Abstract
The hepadnaviral polymerase (P) functions in a complex with viral nucleic acids and cellular chaperones. To begin to identify contacts between P and its partners, we assessed the exposure of the epitopes of six monoclonal antibodies (MAbs) to the terminal protein domain of the duck hepatitis B virus P protein in a partially denaturing buffer (RIPA) and a physiological buffer (IPP150). All MAbs immunoprecipitated in vitro translated P well in RIPA, but three immunoprecipitated P poorly in IPP150. Therefore, the epitopes for these MAbs were obscured in the native conformation of P but were exposed when P was in RIPA. Epitopes for MAbs that immunoprecipitated P poorly in IPP150 were between amino acids (aa) 138 and 202. Mutation of a highly conserved motif within this region (T3; aa 176 to 183) improved the immunoprecipitation of P by these MAbs and simultaneously inhibited DNA priming by P. Peptides containing the T3 motif inhibited DNA priming in a dose-dependent manner, whereas eight irrelevant peptides did not. T3 function appears to be conserved among the hepadnaviruses because mutating T3 ablated DNA synthesis in both duck hepatitis B virus and hepatitis B virus. These results indicate that (i) the conserved T3 motif is a molecular contact point whose ligand can be competed by soluble T3 peptides, (ii) the occupancy of T3 obscures the epitopes for three MAbs, and (iii) proper occupancy of T3 by its ligand is essential for DNA priming. Therefore, small-molecule ligands that compete for binding to T3 with its natural ligand could form a novel class of antiviral drugs.
Hepatitis B virus (HBV) is a small DNA virus that replicates by reverse transcription (reviewed in reference 9). It has a lipid envelope studded with viral glycoproteins that surrounds an icosahedral core particle composed of the core protein. Within the core particle are the viral nucleic acids and reverse transcriptase (P). Other hepadnaviruses infect woolly monkeys, woodchucks, ground squirrels, ducks, geese, and herons (6, 17, 29, 31, 32). Significant differences exist among the hepadnaviruses, but they share a high degree of hepatotropism, follow the same replication cycle, and have a nearly identical genetic organization.
Hepadnaviral reverse transcription (34) occurs within cytoplasmic capsid particles. Reverse transcription begins with binding of P to an RNA stem-loop (ɛ) on the pregenomic RNA, and then this complex is encapsidated. Reverse transcription is primed by P itself, so minus-strand DNA is covalently linked to P (36, 40). Complexes containing P and the viral nucleic acids must be dynamic because three strand transfers are required to produce the mature circular viral DNA (21, 22, 38, 41).
The binding of P to ɛ requires the active participation of a molecular chaperone complex (13). In vitro reconstitution studies with recombinant duck hepatitis B virus (DHBV) P revealed that P-ɛ binding requires HSP90, HSP70, HSP40, HSP23, and HOP (2, 11, 14), although under certain circumstances only HSC70 and HSP40 are necessary (3). Because chaperones modulate protein conformation, the binding of P to ɛ is presumed to involve a conformational change in P, and we have demonstrated that a conformational change in DHBV P following ɛ binding is essential for the activation of P (35, 37). HSP90 and HSP23 appear to remain bound to P following encapsidation, and they may contribute to the function of P during reverse transcription (15). Therefore, P functions as part of a large, dynamic macromolecular complex containing an assortment of cellular proteins and viral nucleic acids.
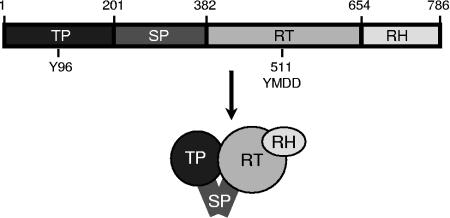
P has four domains (Fig. 1) (5, 28). The terminal protein domain contains the tyrosine residue that primes DNA synthesis and covalently links P to the viral DNA (42, 44). The spacer domain has no known function other than to link the terminal protein domain to the rest of P. The reverse transcriptase and RNase H domains contain the two enzymatic active sites. The crystal structure of P has not been determined, but the HBV reverse transcriptase domain has been modeled based on crystal structures of the human immunodeficiency virus reverse transcriptase (1, 7). Complementation studies with recombinant fragments of HBV P indicate that there are multiple contacts between the terminal protein domain and the reverse transcriptase/RNase H domains (18, 19), but the three-dimensional arrangement of these domains is unknown. Therefore, our knowledge of the arrangement of the P domains is limited to little more than the diagram in Fig. 1.
FIG. 1.
Domain structure of DHBV P. TP, terminal protein; SP, spacer; RT, reverse transcriptase; RH, RNase H; Y96, tyrosine 96, to which the viral DNA is covalently linked; YMDD, a key reverse transcriptase active-site motif beginning at residue 511.
We seek to identify P motifs involved in its enzymatic activity to understand the mechanism of reverse transcription and to guide antiviral drug design. We are especially interested in the terminal protein domain because DNA priming by the terminal protein domain is unique to the hepadnaviruses and hence is an attractive target for antiviral compounds with a high selectivity for HBV. We therefore used a combination of antibody binding experiments, mutagenesis, and peptide competition assays to assess the exposure of sequences in the terminal protein domain and their interactions with other members of the reverse transcription complex.
MATERIALS AND METHODS
Viruses and DNA clones.
pT7DPol contains DHBV3 (32) nucleotides (nt) 170 to 3021 within pBluescript (Promega); the allele employed contains a 33-nt insertion at nt 901 encoding the influenza hemagglutinin epitope (16). D1.5G is an over-length DHBV3 expression vector in pBS(−) containing a 5′ duplication of nt 1658 to 3021. Mutations (Table 1) were inserted into pT7DPol and D1.5G. The plasmid pdɛ contains DHBV3 nt 2526 to 2845 encoding ɛ within pBluescript. pCMV-HBV-LE− contains 1.2 copies of the HBV(ADW2) genome (39) with mutations that ablate surface antigen expression (4) downstream of the cytomegalovirus promoter (from pCDNA3.1 [Invitrogen]) cloned into pBluescript. The K161E and R162E mutations were inserted into the HBV P T3 motif in pCMV-HBV-LE−.
TABLE 1.
Immunoprecipitation of DHBV P in IPP150 and DNA priming activities of P mutants
| Mutation(s) | Region mutated | Amt of immunoprecipitation with indicated MAba
|
Priming activity (%) | |||||
|---|---|---|---|---|---|---|---|---|
| MAb 5 | MAb 6 | MAb 9 | MAb 10 | MAb 11 | MAb 12 | |||
| Wild type | None | ± | ± | ++ | ± | ++ | ++ | 100 |
| K153E | TP | ± | ± | ++ | ± | ++ | ++ | 43 |
| G178E | TP (T3) | ± | + | ++ | + | ++ | ++ | 20 |
| I179D, L180D | TP (T3) | + | ++ | ++ | ++ | ++ | ++ | 0 |
| Y181F | TP (T3) | ± | ± | ++ | ± | ++ | ++ | 100 |
| K182E, R183E | TP (T3) | ++ | ++ | ++ | + | ++ | ++ | 0 |
| K182T, R183T | TP (T3) | + | + | ++ | + | ++ | ++ | 0 |
| P195V | TP | ± | ± | ++ | ± | ++ | ++ | 100 |
| N399D, T400A | RT | ± | ± | ++ | ± | ++ | ++ | 70 |
| T668V, T670V | RH | ± | ± | ++ | ± | ++ | ++ | 88 |
| H693Y | RH | ± | ± | ++ | ± | ++ | ++ | 74 |
| D715V | RH | ± | ± | ++ | ± | ++ | ++ | 104 |
++, 50 to 100% of the WT activity; +, 10 to 50% of the WT activity; ±, <10% of the WT activity.
In vitro transcription and translation.
mRNAs for DHBV P were transcribed with T7 RNA polymerase from pT7DPol. ɛ-containing RNAs were transcribed with T3 RNA polymerase from pdɛ. All RNAs were transcribed by employing Megascript kits (Ambion) according to the manufacturer's instructions. 35S-labeled DHBV P was translated in vitro by employing rabbit reticulocyte lysate (Promega) in a 20-μl total volume containing [35S]methionine (>1,000 Ci/mmol; Amersham) at 30°C for 1 h according to the manufacturer's instructions. The translation of P from RNAs transcribed from pT7DPol initiates at both the 1st and 44th codons of the P open reading frame, leading to the doublet observed in Fig. 2, 4, and 6.
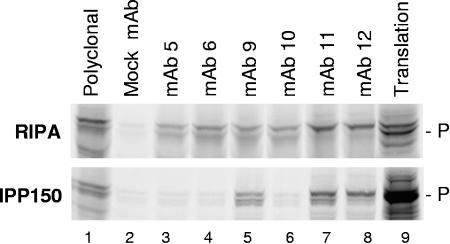
FIG. 2.
Differential immunoprecipitation of P. In vitro-translated P was diluted in RIPA (top) or IPP150 (bottom), and equal aliquots of the translation mixture were immunoprecipitated with saturating amounts of the indicated antibodies. Translation of P in vitro initiates at both the 1st and 44th codons of the P open reading frame, leading to the doublet observed for P. Polyclonal, a rabbit polyclonal anti-DHBV P terminal protein antibody. Lane 9 contains unprecipitated translation products.
FIG. 4.
Mutation of T3 can improve the immunoprecipitation of P in IPP150. P(Y181F) and P(I179D/L180D) were translated in vitro, diluted in IPP150, and immunoprecipitated with the indicated MAbs.
FIG. 6.
Inhibition of DNA priming by MAbs. DHBV P was translated in vitro, the indicated MAbs were added to equal aliquots of the translation mixture and incubated to permit binding to P, and then ɛ, MgCl2, and [α-32P]dGTP were added to permit priming. The 32P signal from priming was quantitated and normalized to the activity of P without a MAb.
Immunoprecipitation.
Antibodies were bound to protein A/G beads (Calbiochem), and the antibody-bead complexes were incubated overnight with in vitro-translated P diluted in 0.5 ml RIPA (20 mM Tris, pH 7.2, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) or IPP150 (10 mM Tris, pH 7.5, 150 mM NaCl, 0.1% NP-40). Immunocomplexes were washed four times with 1 ml of either IPP150 or RIPA, and P was released by boiling in Laemmli buffer. Following SDS-polyacrylamide gel electrophoresis (SDS-PAGE), radioactive P was detected by autoradiography or phosphorimager analysis.
DNA priming assays.
P was translated in vitro in the absence (Fig. 6) or presence (Fig. 7) of ɛ. For the experiments depicted in Fig. 6, 125 ng of ɛ was added to the appropriate samples prior to the priming assay. MgCl2 (to 4 mM) and 10 μCi [α-32P]dGTP were added to the translation reactions, and the samples were incubated at 37°C for 30 min. Reactions were terminated with Laemmli loading buffer, and the products were resolved by SDS-PAGE. The gels were dried, and the 32P priming signal was detected by phosphorimager analysis (the 35S signal from translation was blocked by a layer of exposed X-ray film). Where indicated, 3 μg of monoclonal antibodies (MAbs) or synthetic peptides (0.25 to 1.0 mM) was added during priming; the final dimethyl sulfoxide concentration in all reactions containing peptides was 1%.
FIG. 7.
Inhibition of priming by a peptide containing the T3 motif. P was translated in vitro with ɛ and incubated with increasing amounts of the T3 peptide or the irrelevant MBP peptide, and then MgCl2 and [α-32P]dGTP were added to permit priming. The 32P signal from priming was quantitated and normalized to the activity of P without dimethyl sulfoxide (DMSO) or peptide. Peptide concentrations are in mM, and error bars show the standard deviations from four experiments.
Core particle isolation, endogenous polymerase assay, and Southern blotting.
LMH and Huh7 cells were maintained in Dulbecco's modified Eagle's medium/F12 with 10% fetal bovine serum. Transfections employed FuGENE (Roche Diagnostics) according to the manufacturer's instructions. DHBV and HBV cores were isolated from transfected cells by lysis in core particle preparation lysis buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 0.25% NP-40, 50 mM NaCl, 8% sucrose) followed by sedimentation through a 30% sucrose cushion as described previously (37). The endogenous polymerase assay was performed as described previously (35). Viral DNAs were isolated by proteinase K digestion followed by phenol-chloroform extraction as described previously (10) and then resolved by electrophoresis on 1.2% agarose gels. Southern blotting was performed as described previously (33), with internally 32P-labeled HBV DNA as a probe.
RESULTS
Differential exposure of epitopes for MAbs 5, 6, and 10.
To begin a structural assessment of DHBV P, we raised six immunoglobulin G monoclonal antibodies (MAbs) against the DHBV P terminal protein domain (amino acids 1 to 207) (43) and determined the exposure of their epitopes to in vitro-translated P by immunoprecipitation. In vitro-translated P primes DNA synthesis in the presence of dGTP and ɛ (40) and hence is an excellent source of properly folded, enzymatically active P that is not buried within capsids. The translation of DHBV P in vitro initiates at both the first and second AUGs in the open reading frame, leading to a doublet on SDS-PAGE gels.
35S-labeled P was translated in vitro, diluted in high-detergent buffer (RIPA) or low-detergent buffer (IPP150), divided into equal aliquots, and immunoprecipitated with saturating amounts of the MAbs. RIPA dissociates P-ɛ binding and disrupts many weak protein-protein interactions, whereas IPP150 is a mild buffer that maintains most intermolecular contacts and is used to measure sequence-specific P-ɛ binding by coprecipitation (27). All six MAbs immunoprecipitated P well in RIPA (Fig. 2, top panel), indicating that their epitopes were accessible under these conditions. MAb 12 consistently immunoprecipitated full-length P preferentially over the internal initiation product, whereas MAb 11 occasionally immunoprecipitated the full-length form preferentially (as shown in Fig. 2), but more often immunoprecipitated both forms. MAbs 9, 11, and 12 also immunoprecipitated P well in IPP150, but MAbs 5, 6, and 10 immunoprecipitated P very poorly in this milder buffer (Fig. 2, bottom panel). This effect was not absolute, because weak immunoprecipitation of P in IPP150 by MAbs 5, 6, and 10 could often be detected. Therefore, the epitopes of MAbs 5, 6, and 10 are accessible for antibody binding when P is in RIPA but are largely obscured in IPP150.
Mutations to the T3 motif expose the epitopes for MAbs 5, 6, and 10.
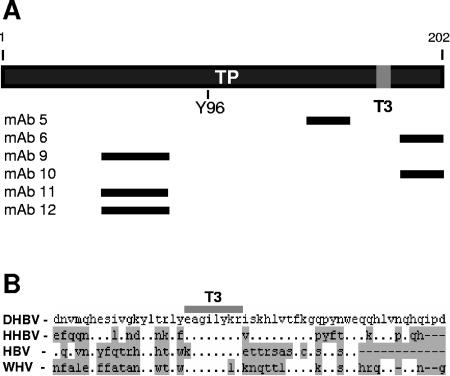
To determine the locations of the terminal protein domain sequences that are obscured from MAb binding in the active conformation of P (in IPP150), we mapped the epitopes for the MAbs by Western blotting fragments of P translated in vitro (Fig. 3A; data not shown). The three MAbs that immunoprecipitated P poorly in IPP150 had epitopes between amino acids (aa) 138 and 160 (MAb 5) or 182 and 202 (MAbs 6 and 10), whereas MAbs that immunoprecipitated P well in IPP150 had epitopes between aa 46 and 77 (MAbs 9, 11, and 12). This led us to hypothesize that a ligand may bind to P between aa 138 and 202 and that this binding may obscure the epitopes for MAbs 5, 6, and 10. We further hypothesized that the putative binding site would be conserved among the hepadnaviruses because the enzymatic activities and mechanism of reverse transcription of the avian and mammalian P proteins are conserved, although the proteins share only about 25% amino acid identity. Therefore, we aligned the terminal protein sequence of DHBV P with those of heron hepatitis B virus, HBV(adw2), and woodchuck hepatitis virus to look for conserved sequences in this region. The alignment revealed a very highly conserved motif at DHBV P aa 176 to 183 that we termed T3 (EAGILYKR) (Fig. 3B).
FIG. 3.
Epitope and T3 motif positions within the terminal protein. (A) Regions of the DHBV terminal protein domain containing the epitopes for the MAbs are indicated by black lines. The T3 motif is indicated as a gray block. (B) Alignment of the DHBV, heron hepatitis B virus (HHBV), HBV, and woodchuck hepatitis virus (WHV) terminal domain sequences around the T3 motif (EAGILYKR). Residues differing from those in DHBV are shaded, and gaps are indicated with dashes.
To determine if T3 contributed to obscuring the epitopes for MAbs 5, 6, and 10, we created five mutants with alterations in T3 and immunoprecipitated the mutant proteins in IPP150. As a control, we tested the ability of six other mutations throughout P to affect the immunoprecipitation of P by the MAbs. 35S-labeled mutant proteins were translated in vitro, diluted in IPP150, and immunoprecipitated. Four of the five mutations in T3 improved the ability of P to be immunoprecipitated in IPP150 by MAbs 5, 6, and 10 (Table 1; data for the I179D/L180D and Y181F mutations are shown in Fig. 4), whereas none of the mutations outside of T3 increased the ability of these MAbs to immunoprecipitate P. The improvement of immunoprecipitation by MAb 5 was less than the improvement seen for MAbs 6 and 10. This is consistent with the different locations of the epitopes for MAb 5 (between aa 138 and 160) and MAbs 6 and 10 (between aa 182 and 202). The ability of the mutations to mimic the effect of RIPA in exposing the epitopes for MAbs 5, 6, and 10 led us to conclude that the T3 motif plays a key role in obscuring the epitopes of these MAbs in IPP150.
Mutations that expose epitopes for MAbs 5, 6, and 10 inhibit DNA synthesis.
We next asked if exposing the epitopes for MAbs 5, 6, and 10 affected DNA priming. We could not expose the epitopes with RIPA because this dissociates P-ɛ binding and hence inhibits DNA priming. Therefore, we assayed our collection of mutant P molecules for DNA priming activity. In the priming reaction, P synthesizes the first nucleotide of the viral genome by covalently attaching dGMP to itself, using dGTP as a substrate, ɛ as a template, and a tyrosine in the terminal protein domain as a primer (Y96 in DHBV) (44). In vitro-translated P primes DNA synthesis efficiently (40) and permits analyses of priming in the absence of encapsidation and the first-strand transfer reaction, which are needed to measure P activity in cells.
We translated 35S-labeled P to monitor the synthesis of P and then initiated priming by adding ɛ, MgCl2, and [α-32P]dGTP to the translation mixture. The products of the priming reaction were resolved by SDS-PAGE, 35S translation signals were blocked by a layer of plastic, and 32P signals from the linkage of [32P]dGMP to P were detected by autoradiography. Mutations that exposed the epitopes for MAbs 5, 6, and 10 strongly inhibited or ablated priming activity, whereas mutations that did not expose the epitopes had little or no effect on priming (Table 1). Therefore, there was a negative correlation between exposure of the epitopes for MAbs 5, 6, and 10 and the ability to prime DNA synthesis.
To determine if altering T3 also inhibited DNA synthesis within viral cores, the T3 mutants were transferred to the DHBV genome in the over-length expression vector D1.5G. Mutant and wild-type plasmids were transfected into LMH cells, and 3 days later intracellular viral core particles were isolated by sedimentation through a sucrose gradient. Extracts containing the core particles were incubated with radioactive nucleotides to monitor DNA synthesis on the endogenous nucleic acids (the endogenous polymerase assay), and then viral nucleic acids were purified and resolved by agarose gel electrophoresis (Fig. 5A). The three sets of mutations in T3 that ablated DNA priming (I179D/L180D, K182E/R183E, and K182T/R183T) also ablated DNA synthesis in DHBV core particles, and the two mutations which retained DNA priming activity in vitro (G178E and Y181F) also retained the ability to synthesize viral DNA in core particles. Therefore, mutations to the T3 domain have similar effects on DNA priming in vitro and DNA synthesis in core particles. These data do not distinguish between a direct inhibition of enzymatic activity or blocking of an essential upstream event, such as the binding of P to ɛ, because both events would lead to a loss of DNA synthesis.
FIG. 5.
Mutating T3 ablates DHBV and HBV DNA synthesis. Expression constructs for DHBV and HBV genomes encoding wild-type P or P proteins with mutations in the T3 motif were transfected into cells, core particles were isolated, and viral DNAs were detected. rcDNA, relaxed circular DNA; ssDNA, single-stranded DNA. (A) DHBV DNA within core particles isolated from LMH cells, as detected by an endogenous polymerase assay. (B) HBV DNA within HBV particles isolated from Huh7 cells, as detected by Southern blotting.
To determine if the T3 motif is also essential for the replication of HBV DNA, the homologs of the DHBV P(K182E/R183E) mutations were inserted into HBV P. Expression constructs for wild-type and HBV P(K161E/R162E) genomes were transfected into Huh7 cells, and 5 days later core particles were isolated. Southern blots of core particle DNA revealed DNA synthesis by wild-type HBV P but not by HBV P(K161E/R162E) (Fig. 5B). Therefore, the T3 motif is essential for DNA synthesis in both DHBV and HBV.
Inhibition of DNA priming by anti-P MAbs.
If the epitopes for MAbs 5, 6, and 10 are occluded in the active form of P, then these MAbs should have little effect on the priming activity of P. In contrast, MAbs whose epitopes are exposed and are located near motifs important for DNA priming should inhibit priming. Therefore, we purified the MAbs by protein A affinity chromatography and measured their effects on priming.
35S-labeled P was translated, purified Abs were added and allowed to bind P, and then ɛ and MgCl2 were added to permit priming (Fig. 6). 32P-labeled P was detected when ɛ was included in the reaction in the absence of any MAb (Fig. 6, lane 3), and this priming signal was set to 100%. The addition of purified MAbs inhibited priming to various degrees. Priming was unaffected by an irrelevant MAb (NM6) against the adenovirus DNA binding protein. The P allele employed for in vitro translation contains an influenza virus hemagglutinin epitope tag (16) between aa 264 and 265 in the spacer domain of P and hence is recognized by MAb 12CA5 (Roche). 12CA5 modestly inhibited priming (41% of the wild-type activity remained). MAb 5 (epitope between aa 138 and 160) and MAb 6 (epitope between aa 188 and 202) also modestly inhibited the priming reaction (57% and 38% of the wild-type activity remained, respectively); this is consistent with their poor ability to immunoprecipitate P under physiological conditions. In contrast, MAbs 9, 11, and 12 recognize an epitope(s) between aa 46 and 77 and immunoprecipitate P well in IPP150. These MAbs all inhibited priming well (to 7 to 15% of wild-type levels), possibly by occluding tyrosine 96, to which dGMP is linked. Therefore, the ability of the MAbs to inhibit priming is consistent with the exposure of their epitopes in low-detergent buffer and with the location of the epitopes on P.
Inhibition of priming by a peptide containing the T3 motif.
The epitopes for MAbs 5, 6, and 10 can be exposed by high-detergent buffer and by mutating the T3 motif. Based on these data, we hypothesized that T3 may be a binding site on P, that binding of a ligand to T3 obscures the MAb epitopes, and that disrupting binding at T3 with either high detergent concentrations in RIPA or mutations to T3 exposes the epitopes. If this were true, a peptide containing the T3 motif should specifically compete with the ligand that binds to P at T3 and should therefore inhibit DNA priming.
To test this hypothesis, we translated 35S-labeled P in the presence of ɛ and 0.25, 0.50, 0.75, or 1.0 mM of a peptide containing the T3 motif (LYEAGILYKRIS; Genscript) or an irrelevant peptide (myelin basic protein peptide [MBP] [APRTPGGRR]; Biosource International). After translation, an aliquot was removed from each reaction to monitor the translation efficiency, and the DNA priming activity of P was assessed in the remaining sample by adding [α-32P]dGTP and MgCl2 and incubating the sample at 37°C for 30 min. The samples were then resolved by electrophoresis, and the 35S signal (translation) and 32P signal (priming) were independently quantitated by phosphorimager analysis. Figure 7 shows priming activities normalized for P translation efficiencies from four competition experiments. A concentration-dependent inhibition of priming by the T3 peptide but not by the MBP peptide was observed.
The specificity of the inhibition of priming by the T3 peptide was further established by synthesizing six additional peptides representing phylogenetically conserved motifs of the DHBV terminal protein domain (T1A [FNPEWKVP], T2A [GVKPKYPDNVMQH], T4A [TFKGQPYNWEQQHLV], T5 [EDVQSPGEGEPL], T6 [HHLGKLSGLY], and T7 [TAKFYPKSISYFP]; all from Genscript) plus one peptide from herpes simplex virus type 2 UL13 (APPAPPSHGGRRR; Genscript). None of these peptides inhibited priming by P (data not shown). Finally, we attempted to use a mutant T3 peptide containing the K182E/R183E substitutions, but this peptide was not soluble under our reaction conditions. Therefore, as predicted, a peptide containing the T3 motif competed specifically with a ligand for P that is needed for priming.
DISCUSSION
P functions in a dynamic complex with cellular chaperones and ɛ. We have identified a molecular contact point on DHBV P, termed T3, that is critical for P priming activity in vitro and DNA synthesis within core particles. The highly conserved T3 motif is located within the region of P containing the epitopes for MAbs that differentially precipitate P in RIPA and IPP150 (Fig. 3). The epitopes for MAbs 5, 6, and 10 are poorly recognized in a mild buffer that maintains the active conformation of P (IPP150) but are recognized much better when P is exposed to detergent concentrations in RIPA that dissociate many weak molecular interactions. The conformation dependence of the epitopes themselves cannot account for these observations because MAbs 5, 6, and 10 recognize linear epitopes (they were raised against a denatured antigen and they recognize denatured P in Western blots) and because mutating T3 and dissolving P in RIPA both improve the recognition of the epitopes. Therefore, we concluded that these epitopes are largely occluded in the native conformation of P that is maintained in IPP150.
The small size of T3 strongly implies that T3 may be a binding site because short conserved linear amino acid sequences are often modular ligand binding sites (e.g., the RB binding motif conserved among the adenovirus E1a, simian virus 40 T antigen, and papillomavirus E7 proteins [20]). Furthermore, the high degree of conservation of T3 despite the limited amino acid identity between avian and mammalian P proteins implies that T3 is important for the function of P. This in turn implies that mutations which expose the epitopes may disrupt binding at T3. This hypothesis is strongly supported by the ability of a peptide containing the T3 motif to specifically inhibit DNA priming in a dose-dependent manner (Fig. 7). Together, the differential exposure of the epitopes in IPP150 and RIPA (Fig. 2), the exposure of the epitopes by mutating T3 (Fig. 4 and Table 1), the differential inhibition of priming by the MAbs (Fig. 6), and the ability of the T3 peptide to specifically inhibit DNA priming (Fig. 7) led us to conclude that the epitopes for MAbs 5, 6, and 10 are occluded by a noncovalent molecular interaction of modest strength involving T3 and that this interaction is necessary for priming.
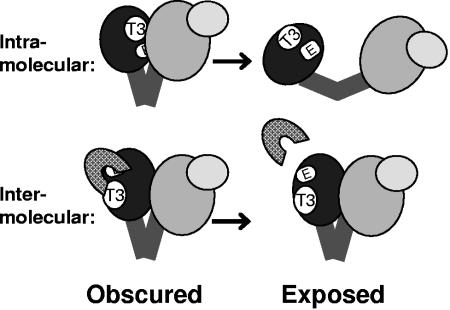
This hypothesis is consistent with two possibilities (Fig. 8). First, binding between T3 and its ligand could be intramolecular, where T3 contacts another portion of P. In this case, disrupting binding at T3 would alter the conformation of P and expose the epitopes for MAbs 5, 6, and 10. Alternatively, binding at T3 could be intermolecular, in which case T3 would bind to another molecule and the disruption of binding would expose the epitopes without necessitating a major change in the conformation of P. Candidate intermolecular ligands for T3 include the chaperones that bind P and ɛ. However, initial experiments have not yielded evidence that HSP23, HSP90, or ɛ is a ligand for T3 (data not shown). Mutating T3 did not alter the sedimentation profile of in vitro-translated DHBV P in IPP150-sucrose gradients (data not shown). This is most consistent with either intermolecular binding of a small ligand or intramolecular binding to another region of P because neither of these binding events would greatly alter the mass of the P complex. However, these experiments are not definitive because P sediments across a broad region of the gradient and detecting a small shift in this distribution is difficult.
FIG. 8.
Model for obstruction of epitopes by ligand binding at T3. Ligand binding at T3 blocks the exposure of the epitopes (E), and disruption of the binding exposes the epitopes. This is compatible with intra- or intermolecular binding. (Top) Intramolecular binding of another region of P to T3. A disruption of binding would induce a conformational change exposing the epitopes. (Bottom) Intermolecular binding of another molecule (hatched shape) to T3. A disruption of binding would expose the epitopes by releasing the ligand.
Regardless of whether interactions involving the T3 motif are intra- or intermolecular, the proper occupancy of T3 is needed for priming in vitro and for DNA synthesis in capsids. This effect is most probably due to a block of P-ɛ binding rather than a direct inhibition of DNA polymerase activity because mutating K182 and R183 within DHBV T3 ablates binding to ɛ (30; data not shown). Formation of the P-ɛ complex is essential for DNA priming, activation of DHBV reverse transcriptase activity, encapsidation of P and the pregenomic RNA, and DNA synthesis within core particles.
HBV P also functions in a dynamic complex with chaperones (12, 24-26) and HBV ɛ (8, 23). It is unknown how similar these interactions are between DHBV and HBV P or if any of these interactions involve T3. However, the high level of conservation of the T3 motif between DHBV and HBV P and the observation that mutating T3 in both DHBV and HBV blocks DNA synthesis within core particles (Fig. 5) strongly imply that the ligand that binds to the DHBV T3 motif will also bind to T3 in HBV P.
Disrupting binding to the T3 motif by competition with the T3 peptide inhibits priming. Therefore, the development of small molecules that compete for binding to T3 may be a novel avenue for antiviral drug design. Drugs targeting the T3 motif would be especially valuable because T3 is in the terminal protein domain that is unique to the hepadnaviruses, and therefore drugs targeting this motif should be highly selective for HBV. Furthermore, mutations conferring resistance to anti-T3 drugs would not generate cross-resistance to nucleoside analog inhibitors such as lamivudine and adefovir because these drugs target the active site of the reverse transcriptase domain.
Acknowledgments
We thank Jianming Hu and Lynda Morrison for helpful discussions.
This work was supported by grant AI38447 from the National Institutes of Health.
REFERENCES
- 1.Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K. A. Walters, D. L. J. Tyrrell, N. Brown, and L. D. Condreay. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology 27:1670-1677. [DOI] [PubMed] [Google Scholar]
- 2.Beck, J., and M. Nassal. 2001. Reconstitution of a functional duck hepatitis B virus replication initiation complex from separate reverse transcriptase domains expressed in Escherichia coli. J. Virol. 75:7410-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck, J., and M. Nassal. 2003. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J. Biol. Chem. 278:36128-36138. [DOI] [PubMed] [Google Scholar]
- 4.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, L. J., R. C. Hirsch, D. Ganem, and H. E. Varmus. 1990. Effects of insertional and point mutations on the functions of the duck hepatitis B virus polymerase. J. Virol. 64:5553-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, S. F., H. J. Netter, M. Bruns, R. Schneider, K. Frolich, and H. Will. 1999. A new avian hepadnavirus infecting snow geese (Anser caerulescens) produces a significant fraction of virions containing single-stranded DNA. Virology 262:39-54. [DOI] [PubMed] [Google Scholar]
- 7.Das, K., X. Xiong, H. Yang, C. E. Westland, C. S. Gibbs, S. G. Sarafianos, and E. Arnold. 2001. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J. Virol. 75:4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fallows, D. A., and S. P. Goff. 1995. Mutations in the E sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J. Virol. 69:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganem, D., and R. J. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2969. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 10.Gong, Y., E. Yao, and J. E. Tavis. 2001. Evidence that the RNAse H activity of the duck hepatitis B virus is unable to act on exogenous substrates. BMC Microbiol. 1:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu, J., and D. Anselmo. 2000. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by HSP90. J. Virol. 74:11447-11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu, J., D. Flores, D. Toft, X. Wang, and D. Nguyen. 2004. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J. Virol. 78:13122-13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, J., and C. Seeger. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 93:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu, J., D. Toft, D. Anselmo, and X. Wang. 2002. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J. Virol. 76:269-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, J., D. O. Toft, and C. Seeger. 1997. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 16:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolodziej, P. A., and R. A. Young. 1991. Epitope tagging and protein surveillance. Methods Enzymol. 194:508-519. [DOI] [PubMed] [Google Scholar]
- 17.Lanford, R. E., D. Chavez, K. M. Brasky, R. B. Burns III, and R. Rico-Hesse. 1998. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc. Natl. Acad. Sci. USA 95:5757-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanford, R. E., Y. H. Kim, H. Lee, L. Notvall, and B. Beames. 1999. Mapping of the hepatitis B virus reverse transcriptase TP and RT domains by transcomplementation for nucleotide priming and by protein-protein interaction. J. Virol. 73:1885-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanford, R. E., L. Notvall, H. Lee, and B. Beams. 1997. Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J. Virol. 71:2996-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, C., and Y. Cho. 2002. Interactions of SV40 large T antigen and other viral proteins with retinoblastoma tumour suppressor. Rev. Med. Virol. 12:81-92. [DOI] [PubMed] [Google Scholar]
- 21.Loeb, D. D., K. J. Gulya, and R. Tian. 1997. Sequence identity of the terminal redundancies of the minus-strand DNA template is necessary but not sufficient for the template switch during hepadnavirus plus-strand DNA synthesis. J. Virol. 71:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeb, D. D., R. C. Hirsch, and D. Ganem. 1991. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses: implications for other reverse transcribing elements. EMBO J. 10:3533-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nassal, M., and A. Rieger. 1996. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J. Virol. 70:2764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, S. G., and G. H. Jung. 2001. Human hepatitis B virus polymerase interacts with the molecular chaperonin Hsp60. J. Virol. 75:6962-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, S. G., S. O. Lim, and G. H. Jung. 2002. Binding site analysis of human HBV Pol for molecular chaperonin, Hsp60. Virology 298:116-123. [DOI] [PubMed] [Google Scholar]
- 26.Park, S. G., J. K. Rho, and G. Jung. 2002. Hsp90 makes the human HBV Pol competent for in vitro priming rather than maintaining the human HBV Pol/pregenomic RNA complex. Arch. Biochem. Biophys. 401:99-107. [DOI] [PubMed] [Google Scholar]
- 27.Pollack, J. R., and D. Ganem. 1994. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J. Virol. 68:5579-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radziwill, G., W. Tucker, and H. Schaller. 1990. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J. Virol. 64:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoedel, F., R. Sprengel, T. Weimer, D. Fernholz, R. Schneider, and H. Will. 1989. Animal hepatitis B viruses. Adv. Viral Oncol. 8:73-102. [Google Scholar]
- 30.Seeger, C., E. H. Leber, L. K. Wiens, and J. Hu. 1996. Mutagenesis of a hepatitis B virus reverse transcriptase yields temperature-sensitive virus. Virology 222:430-439. [DOI] [PubMed] [Google Scholar]
- 31.Sprengel, R., E. F. Kaleta, and H. Will. 1988. Isolation and characterization of a hepatitis B virus endemic in herons. J. Virol. 62:3832-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sprengel, R., C. Kuhn, H. Will, and H. Schaller. 1985. Comparative sequence analysis of duck and human hepatitis B virus genomes. J. Med. Virol. 15:323-333. [DOI] [PubMed] [Google Scholar]
- 33.Staprans, S., D. D. Loeb, and D. Ganem. 1991. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J. Virol. 65:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 35.Tavis, J. E., and D. Ganem. 1996. Evidence for the activation of the hepatitis B virus polymerase by binding of its RNA template. J. Virol. 70:5741-5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavis, J. E., and D. Ganem. 1993. Expression of functional hepatitis B virus polymerase in yeast reveals it to be the sole viral protein required for correct initiation of reverse transcription. Proc. Natl. Acad. Sci. USA 90:4107-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tavis, J. E., B. Massey, and Y. Gong. 1998. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, epsilon. J. Virol. 72:5789-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tavis, J. E., S. Perri, and D. Ganem. 1994. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 68:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valenzuela, P., M. Quiroga, J. Zaldivar, R. Gray, and W. Rutter. 1980. The nucleotide sequence of the hepatitis B viral genome and the identification of the major viral genes. UCLA Symp. Mol. Cell Biol. 18:57-70. [Google Scholar]
- 40.Wang, G. H., and C. Seeger. 1992. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71:663-670. [DOI] [PubMed] [Google Scholar]
- 41.Wang, G. H., and C. Seeger. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber, M., V. Bronsema, H. Bartos, A. Bosserhoff, R. Bartenschlager, and H. Schaller. 1994. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J. Virol. 68:2994-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao, E., Y. Gong, N. Chen, and J. E. Tavis. 2000. The majority of duck hepatitis B virus reverse transcriptase in cells is nonencapsidated and is bound to a cytoplasmic structure. J. Virol. 74:8648-8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zoulim, F., and C. Seeger. 1994. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J. Virol. 68:6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]