Abstract
ARGONAUTE (AGO) RNA-binding proteins are involved in RNA silencing. They bind to short interfering RNAs (siRNAs) and microRNAs (miRNAs) through a conserved PAZ domain, and, in animals, they assemble into a multisubunit RNA-induced silencing complex (RISC). The mammalian AGO2, termed Slicer, directs siRNA- and miRNA-mediated cleavage of a target RNA. In Arabidopsis, there are 10 members of the AGO family, and the AGO1 protein is potentially the Slicer component in different RNA-silencing pathways. Here, we show that AGO1 selectively recruits certain classes of short silencing-related RNA. AGO1 is physically associated with miRNAs, transacting siRNAs, and transgene-derived siRNAs but excludes virus-derived siRNAs and 24-nt siRNAs involved in chromatin silencing. We also show that AGO1 has Slicer activity. It mediates the in vitro cleavage of a mir165 target RNA in a manner that depends on the sequence identity of amino acid residues in the PIWI domain that are predicted by homology with animal Slicer-competent AGO proteins to constitute the RNase catalytic center. However, unlike animals, we find no evidence that AGO1 Slicer is in a high molecular weight RNA-induced silencing complex. The Slicer activity fractionates as a complex of ≈150 kDa that likely constitutes the AGO1 protein and associated RNA without any other proteins. Based on sequence similarity, we predict that other Arabidopsis AGOs might have a similar catalytic activity but recruit different subsets of siRNAs or miRNAs.
Keywords: posttranscriptional regulation, ribonuclease, viral RNA, silencing
ARGONAUTE (AGO) proteins are implicated in RNA-silencing processes that also involve 21- to 26-nt short RNAs (sRNAs) (1) cleaved from double-stranded or partially double-stranded (ds) RNAs by the RNase III enzyme Dicer. There are several types of RNA-silencing mechanisms, including RNA interference (RNAi), the micro RNA (miRNA) pathway, and RNA-directed chromatin silencing (1). RNAi is a type of RNA silencing in which the Dicer substrate is fully double stranded, the sRNA cleavage product is short interfering RNA (siRNA), and the outcome is targeted destruction of siRNA-complementary RNAs. The miRNA pathway is similar except that the Dicer substrate is an inverted repeat RNA with a partially ds structure, the sRNA is referred to as a miRNA, and the target RNAs can be suppressed at the translational level or degraded as in RNAi, depending on the degree of complementarity between the sRNA and its target. Plants possess an additional class of degradative sRNAs called transacting siRNAs (ta-siRNAs) whose formation depends on the miRNA-mediated cleavage of their precursor and its conversion into a dsRNA by RDR6 (2-4). The last pathway, RNA-directed chromatin silencing, is similar to RNAi, but the siRNA targets are either DNA or chromatin-associated RNAs and the outcome is DNA methylation or histone modification at the target locus.
In the best understood of these RNA-silencing mechanisms, the duplex siRNAs or miRNAs produced by Dicer are unwound in an ATP-dependent process. One strand of this RNA is then preferentially assembled with an AGO protein to form an RNA-induced silencing complex (RISC) (5-7). RISC has an associated ribonuclease activity (Slicer) that cleaves its substrate at sRNA-complementary sites. The size and composition of RISC varies (8-10), but AGO proteins are a common component in all systems. The crystal structure of an archaebacterial AGO homologue revealed that the PIWI domain, one of the two signature domains of AGO proteins, adopts a fold similar to RNase H (11). This finding prompted the suggestion that AGO proteins may be the Slicer nuclease, and, consistent with this idea, the human AGO2 has a DDH motif that is essential for Slicer activity and is functionally equivalent to the catalytic metal-coordinating triad DDE of RNase H (12, 13). Final proof that hAGO2 has Slicer activity was from the finding that the bacterially expressed recombinant protein is competent for sRNA-directed RNA cleavage (13). The PAZ domain is a second signature of AGO proteins, and it has been implicated by structural studies as an sRNA-binding feature (14-17).
Many eukaryotes, with the notable exception of fission yeast, have AGO multigene families whose members have specialized biological function, as revealed by the variety of mutant phenotypes (18). For example, mutants defective in RDE1, one of the 23 Caenorhabditis elegans AGO family members, are resistant to RNAi, but the miRNA pathway operates normally (19), whereas AGO-like gene (alg)-1 and alg-2 mutants are competent for RNAi but have heterochronic defects due to defects in a miRNA-silencing pathway (20). Drosophila ago mutants also reveal diversification of siRNA and miRNA pathways: ago2 mutants are RNAi-defective but are able to mediate miRNA-directed RNA cleavage. The ago1 mutants, in contrast, are deficient in miRNA processing and subsequent miRNA-mediated RNA cleavage but not in RNAi (21). The four human ARGONAUTES are equally competent for binding siRNAs and miRNAs, but only AGO2 is able to mediate RNA cleavage of target RNAs (12, 22). Although AGO2 is essential for embryo development, it is still unclear whether this requirement is related to its unique ability to cleave mRNAs (12).
In Arabidopsis, the AGO family comprises 10 members (18, 23) of which two have been unambiguously associated with different forms of RNA silencing. It is therefore likely that, as in animals, the functional diversification of RNA silencing is linked to the variation between AGO family members. AGO1 is associated with the miRNA pathway and transgene-silencing pathway (23, 24), and AGO4 with endogenous siRNAs affecting epigenetic silencing (25, 26). In addition, AGO7 and ZLL/AGO10 have a function in the transition from juvenile to adult phases of plant growth (27) and meristem maintenance (28, 29), respectively. Although a role in sRNA-mediated regulation seems likely, it is not yet supported by evidence.
The most studied Arabidopsis AGO protein is AGO1. It is clearly implicated in miRNA silencing because strong ago1 alleles affect miRNA accumulation and miRNAs target regulation (24), as do dcl1, hen1, and hyl1 (30-33). The ago1 mutants are also impaired in spontaneous silencing of a foreign transgene (cosupression) (23) and exhibit hypersusceptibility to Cucumber Mosaic Virus (CMV) (34), suggesting that, in plants, miRNA-mediated silencing, transgene silencing, and virus induced silencing share a common AGO factor. However, those observations could be explained equally well if AGO1 functions in either the biogenesis of sRNAs or as Slicer.
Here, we investigate the role of AGO1 in RNA silencing. We show that affinity-purified AGO1 is associated with miRNAs, endogenous ta-siRNAs, and transgene-derived siRNAs but not virus-specific siRNAs or siRNAs involved in chromatin silencing. We also show that, dependent on conserved amino acid residues in the PIWI domain, AGO1 mediates the in vitro cleavage of PHAVOLUTA RNA at the mir165 target site. It is therefore likely that AGO1 is a Slicer that selectively recruits sRNAs. The Slicer activity fractionates in a complex of ≈150 kDa that likely constitutes the AGO1 protein and associated RNA without any other proteins.
Materials and Methods
FLAG-AGO Transgenic Arabidopsis. A FLAG-AGO1 construct was generated by fusing the AGO1 cDNA to an N-terminal FLAG sequence under the regulation of the AGO1 promoter in the binary vector pGreen0229. The construct was transformed into heterozygous ago1-36 (Salk_087076) (http://signal.salk.edu) and homozygous ago1-36 transgenic seedlings were identified by PCR. The expression levels of the AGO1 gene and transgene were assessed by RT-PCR on 2-week-old WT, ago1-36, and FLAG-AGO1 seedlings.
Immunoprecipitation and sRNA Analysis. FLAG-AGO1 immunoprecipitation from inflorescences of transgenic FLAG-AGO1 plants or WT plants was performed with α-FLAG M2 agarose beads (Sigma). RNA extracted either from the immunoprecipitate or directly from tissues as control was analyzed by Northern blotting. For analysis of transgene-derived siRNAs, we used F1 plants from crosses between FLAG-AGO1 plants and two genotypes that were silencing GFP. One of these was an RNAi line in which the transgene expressed a GFP inverted repeat (GF-IR) (35). The second genotype (GF-amp) expressed a viral transgene and exhibited sense RNA silencing (previously described as GxA) (36). Virus infections were performed by rub-inoculation on 2-week-old FLAG-AGO1 plants. For the RNA methylation test, GFP synthetic unmethylated siRNAs were added to the AGO1-associated RNA sample, and a β-elimination reaction was performed as described in ref. 37.
Slicer Assay. PHAVOLUTA (PHV) cleavage assays were performed with immunoprecipitated FLAG-AGO1 and 32P-labeled PHV or phv in vitro transcripts (mMESSAGE mMACHINE T7, Ambion, Austin, TX). As a positive control, 20 μl of wheat germ extract was used. Cleavage was tested after 90 min at 25°C by extracting and separating the RNA on an 8 M urea/3% polyacrylamide gel.
Nicotiana benthamiana Transient Expression. Specific mutations in FLAG-AGO1 were introduced by PCR. For transient expression in N. benthamiana, the constructs were transferred into the pBIN61 vector (38) and Agro-infiltrated into N. benthamiana leaves. Immunoprecipitations, Slicer assay and sRNA analysis were as described above.
Size Exclusion Chromatography. Immunoprecipitated and eluted FLAG-AGO1 were concentrated by ultrafiltration and fractionated on a Superose 6 column (Amersham Pharmacia). Fractions were concentrated again and used for the Slicer assay and western analysis.
A more detailed version of Materials and Methods is available as Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Results and Discussion
Epitope Tagging of Arabidopsis AGO1. If AGO1 is an RNA-silencing Slicer, it would physically interact with miRNAs and siRNAs and would cleave mRNA targets that are complementary to these small RNAs. To test these predictions, we constructed an N-terminal FLAG-tagged version of the AGO1 cDNA coupled to 1,648 bp of the AGO1 promoter (Fig. 1A). The construct was transformed into an ago1 mutant (salk_087076, named ago1-36)(Fig. 1B) in which a T-DNA [portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] insertion at the end of the PAZ domain resulted in production of a truncated protein. The ago1-36 alleles are likely null because the encoded protein lacked >50% of the protein sequence, including the PIWI domain and because they show a stunted growth phenotype like that of strong ago1 alleles (Fig. 1C) (39-41). The tagged AGO1 transgene (FLAG-AGO1) fully restored a WT phenotype in ago1-36 homozygous mutants, indicating that the introduced tag does not interfere with AGO1 function (Fig. 1 C and D). The expression level of the transgene assessed by RT-PCR was similar to that of the endogenous AGO1 gene in WT plants (Fig. 1E).
Fig. 1.
Generation of epitope-tagged AGO1 transgenic Arabidopsis. (A) Diagram of the FLAG-AGO1 construct. Positions of the restriction sites used for the cloning are given relative to the start codon. The sequence and position of the FLAG epitope is indicated. Thick lines, regions encoding the PAZ and PIWI domains; thin broken arrow, translation start; black dot, translation stop. (B) Diagram of the AGO1 genomic locus. Gray boxes, exons; triangle, T-DNA insertion in ago1 mutant Salk_087076 line (ago1-36) with left border (Lb) and right border (Rb) orientation. Other symbols as in A.(C) FLAG-AGO1 complements the ago1-36 phenotype. Photographs are taken 2 weeks postgermination. (D) PCR genotyping of the FLAG-AGO1 line. The ago1-36 (Upper) and not the WT allele (Lower) is amplified from the selected FLAG-AGO1 transgenic line. (E) Expression of FLAG AGO1 transcripts. ago1-36 mutants produce a truncated transcript comprising the sequence 5′ (Middle) but not 3′ (Bottom) of the T-DNA insertion. Expression of a full-length AGO1 transcript is restored in the selected FLAG-AGO1 line. Actin primers (Top) were used to confirm equal loading, and reactions without reverse transcriptase were performed to exclude DNA contamination. DNA, control PCR with genomic DNA.
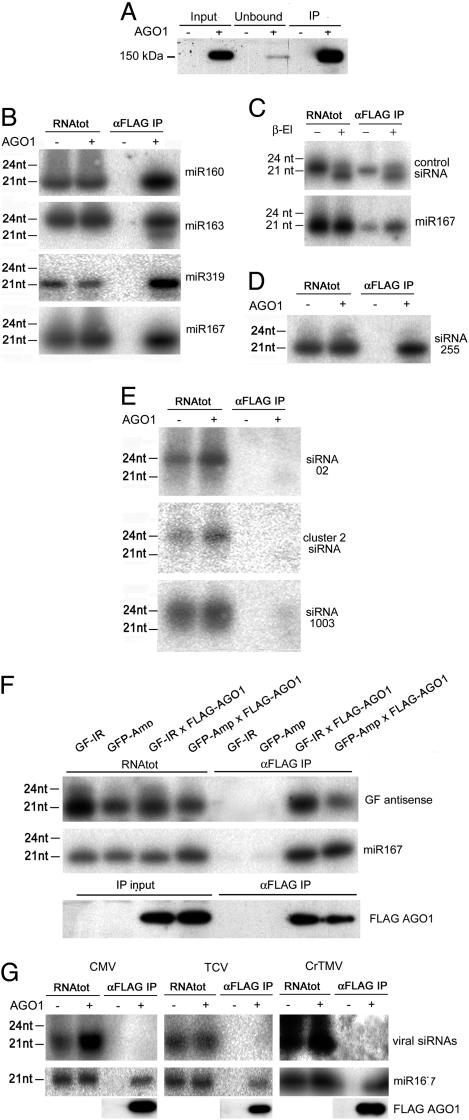
AGO1 Interacts Selectively with miRNAs and siRNAs. FLAG-AGO1 could be immunoprecipitated from young inflorescence extracts, thereby demonstrating that the N terminus of the protein is accessible under native conditions (Fig. 2A). The AGO1-associated RNA was extracted from immunoprecipitated AGO1, and the presence of siRNAs and miRNAs was assessed by Northern blotting.
Fig. 2.
AGO1 recruitment of small RNAs. (A) Immunoprecipitation of FLAG-AGO1. FLAG-AGO1 was immunoprecipitated from crude inflorescence extract as described in Materials and Methods. Input, crude extract before immunoprecipitation; unbound, supernatant after immunoprecipitation; IP, immunoprecipitate. AGO1(+), FLAG-AGO1 transgenic extracts; AGO(-), WT extracts. (B) miRNAs are recruited by AGO1. sRNA blots were hybridized with DNA oligonucleotide probes complementary to the indicated miRNAs. RNAs were either extracted directly from plant material (RNAtot) or from α-FLAG immunoprecipitate (αFLAG IP). (C) AGO1-recruited miRNAs are methylated at the 3′ terminus. A synthetic unmethylated GFP 21-nt RNA oligomer mixed with the immunoprecipitated RNAs (Upper) displays an increased electrophoretic mobility after a β-elimination reaction (β-El +) whereas miR167 (Lower) does not, indicating that its 3′ end is protected by methylation. Both GFP and miR167 hybridization were done on the same blot after stripping. (D) ta-siRNAs are recruited by AGO1. (E) Endogenous chromatin siRNAs are not recruited by AGO1. In B, D, and E, the IP samples were derived from 7-fold more tissue than directly extracted RNA. (F) Transgene-derived siRNAs are associated with AGO1. FLAG-AGO1 plants were crossed with plants coexpressing the GFP together with a GF inverted repeat silencer construct (GF-IR line) or with plants coexpressing the GFP together with a Potato Virus X-GFP silencer construct (GFP-Amp). Blots show GFP-derived siRNAs (Top) or miR167 (Middle) from parent and F1 plants. The IP samples were derived from 20-fold more tissue than directly extracted RNA. (Bottom) A Western blot of FLAG-AGO1. (G) Viral siRNAs are not associated with AGO1. FLAG-AGO1 or WT plants were infected with Cucumber Mosaic Virus strain I17F (CMV), Turnip Crinkle Virus (TCV), and Cruciferae Tobacco Mosaic Virus (CrTMV). siRNAs (Upper) were detected by hybridization with an in vitro transcribed sense probe corresponding to the coat protein sequence of the virus (CMV and TCV) or the full-length cDNA (TMV). miR167 (Lower) was detected with a complementary DNA oligonucleotide probe on the same blot after stripping. The IP samples were derived from 10-fold more tissue than directly extracted RNA.
All tested miRNA species specifically copurified with FLAG-AGO1 and were absent from FLAG immunoprecipitates of nontransformed (WT) extracts (Fig. 2B). The associated RNAs include both 21-nt (miR160, 167, and 319) and 24-nt (miR163) species. The AGO1-associated miRNAs, in common with the total pool of plant miRNAs, are methylated (42). In a β-elimination test, the miRNAs associated with AGO1 remained unmodified, consistent with the presence of an O-methyl group on the 3′ terminal nucleotide either on the 2′ or 3′ carbon of the ribose (37), whereas a synthetic unmethylated siRNA included in the samples as internal control underwent the predicted increase in electrophoretic mobility (Fig. 2C).
The ta-siRNA255 was also physically associated with AGO1 (Fig. 2D), but the 24-nt siRNAs produced by DCL3, including siRNAs 02, cluster 2, and 1003, were not (Fig. 2E) (43). Thus, AGO1 is selective for certain types of endogenous silencing-related sRNAs. We could also demonstrate selectivity of AGO1 with siRNAs of foreign nucleic acids. Transgene-specific siRNAs, either from an inverted repeat transgene (GF-IR) or from a viral amplicon/sense GFP transgene silencing system (GFP-Amp), were associated with AGO1 (Fig. 2F) but virus-specific siRNAs (CMV, Turnip Crinkle Virus, and crucifer Tobacco Mosaic Virus) were not (Fig. 2G).
The AGO1 association of GF-IR siRNAs was unexpected because ago1-27 plants retain the ability to carry out RNAi from inverted repeat transgenes (44). However, the genetic test might not have been conclusive because ago1-27 is a weak allele and might encode a protein still competent for RNAi (34, 44). Conversely, we had anticipated that viral siRNAs would be associated with AGO1 because Arabidopsis ago1 plants were previously reported to be hypersusceptible to CMV (34). This phenotype could have been accounted for by the involvement of AGO1 in virus-induced silencing. However, it is now unlikely that AGO1 is a major cofactor of virus-induced silencing because the viral siRNAs were not associated with AGO1 (Fig. 2G) and we could not reproduce the hypersusceptibility phenotype even with plant genotype (ago1-27) and strain of CMV (I17F) used previously (34) (data not shown). To reconcile the earlier findings with the results presented here, we propose that there could be subtle environmental factors affecting the experiments. For example, there could be an environmentally influenced miRNA that targets the mRNA of a plant-encoded suppressor of viral defense or there could be an environmentally sensitive ago1 phenotype that influences the virulence of CMV.
The Molecular Basis of AGO1 Selectivity. The profile of RNA associated with AGO1 implies that there is selectivity in the mechanism by which this protein recruits sRNA. Among the different factors that might determine this specificity, we have considered sRNA size, subcellular localization, and the effects of virus-encoded suppressors of silencing. Of these factors, sRNA size can be ruled out because 21- and 24-nt sRNAs were present in both the AGO1-associated and AGO1-excluded fractions (Fig. 2). Subcellular location may be involved, but it is unlikely to be the sole determining factor in the AGO1 selection of sRNA because certain classes of both nuclear (chromatin associated siRNAs) (43, 45) and cytoplasmic sRNAs (viral siRNAs) were excluded from AGO1. A third possible factor involves the virus-encoded silencing suppressors produced in virus-infected plants. In principle, the absence of viral siRNAs in the AGO1-associated sRNA could be due to the action of these suppressors. However, this result does not seem likely, because these proteins had no effect on AGO1 association with a miRNA (miR167; Fig. 2G).
An attractive alternative mechanism of selectivity involves linking the mode of sRNA biogenesis with particular AGOs. Perhaps different Dicers dock onto specific AGO proteins so that sRNAs are addressed to specialized effector complexes. Consistent with this idea, the Dicers in animal systems not only are required for the processing of sRNAs from their precursors but they also play a role in the assembly of RISC (8, 46-48). In addition, there is a direct interaction in vitro between a subregion of the PIWI domain of hAGO2 and the RNase III domain of human Dicer (49). Evidence consistent with this channeling model is from the finding that miRNAs and ta-siRNAs associated with AGO1 (Fig. 2 B-D) all require DCL1 for their biogenesis (2, 4, 30) whereas the AGO1 excluded 24-nt siRNAs, and viral siRNAs are instead produced by DCL3 and possibly DCL2 (43).
A prediction from this Dicer-channeling hypothesis is that the Dicer for production of transgene siRNAs would also interact directly with AGO1. In principle, this Dicer could be either DCL1 or DCL4, whose function is unknown, that channels transgene sRNAs into AGO1. The finding that RNAi from inverted repeat transgenes functions in dcl1-9 plants (50) does not necessarily rule out this prediction. Functional redundancy in the DCL family or residual function of the dcl1-9 allele could mask the role of DCL1 in production of transgene siRNAs.
FLAG-AGO1 Has Slicer Activity and Is Not in a High Molecular Weight RISC. To find out whether AGO1 is present in a RISC complex or is itself Slicer, we first examined the ability of immunopurified FLAG-AGO1 to mediate in vitro cleavage of a PHV transcript, a target of miR165 (40, 51). The results, shown in Fig. 3, reveal that PHV 5′ and 3′ RNA cleavage products were specifically formed in extracts from FLAG-AGO1 plants but not from control non transformed plants. No cleavage products were either observed when a G → A mutation was inserted into the PHV RNA sequence at the residue complementary to positions 6 of miR165 (phv, Fig. 3). This alteration induces a dominant mutation phenotype in planta by preventing miRNA-mediated clearing of the PHV and PHB transcripts (52, 53) and abolishes cleavage in a wheat germ in vitro assay (51).
Fig. 3.
AGO1 copurifies with Slicer activity. In vitro-labeled WT PHV or mutant G → A phv target RNAs were incubated with immunoprecipitates from FLAG-AGO1 and WT plants or with wheat germ extracts (Wg) as positive control. The sizes of the predicted 5′ and 3′ PHV RNA cleavage products are indicated.
This miRNA-directed RNA cleavage could result from Slicer activity of either AGO1 or other associated proteins. To investigate these possibilities, we modified the core PIWI domain of AGO1 at residues that are conserved in cleavage-competent AGO proteins from animals (Fig. 4A). In AGO1, the aspartate residue at position 760 is equivalent to the first metal coordinating aspartate D597 of hAGO2. Mutation of this residue to alanine in hAGO2 abolishes in vitro cleavage activity, and we predicted that a similar mutation in AGO1 would yield the same result if AGO1 is Slicer. The Gly-758 and His-798 are also strongly conserved throughout AGOs. Gly-758 is mutated to proline in the hypomorphic ago1-25 mutant allele (34) whereas the H798P mutation in hAGO2 (12) results in loss of RNA cleavage activity.
Fig. 4.
Mutations of conserved residues of the AGO1 PIWI domain affect Slicer activity. (A) Alignment of the catalytic center of the Arabidopsis and human ARGONAUTE PIWI domains. The positions of the Mg2+ coordinating residues in the DDH catalytic triad are indicated above the alignment (arrows), as well as the mutations introduced in FLAG-AGO1 (*). (B and C) Slicer activity of mutant AGO1 proteins. FLAG-AGO1, FLAG-AGO1G758S, and FLAG-AGO1H798P were expressed transiently in N. benthamiana by Agrobacterium infiltration, and Slicer activity of the immunoprecipitated proteins was assayed in vitro (B top). FLAG-AGO1D760A could not be assayed because it was unstable in the transient assay, but it did accumulate and its PHV Slicer activity could be assayed in extracts of transgenic Arabidopsis (C Top). Recruitment of miR165 was verified by Northern blotting of sRNAs extracted from immunoprecipitates (Bottom). The level of immunopurified FLAG-AGO1 proteins was tested by Western blotting (Middle).
The G758S and H798P mutations were introduced into FLAG-AGO1 constructs under the cauliflower mosaic virus (CaMV)35S promoter and expressed transiently in N. benthamiana leaves, whereas the construct carrying the D760A mutation was transformed stably in Arabidopsis under the AGO1 promoter. The immunoaffinity-purified FLAG-AGO1 mutant proteins could be detected by Western blotting, and the WT protein was correctly programmed by endogenous miR165 as it cleaved the PHV RNA in vitro into the predicted 5′ and 3′ fragments (Fig. 4B). The AGO1 G758S, was also able to cleave the PHV RNA target, and it is possible that the hypomorphic phenotype of the corresponding Arabidopsis mutant and its impaired cosuppression ability (34) are due to altered kinetic properties of AGO1 but not to total inactivity. In contrast, D760A and H798P were totally cleavage-deficient (Fig. 4 B and C) despite their ability to recruit miRNAs. We interpret the effect of changes at conserved catalytic site residues as a strong indication that AGO1 is Slicer.
Slicer in animal cells is part of a high molecular weight RISC that includes accessory proteins including TSN nuclease, Gemin, Fragile X syndrome-associated protein, and other proteins (8-10, 54-57). However, if such a large complex exists in plants, the accessory proteins are not required for Slicer activity because size exclusion chromatography revealed that the FLAG-AGO1 and the associated Slicer activity from Arabidopsis inflorescences eluted together close to the 158-kDa molecular mass standard (Fig. 5). The predicted molecular mass of the tagged version of AGO1 is 116 kDa, the associated siRNA would be ≈7 kDa and, given the imprecision of size determination by gel filtration, it seems that, as with hAGO2 (13, 58), the minimal Arabidopsis RISC contains little more than AGO1 and an associated sRNA. These findings do not rule out that a high molecular weight complex, for example including Dicer, HYL1, and other proteins, such as TSN nuclease, is formed during the assembly of a miRNA/siRNA-programmed AGO1 Slicer. Such a complex could be difficult to detect because it exists only transiently or is much less stable in vitro than the equivalent complexes in animals. Alternatively, the high molecular weight complex might have a structure that impairs accessibility of the epitope tag on the N terminus of AGO1 and prevents the purification of the native complex. However, only a minor fraction of the total AGO1 pool would be present in a high molecular weight complex because most AGO1 can be immunoprecipitated from a crude extract (Fig. 2 A).
Fig. 5.
FLAG-AGO1 Slicer is present in low molecular weight complexes. FLAG-AGO1 was immunopurified and eluted from α-FLAG M2 agarose beads by competition with 3XFLAG peptides. The concentrated eluate was fractionated on a Superpose 6 column, and the fractions were tested for cleavage of PHV target RNA (Upper) and for presence of AGO1 by Western blotting (Lower). The elution profile of the molecular weight markers is indicated. Vo, void volume; In, input; Wg, wheat germ.
Selective sRNA Recruitment and Slicer Activity in Other Arabidopsis AGO Proteins? All of the 10 Arabidopsis AGO proteins have PAZ domains and so are potentially able to recruit siRNAs and miRNAs. All 10 also resemble the RNA cleavage-active hAGO2 in that they have conserved aspartates as two of the putative metal-coordinating residues in the catalytic site (Fig. 4A) (11, 12). However, AGO2 and -3 differ from the other AGOs in that they have an additional aspartate as the third coordinating residues in place of the conserved histidine. This third carboxylic acid residue is not likely to impair Slicer activity because two other related enzymes, RNaseH1 and Tn5 integrase, can use aspartate efficiently in that position (13, 59, 60). Most of the AGO proteins, also like hAGO2, have a conserved histidine at the equivalent of AGO1 position 798, and it is likely that they selectively recruit siRNAs or a subset of miRNAs and use them as guides in RNA cleavage reactions. The exceptions are AGO4, -6, -8, and -9. AGO6 and AGO9 have a proline residue aligned with AGO1 position 798 and, based on the in vitro phenotype of the H798P mutant, it is probable that they are not active Slicer proteins. Perhaps these more variant AGO proteins recruit siRNAs but regulate their targets by way of translation interference or DNA methylation. For example, AGO4, which has a serine residue at position 798, has been implicated in chromatin modification and DNA methylation and so may be directly targeted to DNA (25, 26). It will be interesting to find out to what extent selective recruitment of siRNA and miRNA plays a role in the functional diversification of AGO protein function and of RNA silencing pathways.
Supplementary Material
Acknowledgments
We thank the Salk Institute and the Nottingham Arabidopsis Stock Centre for providing the Salk_087076 line and Attila Molnar and Alan Herr for critical reading of the manuscript and fruitful discussions. Hervé Vaucheret (Institut National de la Recherche Agronomique, Versaille, France) is thanked for ago1-27 mutant plants and CMV I17N. The use of imported strains of virus was under license (DEFRA 161A/4391 (01/03). This work was partly supported by a European Molecular Biology Organization fellowship and a Swiss National Science Foundation fellowship (to N.B.). We are grateful to the Gatsby Charitable Foundation for supporting the Sainsbury Laboratory.
Author contributions: N.B. and D.C.B. designed research; N.B. performed research; N.B. analyzed data; and N.B. and D.C.B. wrote the paper.
Abbreviations: sRNA, short RNA; siRNA, short interfering RNA; miRNA, microRNA; AGO, Argonaute; RNAi, RNA interference; ta-siRNA, transacting siRNA; RISC, RNA-induced silencing complex; CMV, Cucumber Mosaic Virus; GF-IR, GFP inverted repeat; PHV, PHAVOLUTA; T-DNA, portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells.
References
- 1.Baulcombe, D. (2004) Nature 431, 356-363. [DOI] [PubMed] [Google Scholar]
- 2.Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A. C., Hilbert, J.-L., Bartel, D. P. & Crete, P. (2004) Mol. Cell 16, 69-79. [DOI] [PubMed] [Google Scholar]
- 3.Allen, E., Xie, Z., Gustafson, A. M. & Carrington, J. C. (2005) Cell 121, 207-221. [DOI] [PubMed] [Google Scholar]
- 4.Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L. & Poethig, R. S. (2004) Genes Dev. 18, 2368-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwarz, D. S., Hutvagner, G., Du, T., Xu, Z., Aronin, N. & Zamore, P. D. (2003) Cell 115, 199-208. [DOI] [PubMed] [Google Scholar]
- 6.Khvorova, A., Reynolds, A. & Jayasena, S. D. (2003) Cell 115, 209-216. [DOI] [PubMed] [Google Scholar]
- 7.Tomari, Y., Matrange, C., Haley, B., Martinez, N. & Zamore, P. D. (2004) Science 306, 1377-1380. [DOI] [PubMed] [Google Scholar]
- 8.Pham, J. W., Pellino, J. L., Lee, Y. S., Carthew, R. W. & Sontheimer, E. J. (2004) Cell 117, 83-94. [DOI] [PubMed] [Google Scholar]
- 9.Nykanen, A., Haley, B. & Zamore, P. D. (2001) Cell 107, 309-321. [DOI] [PubMed] [Google Scholar]
- 10.Hammond, S. M., Bernstein, E., Beach, D. & Hannon, G. (2000) Nature 404, 293-296. [DOI] [PubMed] [Google Scholar]
- 11.Song, J. J., Smith, S. K., Hannon, G. J. & Joshua-Tor, L. (2004) Science 305, 1434-1437. [DOI] [PubMed] [Google Scholar]
- 12.Liu, J., Carmell, M. A., Rivas, F. V., Marsden, C. G., Thomson, M., Song, J. J., Hammond, S. M., Joshua-Tor, L. & Hannon, G. J. (2004) Science 305, 1437-1441. [DOI] [PubMed] [Google Scholar]
- 13.Rivas, F. V., Tolia, N. H., Song, J. J., Aragon, J. P., Liu, J. D., Hannon, G. J. & Joshua-Tor, L. (2005) Nat. Struct. Mol. Biol. 12, 340-349. [DOI] [PubMed] [Google Scholar]
- 14.Ma, J. B., Ye, K. Q. & Patel, D. J. (2004) Nature 429, 318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song, J. J., Liu, J., Tolia, N. H., Schneiderman, J., Smith, S. K., Martienssen, R. A., Hannon, G. J. & Joshua-Tor, L. (2003) Nat. Struct. Mol. Biol. 10, 1026-1032. [DOI] [PubMed] [Google Scholar]
- 16.Yan, K. S., Yan, S., Farooq, A., Han, A., Zeng, L. & Zhou, M.-M. (2003) Nature 426, 1-5. [DOI] [PubMed] [Google Scholar]
- 17.Lingel, A., Simon, B., Izaurralde, E. & Sattler, M. (2003) Nature 426, 465-469. [DOI] [PubMed] [Google Scholar]
- 18.Carmell, M. A., Xuan, Z., Zhang, M. & Hannon, G. J. (2002) Genes Dev. 16, 2733-2742. [DOI] [PubMed] [Google Scholar]
- 19.Tabara, H., Sarkissian, M., Kelly, W. G., Fleenor, J., Grishok, A., Timmons, L., Fire, A. & Mello, C. C. (1999) Cell 99, 123-132. [DOI] [PubMed] [Google Scholar]
- 20.Grishok, A., Pasquinelli, A. E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D. L., Fire, A., Ruvkun, G. & Mello, C. (2001) Cell 106, 23-34. [DOI] [PubMed] [Google Scholar]
- 21.Okamura, K., Ishizuka, A., Siomi, H. & Siomi, M. C. (2004) Genes Dev. 18, 1655-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G. & Tuschl, T. (2004) Mol. Cell 15, 185-197. [DOI] [PubMed] [Google Scholar]
- 23.Fagard, M., Boutet, S., Morel, J.-B., Bellini, C. & Vaucheret, H. (2000) Proc. Natl. Acad. Sci. USA 97, 11650-11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaucheret, H., Vazquez, F., Crete, P. & Bartel, D. P. (2004) Genes Dev. 18, 1187-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zilberman, D., Cao, X., Johansen, L. K., Xie, Z., Carrington, J. C. & Jacobsen, S. E. (2004) Curr. Biol. 14, 1214-1220. [DOI] [PubMed] [Google Scholar]
- 26.Zilberman, D., Cao, X. & Jacobsen, S. E. (2003) Science 299, 716-719. [DOI] [PubMed] [Google Scholar]
- 27.Hunter, C., Sun, H. & Poethig, R. S. (2003) Curr. Biol. 13, 1734-1739. [DOI] [PubMed] [Google Scholar]
- 28.Moussian, B., Schoof, H., Haecker, A., Jurgens, G. & Laux, T. (1998) EMBO J. 17, 1799-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynn, K., Fernandez, A., Aida, M., Sedbrook, J., Tasaka, M., Masson, P. & Barton, M. K. (1999) Development (Cambridge, U.K.) 126, 469-481. [DOI] [PubMed] [Google Scholar]
- 30.Park, W., Li, J., Song, R., Messing, J. & Chen, X. (2002) Curr. Biol. 12, 1484-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen, X., Liu, J., Cheng, Y. & Jia, D. (2002) Development (Cambridge, U.K.) 129, 1085-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han, M.-H., Goud, S., Song, L. & Fedoroff, N. (2004) Proc. Natl. Acad. Sci. USA 101, 1093-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vazquez, F., Gasciolli, V., Crete, P. & Vaucheret, H. (2004) Curr. Biol. 14, 346-351. [DOI] [PubMed] [Google Scholar]
- 34.Morel, J.-B., Gordon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F. & Vaucheret, H. (2002) Plant Cell 14, 629-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwach, F., Vaistij, F. E., Jones, L. & Baulcombe, D. (July 22, 2005) Plant Physiol., 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed]
- 36.Dalmay, T., Hamilton, A. J., Mueller, E. & Baulcombe, D. C. (2000) Plant Cell 12, 369-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alefelder, S., Patel, B. K. & Eckstein, F. (1998) Nucleic Acids Res. 26, 4983-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bendahmane, A., Querci, M., Kanyuka, K. & Baulcombe, D. C. (2000) Plant J. 21, 73-81. [DOI] [PubMed] [Google Scholar]
- 39.Kidner, C. A. & Martienssen, R. A. (2005) Dev. Biol. 280, 504-517. [DOI] [PubMed] [Google Scholar]
- 40.Kidner, C. A. & Martienssen, R. A. (2004) Nature 428, 81-84. [DOI] [PubMed] [Google Scholar]
- 41.Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M. & Benning, C. (1998) EMBO J. 17, 170-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R. W., Steward, R. & Chen, X. (2005) Science 307, 932-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie, Z., Johansen, L. K., Gustafson, A. M., Kasschau, K. D., Lellis, A. D., Zilberman, D., Jacobsen, S. E. & Carrington, J. C. (2004) PLoS Biol. 2, E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beclin, C., Boutet, S., Waterhouse, P. & Vaucheret, H. (2002) Curr. Biol. 12, 684-688. [DOI] [PubMed] [Google Scholar]
- 45.Hamilton, A. J., Voinnet, O., Chappell, L. & Baulcombe, D. C. (2002) EMBO J. 21, 4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee, Y. S., Nakahara, K., Pham, J. W., Kim, K., He, Z., Sontheimer, E. J. & Carthew, R. W. (2004) Cell 117, 69-81. [DOI] [PubMed] [Google Scholar]
- 47.Tabara, H., Yigit, E., Siomi, H. & Mello, C. C. (2002) Cell 109, 861-871. [DOI] [PubMed] [Google Scholar]
- 48.Tomari, Y., Du, T., Haley, B., Schwarz, D. S., Bennett, R., Cook, H. A., Koppetsche, B. S., Theurkauf, W. E. & Zamore, P. D. (2004) Cell 116, 831-841. [DOI] [PubMed] [Google Scholar]
- 49.Tahbaz, N., Kolb, F. A., Zhang, H., Jaronczyk, K., Filipowicz, W. & Hobman, T. C. (2004) EMBO Rep. 5, 189-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finnegan, E. J., Margis, R. & Waterhouse, P. M. (2003) Curr. Biol. 13, 236-240. [DOI] [PubMed] [Google Scholar]
- 51.Tang, G., Reinhart, B. J., Bartel, D. P. & Zamore, P. D. (2003) Genes Dev. 17, 49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallory, A. C., Reinhart, B. J., Jones-Rhoades, M. W., Tang, G., Zamore, P. D., Barton, M. K. & Bartel, D. P. (2004) EMBO J. 23, 3356-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J. & Barton, M. K. (2002) Nature 411, 709-713. [DOI] [PubMed] [Google Scholar]
- 54.Ishizuka, A., Siomi, M. C. & Siomi, H. (2002) Genes Dev. 16, 2497-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caudy, A. A., Myers, M., Hannon, G. J. & Hammond, S. M. (2002) Genes Dev. 16, 2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caudy, A., Ketting, R. F., Hammond, S. M., Denli, A., M., Bathoorn, A. M. P., Tops, B. B. J., Silva, J. M., Myers, M. M., Hannon, G. J. & Plasterk, R. (2003) Nature 425, 411-414. [DOI] [PubMed] [Google Scholar]
- 57.Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M. & Dreyfuss, G. (2002) Genes Dev. 16, 720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rand, T. A., Ginalski, K., Grishin, N. V. & Wang, X. (2004) Proc. Natl. Acad. Sci. USA 101, 14385-14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katayanagi, K., Okumura, M. & Morikawa, K. (1993) Proteins 17, 337-346. [DOI] [PubMed] [Google Scholar]
- 60.Peterson, G. & Reznikoff, W. (2003) J. Biol. Chem. 278, 1904-1909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.