Research Web site at http://www.tigr.org. Tetrahymena CEN1 translated sequence was used to query the Tetrahymena database using the tblastn program. CEN2 was found on sequence #8254664 between positions 498633 and 499370, with possible introns spanning 498790–498909, 499005–499063, and 499256–499320. CEN3 was found on sequence #8253915 between positions 239917 and 240426. CEN4 was found on sequence #8254437 between positions 298144 and 298659. Sequences were compared and analyzed at the Pasteur Bioweb site (bioweb.pasteur.fr). ClustalW (Thompson et al., 1994) was used to align the sequences, and BOXSHADE (written by Hofmann and Baron) was used to create the image of the alignment. Using PHYLIP (Phylogeny Inference Package), protein distances were calculated with PROTDIST (Jones–Taylor–Thornton matrix), and a tree was constructed using NEIGHBORS followed by DRAWGRAM for image generation. For statistical analysis, 1000 replicates were performed and bootstrap values were calculated. (Felsenstein, J. 2004. PHYLIP version 3.6. Distributed by the author, Department of Genome Sciences, University of Washington, Seattle, WA)
Plasmid Construction
As a starting point for the creation of a CEN1 knockout plasmid, pHM74 was constructed. A 2.1-kb fragment containing CEN1 was amplified from genomic Tetrahymena DNA by PCR (Saiki et al., 1988) using primers RM13 and RM14, which were engineered to include unique BamHI and SacII sites at their respective 5′ ends. This fragment, which begins 1 kb upstream of the CEN1 start codon, was cloned into the BamHI and SacII sites of p4T2-1 (Gaertig et al., 1994a) downstream of the NEO2 cassette. Primers RM11 and RM12, which were engineered to contain unique KpnI and XhoI sites at their respective ends, were then used to amplify a 1.6-kb genomic fragment upstream of CEN1. This fragment was cloned into KpnI/XhoI-digested p4T2-1, upstream of the NEO2 cassette, to create pHM74. pHM74 was digested with KpnI and BamHI, eliminating the NEO2 cassette and the 1.6-kb CEN1 upstream region. A KpnI-BamHI adapter from EZCloneSystems (New Orleans, LA) was used to recircularize the plasmid. This construct was digested with EcoRI to remove most of the CENI coding sequence, including the start codon, and blunted. A SmaI, EcoRV fragment from pHM74 containing the NEO2 cassette was inserted into this plasmid, creating pCentrin:NEO 1. To obtain more sequence downstream of the CEN1 gene, a library of 3- to 4-kb PstI, EcoRI-digested genomic DNA was created in pBluescript KSII– (Stratagene, La Jolla, CA) and transformed into Escherichia coli for colony lifts. A PCR fragment generated by amplification of pHM74 DNA with primers RM10 and T7 was used as a probe. Lifts and labeling of the probe were conducted as per instructions for Hybond-N+ nylon membrane and the AlkPhos direct labeling system (Amersham Biosciences, Piscataway, NJ). A clone was recovered with ∼3 kb of sequence downstream of CEN1, and a BglII, SacI fragment was cloned from this into pCentrin:NEO 1 to create pCentrin:NEO1+48. To use all of the upstream sequence available, a PCR fragment from genomic DNA spanning the site where the NEO2 cassette had been inserted in the sequence upstream of CEN1 was amplified with primers NEORemove1 and NEORemove2, digested with BclI and SpeI, and inserted into similarly digested pHM74, creating pHM74-NEORemove. Finally, a BclI, KpnI fragment from this plasmid was ligated into similarly digested pCentrin:NEO1+48 to create pMicCentrinKO1.
For the plasmids encoding the GFP-centrin fusions, genomic DNA was amplified with XhoI and ApaI sites engineered into the primers to allow cloning into similar sites in pVGF-1 (Wiley et al., 2000). Primers were used as follows: for CEN1, primers XhoI-5′Centrin and ApaI-3′Centrin; for CEN2, primers 5′XhoI TtCentrin2 and 3′ApaI TtCentrin2; for CEN3, primers 5′XhoI TtCentrin3 and 3′ApaI TtCentrin3; and for CEN4, primers 5′XhoI TtCentrin4 and 3′ApaI TtCentrin4. Clones were sequenced at Macrogen (Seoul, Korea) to confirm that errors had not been introduced by PCR.
To create a version of CEN1 that could be expressed in E. coli, a PCR fragment containing the coding region of CEN1 was amplified from a Tetrahymena cDNA library (generous gift from Dr. Aaron Turkewitz) using primers 5′BamHI-Centrin and 3′PstI-Centrin, which were engineered to contain the indicated restriction sites. This fragment was cloned into similarly digested pUC18. Because Tetrahymena uses an alternative genetic code, in which UAA and UAG represent glutamine codons, it was necessary to modify the CEN1 sequence to allow expression in E. coli. Four glutamine codons were changed to the canonical CAA or CAG codons using the U.S.E. site-directed mutagenesis system (Amersham Biosciences, Piscataway, NJ) with the following primers: CentSw1, CentSw2, CentSw3, and TMSelect. The resulting plasmid (pUC18-TtCentrinMFE) was digested with BamHI and PstI, and the CEN1 fragment was ligated into the pQE10 6-His fusion expression vector (QIAGEN, Valencia, CA), creating pQE10-CentrinMFE.
To create the CEN1 rescuing construct, a 1.0-kb BamHI, MfeI CEN1 promoter region was excised from pHM74 and cloned into similarly digested pUC18-TtCentrinMFE. CEN1, with its promoter, was then excised from this construct with SmaI and SphI, blunted, and ligated into the blunted BsmBI site of grl3/4:BLAST (a construct that replaces the GRL3 and GRL4 promoter region with the Bsr gene, which provides resistance to Blasticidin S, a generous gift from Aaron Turkewitz). Finally, the CEN1 3′ untranslated region (UTR) was amplified by PCR of genomic DNA using primers Centrin_BstXI and BsmBI-dsCentrin and ligated into BstXI (partial), BsmBI-digested grl3/4:BLAST-PrCEN1 to create grl3/4:BLAST-PrCEN1–3′UTR. The resulting plasmid was sequenced to ensure no errors had been introduced.
Protein Expression and Antibody Generation
pQE10-CentrinMFE was transformed into the E. coli strain M15, and a culture was grown in 500 ml of Luria Broth + 0.2% glucose with 50 μg/ml kanamycin and 100 μg/ml ampicillin. At an OD600 of 0.4, cells were cooled to room temperature for 1 h. Isopropyl β-d-thiogalactoside was added to 0.3 mM and induction was allowed to proceed for 2.5 h. Cells were pelleted, washed once with phosphate-buffered saline (PBS), and stored frozen. Pellets were thawed, and resuspended in 10 ml of PBS containing 1 mM phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, 1 μg/ml aprotinin, and 1 μg/ml pepstatin A. Lysozyme was added to 200 μg/ml before a 10-min incubation on ice. Cells were sonicated five times for 15 s each with 1-min intervals of rest on ice and centrifuged 15 min at 10,000 × g at 4°C. The supernatant was loaded onto a column containing Talon resin (BD Biosciences Clontech, Palo Alto, CA) equilibrated in PBS, and fractions were eluted in PBS + 200 mM imidozole. Fractions with Cen1 were pooled, and 1 mg was sent to Animal Pharm (Healdsburg, CA), for antibody generation in rabbits.
Northern Blot Analysis
RNA was isolated using the TRI Reagent (Molecular Research Center, Cincinnati, OH) as per the manufacturer's instructions. Samples (each containing 10 μg of RNA) were run on formaldehyde gels and transferred to Hybond N+ (Amersham Biosciences) in 20× SSC (Ausubel et al., 1997). Northern blots were probed with randomly primed [α-32P]dATP-labeled fragments (Feinberg and Vogelstein, 1983), which were obtained by amplification by PCR of genomic DNA with the primers used to create the GFP fusions described above. Probes were specific for the target sequences as determined by Southern blot analysis of total DNA (our unpublished data).
Western Blot Analysis
Cells were lysed in buffer as described previously (Williams, 2000), and whole cell extracts were run on 12.5% or 11.5% standard Laemmli acrylamide gels (Ausubel et al., 1997) along with BenchMark prestained molecular weight standards (Invitrogen, Carlsbad, CA). Proteins were electrophoretically transferred to Immobilon-P membrane and blotted according to the manufacturer's instructions (Millipore, Billerica, MA). Membranes were incubated with antibodies diluted in Tris-buffered saline containing 0.05% Tween and 1% bovine serum albumin for 1 h at room temperature. Primary antibodies (either serum from bleed 4 of a Cen1-injected rabbit, an anti-GFP monoclonal antibody [mAb] [B34 from Abcam, Cambridge, MA]), or an anti-α-tubulin mAb (12G10; a kind gift of Joseph Frankel, University of Iowa, Iowa City, IA) were diluted 1:2000, 1:5000, and 1:1000, respectively. Secondary antibodies (anti-rabbit IR800 [Li-Cor Biosciences, Lincoln, NE], anti-mouse Alexa680 (Molecular Probes, Eugene, OR), anti Mouse horseradish peroxidase [HRP], and anti-rabbit HRP [Pierce Chemical, Rockford, IL]) were diluted 1:10,000. Blots were visualized either on a Li-Cor Odyssey scanner or by SuperSignal West Pico chemiluminescent substrate (Pierce Chemical) and exposure to film.
Macronuclear Transformations
Plasmids encoding GFP-centrin fusions were transformed into the developing macronucleus during late stages of conjugation of strains B2086 and CU428 by electroporation (Gaertig and Kapler, 2000). Paromomycin-resistant cells were selected and viewed on the fluorescence microscope. The cen1Δ rescuing construct was transformed into the developing macronucleus of the strains UCB8 and UCB9 during conjugation by electroporation, using KpnI, XhoI-digested grl3/4:BLAST-prCEN1–3′UTR, followed by selection with Blasticidin S (InvivoGen, San Diego, CA).
Micronuclear Knockout and Heterokaryon Generation
pMicCentrinKO1 was digested with SacI and introduced into the micronucleus of B2086- and CU428-conjugating cells by biolistic bombardment as described previously (Bruns and Cassidy-Hanley, 2000a). To check for proper integration of the CEN1 knockout construct, total Tetrahymena DNA was isolated (Gaertig et al., 1994b) and digested with NsiI and PstI. Standard Southern blot analysis was performed using a probe representing sequence upstream of the transforming fragment. The probe was generated by PCR amplification with primers CentrinUS1 and CentrinUS2 and labeled by the AlkPhos Direct (Amersham Biosciences) labeling system. Crosses to confirm that the transformation occurred in the micronucleus were performed, and the knockout heterokaryon was constructed, as described previously (Hai et al., 2000) with the modification that an outcross was performed on the original transformant strains to rid them of an apparent micronuclear aneuploidy before mating with B*VI.
Analysis of cen1Δ Cells
Cultures containing equal numbers of UCB8 and UCB9 cells were starved and mixed. Ten hours after mixing, an equal volume of 2× SPP (1% proteose peptone, 0.1% yeast extract, 0.2% glucose, 0.003% FeEDTA final) was added. Seventeen hours after mixing, paromomycin was added to 120 μg/ml to select against cells that had not gone through conjugation. Samples were removed for Western blotting at 6-, 25-, 50-, 75-, and 96-h time points. Samples were removed from separate mating cultures and prepared for immunofluorescence microscopy at 17-, 24-, 48-, and 72-h time points. Wild-type (WT) cells were treated with paromomycin as a control. To determine whether cells could continue to grow without Cen1, individual conjugating pairs were isolated into 30-μl drops of SPPA media, and cells were counted at 50 and 75 h.
Fluorescence Microscopy
Cells containing the GFP-Centrin fusions were placed on slides and allowed to dry just until cells no longer swam. GFP-Cen1 cells were from logarithmic cultures, GFP-Cen2 cells were from dense cultures, and GFP-Cen3 and GFP-Cen4 were from starved cultures, as dense and starved cultures had less nonspecific background signal. Immunofluorescence microscopy was performed as described previously (Stuart and Cole, 2000). Cells were fixed with both formaldehyde and EtOH as described previously. Fixed cells were visualized on Antibody slides (Bellco Glass, Vineland, NJ). Antibodies were diluted in PBS + 1% bovine serum albumin. Tetrahymena Cen1 antibody was diluted 1:1000, the 20H5 monoclonal centrin antibody raised against Chlamydomonas centrin (a generous gift from Dr. J. Salisbury, Mayo Clinic, Rochester, MN) was diluted 1:2500, the antibody against polyglutamylation (a generous gift from Dr. M. Gorovsky) (Shang et al., 2002) was diluted 1:500, and the 12G10 mAb against α-tubulin (a generous gift of Dr. J. Frankel, University of Rochester, Rochester, NY) was diluted 1:50. Secondary antibodies were used at a 1:1000 dilution and included goat anti-rabbit Alexa594, goat anti-rabbit Alexa488, and donkey anti-mouse Alexa488 (Molecular Probes). Antibody incubations were carried out at room temperature for 1 h, or alternatively, overnight at 4°C. To visualize DNA, 4,6-diamidino-2-phenylindole (DAPI) was applied to the cells at 1 μg/ml for 3 min. After each antibody incubation, cells were washed six times with PBS + 0.1% bovine serum albumin. Standard fluorescence microscopy was carried out using a Leica DMRXA/RF4/V automated microscope with a Cooke Sensi-Cam digital camera. Images were collected and subjected to no neighbors or nearest neighbors deconvolution algorithms using the Slidebook software package (Intelligent Imaging Innovations, Denver, CO).
Immunoelectron Microscopy
Tetrahymena cells were pelleted, frozen under high pressure in a BAL-TEC HPM-010 high-pressure freezer, freeze substituted in 0.25% gluteraldehyde/0.1% uranyl acetate, and embedded in Lowicryl HM20. Serial sections (55 nm) were taken and labeled with 1:200 dilutions of primary antibody in 1% nonfat milk phosphate-buffered saline/Tween 20, which included an affinity-purified rabbit anti-GFP primary antibody (a gift from Jason Kahana, Ludwig Institute for Cancer Research, New York, NY, and Pamela Silver, Dana-Farber Cancer Institute, Boston, MA) and the Cen1 antibody described above. A 1:20 dilution of 15-nm gold-conjugated goat anti-rabbit secondary antibody (Ted Pella, Redding, CA) was used. Grids were stained with uranyl acetate and lead citrate and were visualized in a Philips CM10 electron microscope.
RESULTS
Identification of a Tetrahymena Centrin Gene Family
We wished to isolate the T. thermophila centrin gene for analysis with respect to its potential role at basal bodies. A fragment was amplified by PCR from Tetrahymena genomic DNA using degenerate primers designed based on the alignment of several published centrins (see Materials and Methods). Flanking sequences were cloned by inverse PCR, and DNA sequencing revealed the fragment to be a centrin gene. During the course of this work, this centrin gene was independently reported by Guerra et al. (2003). We refer to this gene as CEN1.
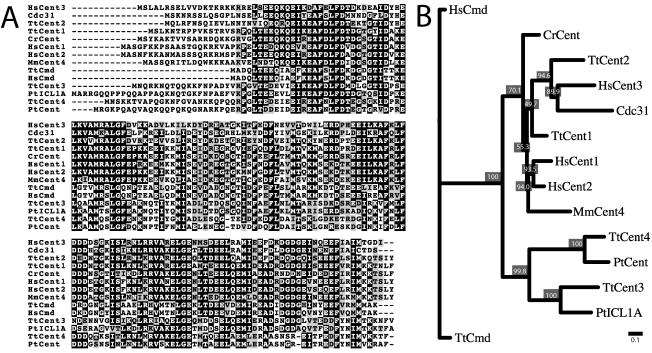
The corresponding protein sequence of CEN1 was used as a query to search the recently sequenced Tetrahymena genome at The Institute for Genomic Research (TIGR, www.tigr.org), and three additional potential centrin genes were identified. Two of these genes, called CEN3 and CEN4 herein, contain 510 and 515 base pairs, respectively, of uninterrupted protein coding sequence. The remaining gene, CEN2, contains 492 base pairs of coding sequence with three introns predicted by the protein alignment and standard splicing rules (Alberts et al., 2002). All three centrins had predicted molecular masses of 19.5 kDa, which is similar to Cen1's molecular mass of 19.4 kDa. The translated sequences were aligned with a variety of characterized centrins as well as the Tetrahymena and human calmodulins and placed in a phylogenetic tree (see Materials and Methods) (Figure 1). The phylogenetic analysis revealed three distinct branches: one for the calmodulins, one for two centrins identified in the ciliate Paramecium tetraurelia and CEN3 and CEN4 from Tetrahymena, and the third for all the remaining centrins included in the analysis. Of the Tetrahymena centrins, Cen1 is most similar to the Chlamydomonas centrin and human Centrin2, which are involved in duplication of basal bodies and centrioles, respectively (Salisbury et al., 2002; Koblenz et al., 2003). Cen2 is most closely related to budding yeast Cdc31, which is essential for spindle pole body duplication, and to human Centrin3. Cen3 and Cen4 are similar to centrins found in the infraciliary lattice of the ciliate P. tetraurelia (Klotz et al., 1997), and this more distantly related branch of the centrin family may be unique to the ciliates.
Figure 1.
Comparison of centrins from different organisms. Tt, T. thermophila; Hs, Homo sapiens; Mm, Mus musculus; Cr, Chlamydomonas reinhardtii; Pt, P. tetraurelia; Cdc31 from S. cerevisiae; Cmd, calmodulin. (A) Twelve representative centrins were aligned along with two calmodulin proteins. Residues where 50% or more are identical between the proteins are shaded black, and residues where 50% or more are similar are shaded gray. (B) The centrin and calmodulin sequences were subjected to phylogenetic analysis using the Neighbor-Joining method; 1000 replicates were performed, and a representative unrooted tree is displayed as a dendogram. Bootstrap values are displayed as percentages at each node and indicate how often the separation delineated by the branching occurred. Bar = 0.1 substitutions per one amino acid (see Materials and Methods).
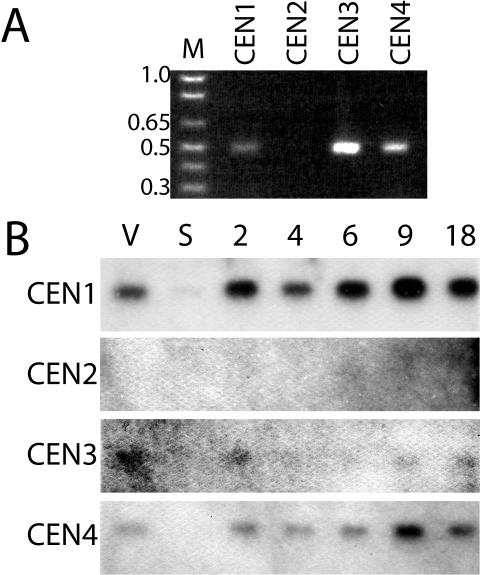
We checked for expression of these sequences to confirm that they encoded genes. Initially, PCR was carried out on a cDNA library generated from vegetatively growing cells, and an uncloned cDNA preparation also from vegetatively growing cells. In both cases, amplification of sequence resulting from primers specific to CEN1, CEN3, and CEN4 was observed, but not for primers specific to CEN2, which are capable of amplification of CEN2 from the genome (Figure 2A; our unpublished data). Five clones from each of these PCR products were sequenced, confirming that the primers used were specific for the targeted sequences. To determine whether CEN2 was expressed during other stages of the Tetrahymena life cycle and to examine expression patterns for the three other centrins, RNA was isolated from vegetatively growing cells, cells in starvation media, and cells at various time points through the mating cycle (conjugation) for Northern analysis with probes derived from each of the four centrin genes (Figure 2B). CEN1, CEN3, and CEN4 have similar expression profiles: bands of the expected size are observed during vegetative growth, and the intensity of these bands decreases in cells in starvation media, which were no longer dividing. During conjugation, which occurred in a starvation medium, expression increases to levels approximately twice those seen during logarithmic growth, peaking 9 h after initiation of mating. During conjugation, cells lose the oral apparatus, and its reformation is an early event after conjugants separate (Kiersnowska et al., 1993); the increase in the centrins's expression during later stages of conjugation could play a role in this process. Among the centrin genes, CEN1 seems to have the highest expression levels, followed by CEN4 and then CEN3, although it is possible that differences in band intensity may be the result of the ability of each probe to recognize the appropriate message. Because mRNA was detected for CEN1, CEN3, and CEN4, these sequences represent three bona fide members of a Centrin family in Tetrahymena. CEN2 expression was not detected for this limited set of conditions, although this result does not preclude expression during other untested growth conditions.
Figure 2.
Expression of genes in the Tetrahymena centrin family. (A) PCR products using primers specific for CEN1, CEN2, CEN3, and CEN4 on cDNA from cells during vegetative growth displayed on an agarose gel. Sizes of select marker (M) bands are displayed in kilobase pairs. (B) Northern blot analysis of RNA from vegetatively growing cells (V), starved cells (S), and conjugating cells at various time points after initiation of mating (2, 4, 6, 9, 18 h) (see Materials and Methods). Similar amounts of total RNA were loaded as assessed by absorbance spectroscopy and ethidium bromide staining our unpublished data).
GFP Tagging and Localization of Tetrahymena Centrin Family Members
To understand more about the functions of the centrin genes, we attempted to localize their products within the cell. Each gene was fused at its N terminus to the GFP (Chalfie et al., 1994) in the vector pVGF1 (Wiley et al., 2000). This vector is based on a ribosomal DNA processing vector in which rDNA sequences on the plasmid, into which the GFP fusion is inserted, are processed in the developing macronucleus, resulting in amplification and strong expression (Karrer, 2000). An N-terminal GFP-centrin fusion protein has been reported previously to localize correctly to centrioles in cell culture, and a C-terminal centrin-GFP fusion protein localized to Chlamydamonas basal bodies (White et al., 2000; Ruiz-Binder et al., 2002), although these fusions are probably not fully functional (Ruiz-Binder et al., 2002; Kilmartin, 2003). Although Cen2 mRNA is not normally detectable during vegetative growth, we tagged this protein along with the others to gain some understanding of where it might function if expressed. Transformants were recovered for each construct, and live cells were imaged by fluorescence microscopy. GFP-Cen1 and GFP-Cen2 both localized to the oral apparatus, a complex cytoskeletal structure rich in basal bodies, microtubule structures, and other nonmicrotubule-based filaments (Sattler and Staehelin, 1976; Frankel, 2000) and to cortical rows in a pattern similar to that observed for basal bodies. Interestingly, transformation of both constructs resulted in an apparent disorganization of the cortical rows, such that the rows sometimes ended prematurely, or curled around in unusual patterns, suggestive of a dominant-negative phenotype (Figure 4A, a and b). Consistent with these constructs being deleterious alleles, the transformants grew slowly, and neither construct was stably maintained over the course of extended vegetative growth. Nonetheless, Cen1 and Cen2 seem to localize to basal bodies.
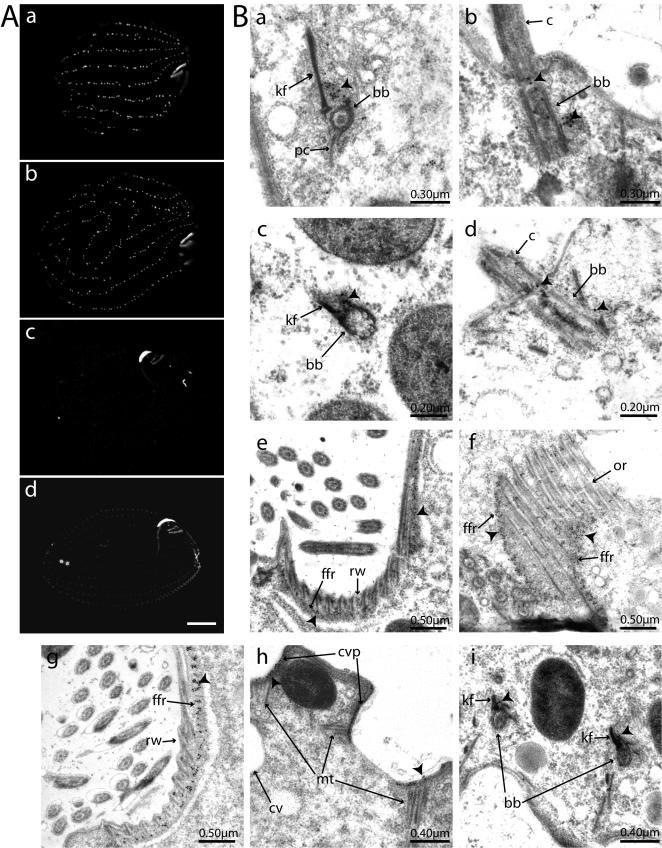
Figure 4.
Localization of GFP-centrin fusion proteins. (A) Fluorescence signal of live cells: a, GFP-Cen1; b, GFP-Cen2; c, GFP-Cen3; and d, GFP-Cen4. The oral apparatus is on the right side in each image. In a and to a greater extent b, cortical rows are affected by the overexpression of the GFP fusion protein. The strong signal in c is at the oral apparatus primarily in the oral crescent, internal to the undulating membrane, and in the apical band. These same regions are visible in d. The contractile vacuole pores are the two small circles near the posterior of the cell. Bar, 10 μm. (B) Immunoelectron microscopy with a GFP antibody and a 15-nm gold-conjugated secondary antibody. Arrowheads indicate representative concentrations of gold particles. a and b, GFP-Cen1 localization; c and d, GFP-Cen2 localization. kf, kinetodesmal fiber; pc, postciliary microtubules; bb, basal body; c, cilia. Representative cross-sectional (a and c) and longitudinal views (b and d) of basal bodies are shown. e and f, localization to the fine-filamentous reticulum of GFP-Cen3, with f also showing some localization along the oral ribs. Ffr, fine-filamentous reticulum; rw, ribbed wall; or, oral-ribs. GFP-Cen4 localization is shown in g, h, and i. g, localization in the fine filamentous reticulum of the oral apparatus. h, sections of both contractile vacuole pores as well as some associated microtubules. cvp, contractile vacuole pore; mt, microtubules; cv, contractile vacuole. h, two basal bodies in cross section with signal apparent along the kinetodesmal fibers.
In contrast, GFP-Cen3 and GFP-Cen4 transformants did not have any obvious growth defects. The fluorescence signal for Cen3 and Cen4 fusion proteins also was strikingly different from that of the other centrin fusions. GFP-Cen3 localized primarily to the oral apparatus. However, this fluorescence signal was distinct from the GFP-Cen1 and GFP-Cen2 signal at the oral apparatus, which was punctate and in all likelihood originated from GFP signal at the oral apparatus basal bodies. Rather, the GFP-Cen3 signal was found in a crescent-shaped region to the cell's left of the undulating membrane and extended along the deep rootlet. Signal also was present at the three membranelles of the oral apparatus, and in a crescent-shaped band at the apical tip of the cell (Figures 3B and 4A, c). GFP-Cen4 also localized to the structures where GFP-Cen3 was observed as well as to the contractile vacuole pores (CVPs) through which the contractile vacuole expels fluid at the posterior of the cell (Frankel, 2000). Additionally, faint signal was observed along the cortical rows, reminiscent of basal body localization (Figure 4A, d).
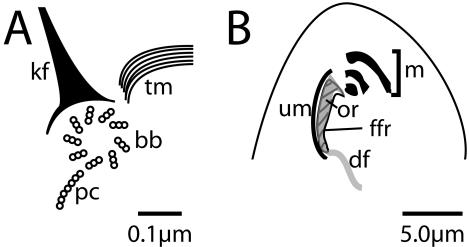
Figure 3.
Schematic drawings of centrin containing structures. (A) A basal body from a cortical row is represented along with some of the structures associated with it. bb, basal body; kf, kinetodesmal fiber; tm, transverse microtubules; pc, postciliary microtubules. (B) Some of the structures within the oral apparatus are represented. Um, undulating membrane; m, membranelles, of which there are three; or, oral-rib (gray in diagram); df, deep-fiber; ffr, fine-filamentous reticulum (striped in diagram). The undulating membrane and membranelles contain basal bodies and cilia. The oral-rib is a sheet of microtubules along the inside of the oral apparatus cavity beneath the undulating membrane, edged with ribbed walls (not labeled). The deep-fiber consists of a subset of microtubules that continue inward from the oral-rib into the cell. The fine-filamentous reticulum lies beneath the oral ribs.
We examined cells by immunoelectron microscopy to better resolve the localization of the four centrin fusions. Cells were prepared for electromicroscopy by high-pressure freezing and freeze substitution, and sections were stained using an antibody that recognizes GFP followed by a secondary antibody conjugated to 15-nm gold particles (see Materials and Methods). GFP-Cen1 is abundant at the base of the basal body adjacent to the kinetodesmal fiber, a nonmicrotubule-based striated rootlet that extends from the basal body toward the anterior of the cell, at the location where new basal body formation occurs (Figures 3A and 4B, a and b) (Allen, 1969). Additionally, signal was observed along the length of the basal body, with a second concentration at the transition zone between the basal body and the ciliary axoneme. Transition zone localization for centrin has been reported in Chlamydomonas (Sanders and Salisbury, 1994). The overall signal from GFP-Cen2 was significantly weaker than that for GFP-Cen1 in this experiment, but when multiple images were analyzed signal was detected in a pattern similar to that of GFP-Cen1 (Figure 4B, c and d). GFP-Cen3 and GFP-Cen4 both localized primarily to the region directly underneath the ribbed wall and the oral-ribs within the oral apparatus that corresponds to the fine filamentous reticulum (Williams and Luft, 1968; Williams and Bakowska, 1982) (Figures 3B and 4B, e–g), and GFP-Cen4 signal was clearly detected at the contractile vacuole pores (Figure 4B, h). Weak GFP-Cen4 signal also was observed at the basal bodies, primarily on the kinetodesmal fiber (Figure 4B, i). When a strain not expressing GFP was submitted to identical antibody incubations, there was almost no background signal apparent, supporting the authenticity of the localization data (our unpublished data).
Together, the localization of the GFP fusion proteins and the expression patterns of the different centrins suggest that each member of the Tetrahymena centrin family has a unique role within the cell. The possibility remains that there is some redundancy among the centrin genes, particularly between CEN3 and CEN4, because their protein products have similar distributions within the oral apparatus. Because our primary interest is the role of centrin in basal body duplication during vegetative growth, we chose to focus on Cen1. Of the four Tetrahymena centrins, Cen1 is most similar to human centrin2, which is required for centriole duplication. Furthermore, we have shown that Cen1 is expressed strongly during logarithmic growth, and a GFP fusion protein localizes to basal bodies at the precise location where initiation of basal body duplication is thought to occur (Allen, 1969).
Basal Body Localization of Endogenous Cen1
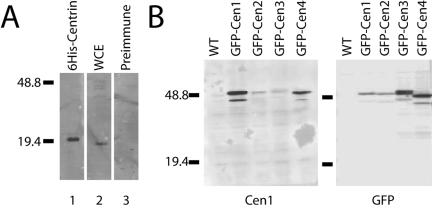
A polyclonal antibody was raised against full-length 6-histidine–tagged Cen1 (see Materials and Methods) and initially characterized by Western blot analysis. The antibody detected recombinant 6-histidine–tagged Cen1 (Figure 5A, lane 1), and a band the size expected for centrin (19.5 kDa) in a whole cell Tetrahymena extract (lane 2). We then took advantage of the GFP-centrin fusion strains, to examine the antibody's reactivity with each individual Tetrahymena centrin family member. GFP adds approximately 27 kDa to the molecular mass of the protein, thus allowing for easy separation of the tagged allele from the endogenous proteins, which all have a molecular mass of 19.5 kDa. To control for the individual expression level of each protein, antibody recognizing the GFP tag was used as a control. As expected, the Cen1 antibody strongly recognized GFP-Cen1 (Figure 5B, left). The antibody had minimal cross-reactivity with GFP-Cen3 and slightly more cross reactivity with GFP-Cen2 and GFP-Cen4 compared with the GFP probed immunoblot. Endogenous centrin was clearly visible with the Cen1 antibody on longer exposures (our unpublished data). We therefore concluded that our antibody is largely specific to Cen1, but it may interact with the other centrins at a low-to-moderate level.
Figure 5.
Reactivity of the Cen1 antibody. (A) Western immunoblot analysis using the Cen1 antibody as a probe on recombinant 6-histidine–tagged Centrin (lane 1) and Tetrahymena whole cell extract (wce) from cells in logarithmic growth (lane 2). The reactivity of the preimmune serum on wce also is shown (lane 3). (B) Reactivity of the Cen1 antibody with the overexpressed GFP-centrin fusion proteins. Whole cell extracts from strains expressing different fusion proteins were probed with either the Cen1 antibody (left) or a GFP antibody (right). Because the fusion proteins are overexpressed, the signal overwhelms the endogenous Centrin signal, which is visible with longer exposures. Select marker bands are noted and sizes are in kilodaltons.
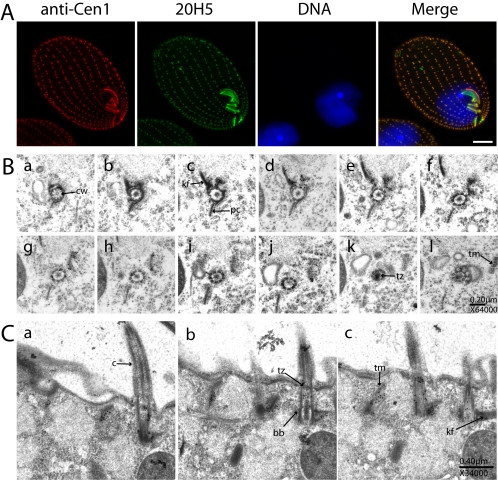
To examine localization of endogenous Cen1, we used the Cen1 antibody for immunofluorescence and immunoelectron microscopy. Immunofluorescence was performed on cells from a logarithmic culture (Figure 6A) using our rabbit polyclonal antibody (anti-Cen1), and a mAb (20H5) that recognizes centrins from a variety of species and has previously been used to localize centrin in Tetrahymena (Jerka-Dziadosz et al., 1995; Guerra et al., 2003). Staining patterns from each antibody were very similar but not identical. Both recognized the basal bodies along the cortical rows and in the oral apparatus, but the 20H5 antibody recognized additional structures that the Cen1 antibody did not. The signal from 20H5 was observed at the oral crescent (corresponding to the fine filamentous reticulum) and also at the apical filamentous band (Jerka-Dziadosz, 1981; Jerka-Dziadosz et al., 1995). Both these structures were revealed by GFP-Cen3 and GFP-Cen4. However, the 20H5 antibody did not recognize the contractile vacuole pores, raising the possibility that this antibody can recognize Cen1 and Cen3, but not Cen4, or that Cen4 is not accessible to the antibody at the CVPs. Consistent with this interpretation, the 20H5 antibody strongly recognized Cen1 and Cen3 by Western blot analysis, whereas showing moderate affinity to Cen4 and virtually no affinity for Cen2 (our unpublished data). Like the GFP-Cen1 data, data from the Cen1 antibody indicates that Cen1 localizes specifically to basal bodies.
Figure 6.
Cen1 localization at basal bodies. (A) Immunofluorescence microscopy using the Cen1 antibody and the monoclonal centrin antibody 20H5 raised against Chlamydamonas centrin. DNA is visualized with DAPI. Bar, 10 μm. (B) Serial section immunoelectron microscopy of a basal body in cross section with the Cen1 antibody. Panels proceed from the axoneme distal part of the basal body (a) and track upwards through the structure. Figure 3A serves as a reference for some of the basal body associated structures that are present. Certain basal body associated structures are identified in sections where they are most prominent, although this does not preclude these same structures from being visible in other sections. cw, cartwheel; kf, kinetodesmal fiber; pc, postciliary microtubules; tz, transition zone; tm, transverse microtubules; c, cilia; bb, basal body. Centrin concentrates on the transverse microtubules that are seen obliquely, on the kinetodesmal fiber, and at the transition zone (k). (C) Serial section immunoelectron microscopy of three basal bodies in longitudinal section.
To map Cen1 localization within basal bodies, immunoelectron microscopy was performed on serial sections of cells from a logarithmic culture using the Cen1 polyclonal antibody. This analysis on endogenous levels of Cen1 revealed a similar localization pattern to that observed with the GFP-Cen1 strain. Serial sections through six basal bodies where examined. At the basal body, there are two major centers of Cen1 concentration. The first is at the base of the basal body, along and adjacent to the kinetodesmal fiber (Figures 3A, 6B, a–e, and 6C, a–c). At the very base of the basal body (most distal to the axoneme), this signal is immediately adjacent to the basal body (Figure 6B, a), the site of new basal body formation (Allen, 1969). As the cross-sectional serial sections are tracked through the basal body, this signal moves away from the basal body, following a path along the transverse microtubules that extend to the adjacent cortical row (for reference, see Figures 3A and 6B, c–j). There is also a moderate amount of signal from the Cen1 antibody within the basal body cylinder that extends the length of the basal body immediately adjacent to basal body microtubules (Figure 6B, g–j). The second concentration of Cen1 occurs at the transition zone between the basal body and the axoneme (Figure 6B, k). These localization patterns also were observed in longitudinal sections (Figure 5C, a–c).
Cen1 Is Required for Basal Body Duplication and Maintenance in Tetrahymena
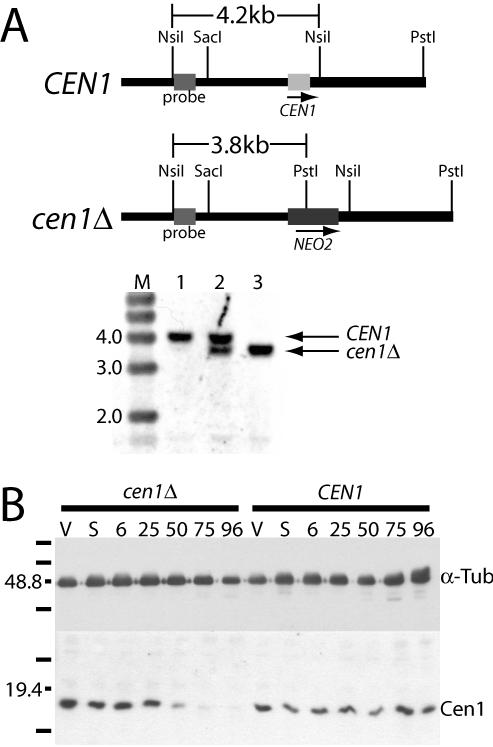
Both Cen1's localization to the base of the basal body where duplication is initiated, and the disruption of the cortical rows caused by overexpression of the GFP-Cen1 fusion protein, are suggestive that Cen1 has an important role at the basal body. To assess this more directly, a null strain was created. We replaced CEN1 with the NEO2 drug resistance cassette, which provides resistance to paromomycin (Gaertig et al., 1994a), in the diploid micronucleus by homologous recombination (Bruns and Cassidy-Hanley, 2000a), as confirmed by Southern analysis (Figure 7A). This strain was then used to create a knockout heterokaryon (Hai et al., 2000), a strain homozygous for the knockout in the micronucleus, but containing only wild-type CEN1 in the expressed macronucleus. When two such strains with different mating types are mated to each other, a new macronucleus is formed based on the DNA of the germline micronuclei of the parent strains. The resulting progeny lack the CEN1 gene, having the NEO2 gene cassette in its place (Figure 7A). CEN1 knockout heterokaryon strains UCB8 and UCB9 were mated to each other, and 120 μg/ml paromomycin was added to select for cells that had proceeded successfully through mating and development of progeny. The cell density of the culture increased only slightly by 72 h after initiation of mating (starting at 2.5 × 105 cells/ml, and increasing to 2.8 × 105 cells/ml), and eventually all cells died, either due to sensitivity to paromomycin (nonmating, parental cells) or due to the lack of Cen1 (progeny). No “survivors” were observed in these cultures, indicating that none of the other centrins can substitute for Cen1 in its essential function. Western blot analysis confirmed that Cen1 protein levels were reduced significantly by 50 h after mating and that the reduction in Cen1 protein levels continued over time (Figure 7B). These Western blot results further indicate that either the antibody is very specific for Cen1 and/or Cen1 is the predominantly expressed centrin in cells, because the decrease in Cen1 was not masked by the other centrins.
Figure 7.
Cen1 protein levels decrease when CEN1 is deleted. (A) Southern blot analysis of the CEN1 locus. The fragment used for transformation extends from the SacI site to the PstI site with the NEO2 cassette replacing CEN1. Total Tetrahymena DNA from wild-type cells (lane 1), the micronuclear transformant (lane 2), and progeny from the knockout heterokaryon strains rescued by CEN1 transformed at a second genomic location (lane 3) was digested with NsiI and PstI. The sizes of select marker bands (M) are noted in kilobase pairs. (B) Western blot analysis of progeny from the cen1Δ knockout heterokaryon strains UCB8 and UCB9 (cen1Δ) and of the progeny from wild-type strains B2086 and CU428 (CEN1). Cen1 is visualized with the Cen1 polyclonal antibody, and a mAb to α-tubulin is used as a loading control. V, vegetative growth; S, starvation; numbers correspond to hours after parental cells were mixed. Marker bands are noted, and sizes are in kilodaltons.
The ability of cen1Δ cells to divide was tested by mating UCB8 and UCB9 cen1Δ knockout heterokaryon strains and isolating individual conjugating pairs into small drops of media. As a control, B2086 and CU428 cells (both wild-type for CEN1) were subjected to the same procedure. Cells were counted after 50 h of growth and checked again at 75 h, and an estimate as to how many divisions each cell had completed was made. Both wild-type and cen1Δ cells had a significant population of cells that were completely unable to divide after conjugation (29 and 55%, respectively). Most wild-type cells went on to form stable vegetative cultures (62%). In contrast, none of the cen1Δ cells progressed to form a stable vegetative culture. However, of the 45% that did divide after conjugation, more than half divided approximately three times. Therefore, at least some of the cen1Δ strains were able to undergo several cell divisions before arresting and eventually dying.
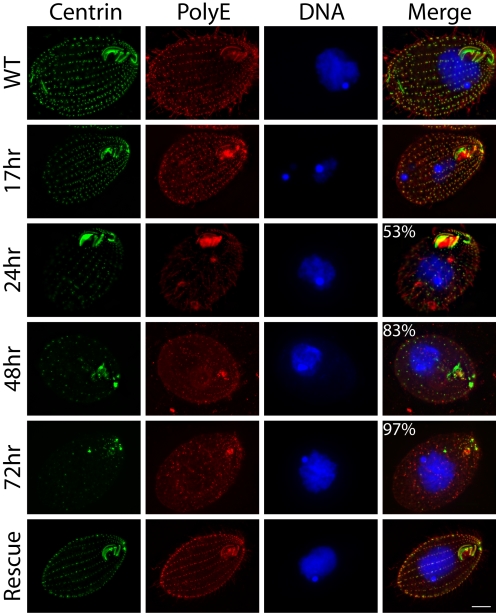
The effect of the lack of Cen1 on basal body integrity was observed by fixing and staining cells at various time points after mating with both the 20H5 monoclonal centrin antibody and an antibody raised against a polyglutamic acid peptide that recognizes a tubulin modification, polyglutamylation, that is abundant at basal bodies (Shang et al., 2002) (Figure 8). The number of cells with an obviously reduced number of basal bodies compared with wild-type was counted at each time point. Cells proceeded through conjugation without apparent problems, and 17 h after initiation of mating the cells had separated and looked normal. However, 24 h after mating, some cells displayed reduced centrin signal and fewer basal bodies along the cortical rows. By 48 h, cells had fewer basal bodies, and by 72 h most of the cells showed a severe phenotype, including loss of most basal bodies, disorganization of the remainder along the cortical rows, and loss of the oral apparatus. Cell division intermediates were observed in the samples, and although these cells looked capable of completing the division cycle, because they did not accumulate at later time points, they were unable to form a new oral apparatus and had fewer basal bodies, especially in the posterior half of the cell (Supplemental Figure 1A). Additionally, there was signal from the polyglutamic acid antibody at locations where there was no centrin signal, indicating that there was no longer enough Cen1 to localize to all of the remaining basal bodies.
Figure 8.
Deletion of CEN1 results in a failure to duplicate basal bodies. cen1Δ knockout heterokaryon strains UCB8 and UCB9 were mated, and their progeny were analyzed at 17-, 24-, 48-, and 72-h time points by immunofluorescence microscopy. The monoclonal centrin antibody 20H5 was used along with an antibody that recognizes polyglutamic acid, which is prominent on modified tubulin at basal bodies. DNA is visualized with DAPI. WT is strain CU428 in logarithmic growth. Percentages refer to the proportion of cells with reduced numbers of basal bodies, with the images shown being representative (n = 200). Rescue refers to UCB8 × UCB9 progeny that were rescued by CEN1 transformed into a second site in the genome. In the WT and 17-h samples, 100% of cells had an apparently normal number of basal bodies. The oral apparatus is oriented on the right side of each panel. Bar, 10 μm.
To ensure that the loss of basal bodies was due to the lack of Cen1, and not the result of the introduction of paromomycin to the cultures, three controls were performed. First, UCB8 and UCB9 cen1Δ knockout heterokaryon strains were mated to each other, and the resulting culture was allowed to progress in the absence of drug, allowing parental, unmated cells to largely overtake the population. Nevertheless, the loss of centrin signal and basal bodies was still observed in some cells, whereas in a wild-type control, cross-centrin signal remained high throughout the population (Supplemental Figure 1B; our unpublished data). In the second control, a wild-type strain was subjected to 120 μg/ml paromomycin and observed at time points after addition of drug similar to those used in the experiment with the knockout strains. Although dying cells often had disorganized basal bodies, these cells were distinct from cen1 knockout cells in that centrin signal remained strong, and these cells were small and rounded (Supplemental Figure 1C). In the third control, a construct that reintroduces CEN1 at the GRL3/4 locus (Verbsky and Turkewitz, 1998) rescued cen1Δ cells. UCB8 and UCB9 cells were mated, and this construct was transformed into the developing macronucleus by electroporation. Transformants were recovered that were resistant both to blasticidin S, which selects for the bsr gene used in this construct, and to paromomycin, which selects for the NEO2 cassette used to disrupt CEN1 (Figure 7A; our unpublished data). Rescued cultures grew normally and were subjected to immunofluorescence microscopy (Figure 8, bottom). The cortical rows and oral apparatus in these cells were indistinguishable from wild type, indicating that when Cen1 is restored to these cells, they no longer have basal body defects.
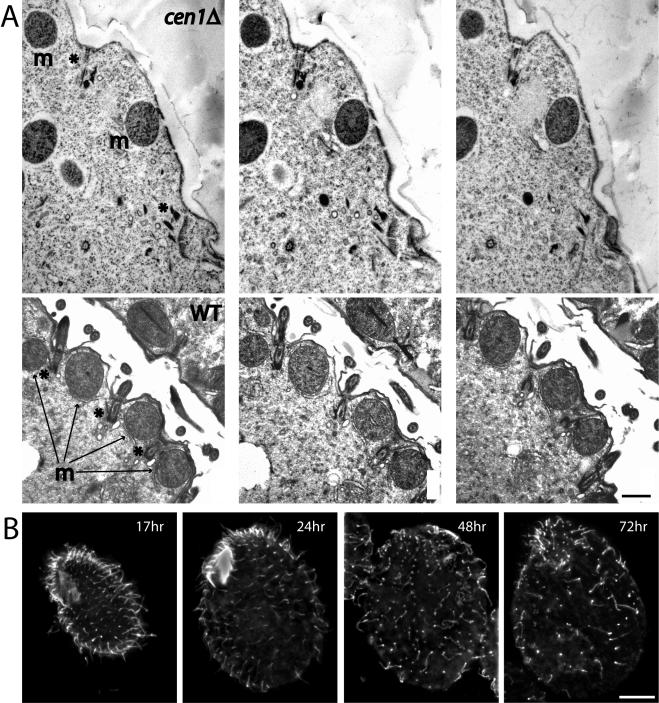
The immunofluorescence data on the cen1Δ strain strongly suggests that basal bodies are absent from these cells at later time points. However, fluorescence signal at late time points was still observed, more so with the antibody to polyglutamic acid, and the question remained whether this signal was at basal bodies, or some identifiable remnant of basal bodies. To address this issue, electron microscopy was performed on cells processed 68 h after the initial mixing of the two knockout heterokaryon strains. Fewer basal bodies were observed in sections from the cen1 knockout strains compared with a wild-type control cross. When basal bodies were detected, they often seemed incomplete without cilia attached (Figure 9A). Occasionally the electron dense structures associated with the base of the basal body were present without all of the microtubules that normally comprise a basal body (our unpublished data). Immunoelectron microscopy with the Cen1 antibody revealed that basal bodies with more Cen1 were better preserved than basal bodies with less antibody signal (our unpublished data). The possibility that Cen1 is important for maintenance of basal bodies was addressed further by immunofluorescence microscopy using an antibody to α-tubulin, which detects cilia, a marker for a functioning basal body, as well as basal bodies. By 48 h after the initiation of mating of cen1Δ cells, many punctate spots reminiscent of basal bodies remained but a large percentage of these did not have attached cilia, indicating the absence of functional basal bodies at these sites (Figure 9B). Together, the results from the micronuclear knockout strains indicate that centrin is important for both the duplication and stability of basal bodies.
Figure 9.
Cen1 is required for basal body maintenance. (A) Serial sections of a cen1Δ cell (top) and a cell from a wild-type control cross (bottom). The asterisks (*) (top left) indicate the location of one incomplete basal body and a second site of interest. The basal body (top left) does not nucleate a cilium and seems disorganized at its base. The material at the bottom right is unrecognizable as a basal body, although it is present where a basal body would be expected given the location of the mitochondrion on the right (m) and the indentation in the cell surface. In the wild-type panels, three basal bodies (*), two with cilia clearly attached, are visible. Basal bodies are between mitochondria (m), and the cilia emerge from indentations in the cell surface. Bar, 0.5 μm. (B) α-Tubulin localization in cen1Δ cells. Times are after mating initiation of the cen1Δ knockout heterokaryon strains, and cells were stained with a mAb raised against α-tubulin (12G10). By 48 h, many apparent basal bodies do not have cilia attached. Bar, 10 μm.
DISCUSSION
We report the initial characterization of a family of four centrin genes in the ciliate T. thermophila. Two of these genes are closely related to the majority of the centrin genes found in other organisms, whereas the other two are related to the centrin genes identified in another ciliate, P. tetraurelia. Ciliates contain many distinct and highly specialized cytoskeletal structures. In Tetrahymena, these include basal bodies, cilia, micronuclear spindles, macronuclear microtubule arrays, a cortical network organized around the basal bodies of the ciliary rows, the contractile vacuole pores, and the complex microtubule and filamentous structures that comprise the oral apparatus (Gaertig et al., 1993; Frankel, 2000). Because centrins play multiple roles at structures associated with microtubules (Taillon et al., 1992), it would not be surprising if specialized centrins evolved along with some of these structures unique to ciliates. For example, GFP-Cen3 localizes predominantly to the fine filamentous reticulum of the oral apparatus and a band at the apex of the cell, and GFP-Cen4 localizes primarily to these structures and to the contractile vacuole pores. These structures are involved in dynamic processes: the undulating membrane of the oral apparatus sweeps media (food) into the oral cavity, whereas when the contractile vacuole expands, contracts, and ruptures, fluid is expelled through the CVPs maintaining the cell's proper osmolarity. The function of the apical band is not known; it is distinct from the contractile division furrow ring, although it, too, may be contractile and involved in regulating cell shape (Jerka-Dziadosz, 1981). In Paramecium multimicrnucleatum, the number of contractile vacuoles increases in response to an increased amount of calcium in the environment (Iwamoto et al., 2003). The assumption is that this is a response with which the cell rids itself of excess calcium, but it is possible that calcium, through centrin, more directly stimulates the assembly of this structure. Although the roles for Cen3 and Cen4 at the fine filamentous reticulum and the contractile vacuole pore are not known, it is possible the contractile properties of centrin affect the dynamic properties of these structures (Salisbury et al., 1984). Additionally, Tetrahymena Cen1 along with inner arm dynein is involved in ciliary beat changes in response to calcium (Hennessey et al., 2002; Guerra et al., 2003), and perhaps centrins at the fine filamentous reticulum perform a similar task for this structure.
Tetrahymena CEN1 and CEN2 are on branches of the centrin tree that are most widely conserved throughout the eukaryotic world, and here we examined the function of Cen1 in MTOC duplication. Centrin is required for this process in species as diverse as yeast, green algae, and humans (Baum et al., 1986; Salisbury et al., 2002; Koblenz et al., 2003). Although GFP-Cen1 and GFP-Cen2 both localize to basal bodies, CEN2 is not normally expressed during vegetative growth, or conjugation. This situation may be similar to that in humans, in which centrin1 expression is limited to the testes and retinal cells (Hart et al., 1999). We have not determined whether CEN2 is expressed under different conditions, or not at all, although the similarity to other centrins, in particular yeast Cdc31 and human centrin3, leads us to suspect that it is a functional gene. CEN1, however, is highly expressed in vegetative and mating cells. Localization of the endogenous protein places the majority of Cen1 at two positions on the basal body. One is at the transition zone between the basal body and the axoneme of the cilium, as has been reported in Chlamydomonas (Sanders and Salisbury, 1994). The second is at the base of the basal body adjacent to the kinetodesmal fiber where probasal body formation is initiated (Allen, 1969). Disruption of CEN1 led to the progressive loss of basal bodies. Of the basal bodies that remained, many were defective and unable to act as MTOCs for the cilia. Cen1 is a stable molecule, and small amounts of Cen1 remained even 72 h after mating initiation of the knockout heterokaryon strains. Therefore, our experiments examine the results of a slow depletion of Cen1. As Cen1 decreased, cells were unable to fill their cortical rows with the proper number of basal bodies, or to form a new oral apparatus, suggesting the requirement for Cen1 in the duplication of basal bodies.
The deterioration of the remaining basal bodies indicates that Cen1 also is required for proper basal body maintenance. The depletion of γ-tubulin yielded a related phenotype, demonstrating the necessity for this protein in basal body maintenance as well as basal body duplication. When γ-tubulin is depleted, oral apparatus basal bodies are lost before cortical basal bodies, suggesting that turnover of γ-tubulin is more rapid in the oral apparatus than at the basal bodies of the cortical rows, and required for these organelles' stability (Shang et al., 2002). The CEN1 knockout strain had no such hierarchy of loss, indicating that turnover of Cen1 is not significantly different at basal bodies along the cortical rows and basal bodies at the oral apparatus. Instead, Cen1 seems to be very stably associated with basal bodies and plays some role in preserving these organelles. Hence, a cen1Δ cell preparing for division can have one existing oral apparatus that seems normal and a highly aberrant developing oral apparatus (Supplemental Figure 1A). We do not know whether Cen1's roles in basal body duplication and maintenance can be separated, but it is tempting to speculate that Cen1 localization to the base of the basal body is important for duplication, whereas localization to the transition zone is important for basal body maintenance and function. Future work on Cen1 binding partners will shed light on this issue.
Our experiments did not reveal any association between the Tetrahymena centrin proteins and the micronucleus. This is somewhat surprising because centrins are found at mitotic spindle poles in a wide range of organisms. This supports the previous finding that centrin was not detected with γ-tubulin at the micronucleus (Shang et al., 2002). However, centrin antibodies tested to date have not revealed centrin localization at the contractile vacuole pores, and our analysis of Cen4 indicates that it is likely present at this structure. Thus, the possibility remains that the experiments have not been sensitive enough to reveal centrin at spindle poles. Also, the GFP tag could interfere with centrin localization to the spindle pole in Tetrahymena. Another possibility is that there are yet more Tetrahymena centrin genes encoding isoforms that are at the spindle pole. Finally, it is possible that Tetrahymena does not require centrin for its mitotic apparatus. Few components of the Tetrahymena mitotic apparatus have been identified, and this issue may take some time to resolve.
We have shown there exists a family of Tetrahymena centrin genes with at least three bona fide members based on expression. Two of the centrins likely function at specialized fibrillar structures in the oral apparatus and the apical tip, and one of these also may act at the contractile vacuole pore. The predominant centrin, encoded by CEN1, localizes to basal bodies, and is required for their duplication and maintenance. The genetic malleability of these cells will likely lead to further insights into the mechanisms by which Cen1 functions at the basal body, and Cen3 and Cen4 function at their respective locations in the cell.
Supplementary Material
Acknowledgments
This manuscript and our understanding of the Tetrahymena cytoskeleton were greatly enhanced by Joe Frankel's careful reading and advice. We thank Chandler Fulton, Joe Frankel, Jeffrey Salisbury, and Marty Goroversusky for antibodies; Peter Bruns and Aaron Turkewitz for strains; and Jacek Gaertig and Aaron Turkewitz for cDNA preparations. David Prescott, Eric Cole, Aaron Turkewitz, Meng Chao Yao, Joe Frankel, and Jacek Gaertig were very helpful with advice on the molecular biology, genetics, and/or cytology of Tetrahymena. Preliminary sequence data were obtained from The Institute for Genomic Research Web site at http://www.tigr.org. Ed Orias and Jonathan Eisen helped with genome sequence issues. We are particularly indebted to Craig Weaver at Martek Biosciences (Boulder, CO) for help with their Bio-Rad PDS-1000/He Biolistics Apparatus. Danielle Perry was instrumental in expressing Cen1 in E. coli. Catherine Lozupone helped with the phylogenetic analysis of the centrin family. We also thank Michele Jones and Chad Pearson for helpful comments on the manuscript, and the members of the laboratory for support. This work has been supported by National Institutes of Health Grant GM-067898 (to M. W.). Early phases of the project at Colgate University were supported by National Science Foundation Grant MCB-9727240 (to H.B.M.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–10–0919) on June 8, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adams, I. R., and Kilmartin, J. V. (2000). Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 10, 329–335. [DOI] [PubMed] [Google Scholar]
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. (2002). Molecular Biology of the Cell, New York: Garland Science.
- Allen, R. D. (1969). The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J. Cell Biol. 40, 716–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smoth, J. A., and Struhl, K. (1997). Current Protocols in Molecular Biology, New York: John Wiley & Sons, Inc.
- Baum, P., Furlong, C., and Byers, B. (1986). Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc. Natl. Acad. Sci. USA 83, 5512–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson, J., and Wright, M. (2003). Basal body/centriole assembly and continuity. Curr. Opin. Cell Biol. 15, 96–104. [DOI] [PubMed] [Google Scholar]
- Bruns, P. J., and Cassidy-Hanley, D. (2000a). Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 62, 501–512. [DOI] [PubMed] [Google Scholar]
- Bruns, P. J., and Cassidy-Hanley, D. (2000b). Methods for genetic analysis. Methods Cell Biol. 62, 229–240. [DOI] [PubMed] [Google Scholar]
- Byers, B. (1981). Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae. In: Molecular Genetics in Yeast, ed. D. Von Wettstein, J. Friis, M. Kielland-Brandt, and A. Stenderup, Copenhagen, Denmark: Munksgaard, 119–131.
- Chalfie, M., Tu, Y., Euskirchen, G., Ward, W., and Prasher, D. (1994). Green fluorescent protein as a marker for gene expression Science 263, 802–805. [DOI] [PubMed] [Google Scholar]
- Chapman, M. J., Dolan, M. F., and Margulis, L. (2000). Centrioles and kinetosomes: form, function, and evolution. Q. Rev. Biol. 75, 409–429. [DOI] [PubMed] [Google Scholar]
- Errabolu, R., Sanders, M. A., and Salisbury, J. L. (1994). Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J. Cell Sci. 107, 9–16. [DOI] [PubMed] [Google Scholar]
- Feinberg, A. P., and Vogelstein, B. (1983). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Fischer, T., Rodriguez-Navarro, S., Pereira, G., Racz, A., Schiebel, E., and Hurt, E. (2004). Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat. Cell Biol. 6, 840–848. [DOI] [PubMed] [Google Scholar]
- Frankel, J. (2000). Cell biology of Tetrahymena thermophila. Methods Cell Biol. 62, 27–125. [DOI] [PubMed] [Google Scholar]
- Frankel, J., and Nelsen, E. (2001). The effects of supraoptimal temperatures on population growth and cortical patterning in Tetrahymena thermophila: a comparison. J. Eukaryot. Microbiol. 48, 135–146. [DOI] [PubMed] [Google Scholar]
- Gaertig, J., Gu, L., Hai, B., and Gorovsky, M. A. (1994a). High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 22, 5391–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig, J., and Kapler, G. (2000). Transient and stable DNA transformation of Tetrahymena thermophila by electroporation. Methods Cell Biol. 62, 485–500. [DOI] [PubMed] [Google Scholar]
- Gaertig, J., Thatcher, T. H., Gu, L., and Gorovsky, M. A. (1994b). Electroporation-mediated replacement of a positively and negatively selectable β-tubulin gene in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 91, 4549–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig, J., Thatcher, T. H., McGrath, K. E., Callahan, R. C., and Gorovsky, M. A. (1993). Perspectives on tubulin isotype function and evolution based on the observation that Tetrahymena thermophila microtubules contain a single α- and β-tubulin. Cell Motil. Cytoskeleton 25, 243–253. [DOI] [PubMed] [Google Scholar]
- Guerra, C., Wada, Y., Leick, V., Bell, A., and Satir, P. (2003). Cloning, localization, and axonemal function of Tetrahymena centrin. Mol. Biol. Cell 14, 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai, B., Gaertig, J., and Gorovsky, M. A. (2000). Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 62, 513–531. [DOI] [PubMed] [Google Scholar]
- Hart, P. E., Glantz, J. N., Orth, J. D., Poynter, G. M., and Salisbury, J. L. (1999). Testis-specific murine centrin, Cetn 1, genomic characterization and evidence for retroposition of a gene encoding a centrosome protein. Genomics 60, 111–120. [DOI] [PubMed] [Google Scholar]
- Hennessey, T. M., Kim, D. Y., Oberski, D. J., Hard, R., Rankin, S. A., and Pennock, D. G. (2002). Inner arm dynein 1 is essential for Ca2+-dependent ciliary reversals in Tetrahymena thermophila. Cell Motil. Cytoskeleton 53, 281–288. [DOI] [PubMed] [Google Scholar]
- Iwamoto, M., Allen, R. D., and Naitoh, Y. (2003). Hypo-osmotic or Ca2+-rich external conditions trigger extra contractile vacuole complex generation in Paramecium multimicronucleatum. J. Exp. Biol. 206, 4467–4473. [DOI] [PubMed] [Google Scholar]
- Jensen, C. G., Davison, E. A., Bowser, S. S., and Rieder, C. L. (1987). Primary cilia cycle in PtK1 cells: effects of colcemid and taxol on cilia formation and resorption. Cell Motil. Cytoskeleton 7, 187–197. [DOI] [PubMed] [Google Scholar]
- Jerka-Dziadosz, M. (1981). Cytoskeleton-related structures in Tetrahymena thermophila: microfilaments at the apical and division-furrow rings. J. Cell Sci. 51, 241–253. [DOI] [PubMed] [Google Scholar]
- Jerka-Dziadosz, M., Jenkins, L. M., Nelsen, E. M., Williams, N. E., Jaeckel-Williams, R., and Frankel, J. (1995). Cellular polarity in ciliates: persistence of global polarity in a disorganized mutant of Tetrahymena thermophila that disrupts cytoskeletal organization. Dev. Biol. 169, 644–661. [DOI] [PubMed] [Google Scholar]
- Karrer, K. M. (2000). Tetrahymena genetics: two nuclei are better than one. Methods Cell Biol. 62, 127–186. [DOI] [PubMed] [Google Scholar]
- Kiersnowska, M., Kaczanowski, A., de Haller, G. (1993). Inhibition of oral morphogenesis during conjugation of Tetrahymena thermophila and its resumption after cell separation. Eur. J. Protistol. 29, 359–369. [DOI] [PubMed] [Google Scholar]
- Kilmartin, J. V. (2003). Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 162, 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz, C., Garreau de Loubresse, N., Ruiz, F., and Beisson, J. (1997). Genetic evidence for a role of centrin-associated proteins in the organization and dynamics of the infraciliary lattice in Paramecium. Cell Motil. Cytoskeleton 38, 172–186. [DOI] [PubMed] [Google Scholar]
- Koblenz, B., Schoppmeier, J., Grunow, A., and Lechtreck, K. F. (2003). Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J. Cell Sci. 116, 2635–2646. [DOI] [PubMed] [Google Scholar]
- Laoukili, J., Perret, E., Middendorp, S., Houcine, O., Guennou, C., Marano, F., Bornens, M., and Tournier, F. (2000). Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J. Cell Sci. 113, 1355–1364. [DOI] [PubMed] [Google Scholar]
- LeDizet, M., and Piperno, G. (1995). The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol. Biol. Cell 6, 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, V. D., and Huang, B. (1993). Molecular cloning and centrosomal localization of human caltractin. Proc. Natl. Acad. Sci. USA 90, 11039–11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeddu, L., Klotz, C., Le Caer, J. P., and Beisson, J. (1996). Characterization of centrin genes in Paramecium. Eur. J. Biochem. 238, 121–128. [DOI] [PubMed] [Google Scholar]
- Middendorp, S., Paoletti, A., Schiebel, E., and Bornens, M. (1997). Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc. Natl. Acad. Sci. USA 94, 9141–9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias, E., Hamilton, E. P., and Orias, J. D. (2000). Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. Methods Cell Biol. 62, 189–211. [DOI] [PubMed] [Google Scholar]
- Ruiz-Binder, N. E., Geimer, S., and Melkonian, M. (2002). In vivo localization of centrin in the green alga Chlamydomonas reinhardtii. Cell Motil. Cytoskeleton 52, 43–55. [DOI] [PubMed] [Google Scholar]
- Saiki, R. K., Gelfand, D. H., Stoffel, S., Scharf, S. J., Higuchi, R., Horn, G. T., Mullis, K. B., and Erlich, H. A. (1988). Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239, 487–491. [DOI] [PubMed] [Google Scholar]
- Salisbury, J. (1995). Centrin, centrosomes, and mitotic spindle poles. Curr. Biol. 7, 39–45. [DOI] [PubMed] [Google Scholar]
- Salisbury, J., Suino, K., Busby, R., and Springett, M. (2002). Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12, 1287. [DOI] [PubMed] [Google Scholar]
- Salisbury, J. L., Baron, A., Surek, B., and Melkonian, M. (1984). Striated flagellar roots: isolation and partial characterization of a calcium-modulated contractile organelle. J. Cell Biol. 99, 962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, M., and Salisbury, J. (1994). Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 124, 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, M. A., and Salisbury, J. (1989). Centrin-mediated microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 108, 1751–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler, C. A., and Staehelin, L. A. (1976). Reconstruction of oral cavity of Tetrahymena pyriformis utilizing high voltage electron microscopy. Tissue Cell 8, 1–18. [DOI] [PubMed] [Google Scholar]
- Selvapandiyan, A., Debrabant, A., Duncan, R., Muller, J., Salotra, P., Sreenivas, G., Salisbury, J. L., and Nakhasi, H. L. (2004). Centrin gene disruption impairs stage-specific basal body duplication and cell cycle progression in Leishmania. J. Biol. Chem. 279, 25703–25710. [DOI] [PubMed] [Google Scholar]
- Shang, Y., Li, B., and Gorovsky, M. A. (2002). Tetrahymena thermophila contains a conventional γ-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J. Cell Biol. 158, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, K. R., and Cole, E. S. (2000). Nuclear and cytoskeletal fluorescence microscopy techniques. Methods Cell Biol. 62, 291–311. [DOI] [PubMed] [Google Scholar]
- Sullivan, D. S., Biggins, S., and Rose, M. D. (1998). The yeast centrin, Cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J. Cell Biol. 143, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon, B. E., Adler, S. A., Suhan, J. P., and Jarvik, J. W. (1992). Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J. Cell Biol. 119, 1613–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn, K. C., and Harper, J. D. (1998). Microtubule-organizing centers and nucleating sites in land plants. Int. Rev. Cytol. 181, 75–149. [DOI] [PubMed] [Google Scholar]
- Verbsky, J. W., and Turkewitz, A. P. (1998). Proteolytic processing and Ca2+-binding activity of dense-core vesicle polypeptides in Tetrahymena. Mol. Biol. Cell 9, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R., Pan, Z., and Salisbury, J. (2000). GFP-Centrin as a marker for centriole dynamics in living cells. Microsc. Res. Tech. 49, 451–457. [DOI] [PubMed] [Google Scholar]
- Wiley, E. A., Ohba, R., Yao, M. C., and Allis, C. D. (2000). Developmentally regulated rpd3p homolog specific to the transcriptionally active macronucleus of vegetative Tetrahymena thermophila. Mol. Cell. Biol. 20, 8319–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, N. E. (2000). Preparation of cytoskeletal fractions from Tetrahymena thermophila. Methods Cell Biol. 62, 441–447. [DOI] [PubMed] [Google Scholar]
- Williams, N. E., and Bakowska, J. (1982). Scanning electron microscopy of cytoskeletal elements in the oral apparatus of Tetrahymena. J. Protozool. 29, 382–389. [Google Scholar]
- Williams, N. E., and Luft, J. H. (1968). Use of a nitrogen mustard derivative in fixation for electron microscopy and observations on the ultrastructure of Tetrahymena. J. Ultrastruct. Res. 25, 271–292. [DOI] [PubMed] [Google Scholar]
- Wright, R. L., Salisbury, J., and Jarvik, J. W. (1985). A nucleus-basal body connector in Chlamydomonas reinhardtii that may function in basal body localization or segregation. J. Cell Biol. 101, 1903–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.