Abstract
Positive-sense single-stranded RNA viruses have developed strategies to exploit cellular resources at the expense of host mRNAs. The genomes of these viruses display a variety of structures at their 5′ and 3′ ends that differentiate them from cellular mRNAs. Despite this structural diversity, viral RNAs are still circularized by juxtaposition of their 5′ and 3′ ends, similar to the process used by cellular mRNAs. Also reminiscent of the mechanisms used by host mRNAs, translation of viral RNAs involves the recruitment of translation initiation factors. However, the roles played by these factors likely differ from those played by cellular mRNAs. In keeping with the general parsimony typical of RNA viruses, these host factors also participate in viral RNA replication. However, the dual use of host factors requires that viral RNA template utilization be regulated to avoid conflict between replication and translation. The molecular composition of the large ribonucleoprotein complexes that form the viral RNA replication and translation machineries likely evolves over the course of infection to allow for switching template use from translation to replication.
For positive-sense single-stranded RNA [(+) ssRNA] viruses, the largest class of viruses, the earliest events following cell entry and capsid disruption are translation and replication of the virus genome. Viral RNAs share characteristics with host cell mRNAs, but must have differential features to allow for preferential translation. Viral RNAs are also distinct from host mRNAs in that they are replicated as well as translated. The inherent conflict between the two processes must be resolved by molecular control switches for template use. (+) ssRNA viruses use host proteins in coordination with virus proteins to accomplish replication and translation of their genome.
The ability of a virus to invade a host relies on the occurrence of a compatible interaction that depends, among others, on the formation of functional heterocomplexes between host and virus proteins. In several cases, the ability of a virus to interact with a host can be mapped to single nucleotide differences in the genome of either host or virus, underlining the specificity of the interactions required. Reports on interactions between virus and host proteins have generated new insight into how host and (+) ssRNA viruses interact but has also left questions unanswered concerning interactions observed in one set of host-pathogen but not yet in others. This review focuses on the features that differentiates viral RNA from host mRNA and the processes related to RNA translation and replication that show conservation of function across a broad range of (+) ssRNA viruses infecting prokaryotes, animals, and plants. The premise used here is that the identification of differences between viral and host RNAs will point to different strategies for replication and translation and should lead to hypotheses on the mechanisms that control the outcome of host-virus interactions.
HOST mRNAs AND THEIR TRANSLATION
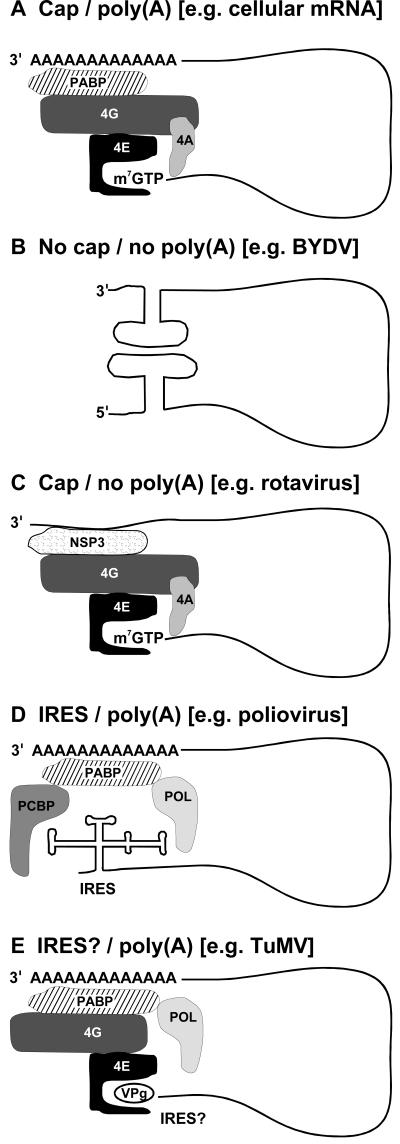
To identify elements unique to viral RNAs, we must first briefly review the canonical model of translation of cellular mRNAs to then identify how viral RNAs deviate from this model to control the host cell to their advantage. Most eukaryotic mRNAs have a 5′ untranslated region (UTR), a modified nucleotide (cap structure) at the 5′ end, and a poly(A) tail at the 3′ end. Both the 5′ UTR secondary structure and the 3′ end poly(A) tail affect translation initiation. The scaffold protein eIF4G (eukaryotic translation initiation factor 4G) is the cornerstone of translation initiation. By interacting with the 5′ cap-binding protein eIF4E (within the eIF4F complex) and the 3′ bound poly(A)-binding protein (PABP), eIF4G acts as a molecular bridge between the 5′ and 3′ ends of the mRNA resulting in mRNA circularization (the closed-loop model for mRNA configuration at translation initiation; Fig. 1A; see Gingras et al., 1999 for a detailed review of translation initiation factors). Circularization increases translation efficiency by promoting de novo initiation by ribosomes and recycling of terminating ribosomes on the same mRNA (Welch et al., 2000). A helicase, eIF4A, resolves secondary structure present in the 5′ UTR, facilitating access by translation factors. By its association with eIF4G, eIF3 then recruits the 40S ribosomal subunit and creates a link between the mRNA/eIF4F complex and the ribosome. Upon recognition of the AUG codon, most translation initiation factors are released, followed by recruitment of the 60S ribosomal subunit and the beginning of translation. In plants, two isoforms of eIF4F are present and seem to have complementary roles: eIF4F, containing eIF4E and eIF4G, and eIF(iso)4F, containing eIF(iso)4E and eIF(iso)4G (Browning, 2004).
Figure 1.
Interaction of the 3′ and 5′ ends of the RNA of (+) ssRNA viruses is mediated by RNA-RNA interactions or through the recruitment of host translation initiation factors. A, Simultaneous interactions between eIF4G-eIF4E and eIF4G-PABP result in a closed-loop conformation typical of cellular mRNAs. B, Direct RNA-RNA interactions circularize the uncapped and nonpolyadenylated RNA of viral genomic RNAs (e.g. BYDV). C, Rotaviral NSP3 interacts simultaneously with the nonpolyadenylated 3′ end of the viral RNA and with the cap-bound eIF4G. NSP3 is present in a complex with eIF4A and eIF4E. D, End-to-end interactions for the uncapped, polyadenylated poliovirus RNA occur via the host proteins PABP and PCBP and the viral poly(A) tail and IRES structure. E, RNA circularization of the RNA plant potyviruses might be mediated by eIF4G, eIF4E, PABP, and VPg interactions.
THE (+) ssRNA VIRUSES
By contrast to cellular mRNAs, the 5′ termini of viral RNAs can take one of several forms: a 5′ phosphate group, a cap or a virus-encoded polypeptide called VPg (Viral Protein genome-linked) covalently attached to the first nucleotide of the RNA. Some viral 5′ UTRs differ substantially from those of cellular mRNAs by their length and/or secondary structure. They can also contain an internal ribosome entry sequence (IRES) that allows for direct entry of ribosomes on the RNA, therefore bypassing the requirement for the eIF4F initiation complex resulting in cap-independent translation. The 3′ end of the RNA can feature a poly(A) tail, a tRNA-like structure, or simply a 3′ OH group. Although (+) ssRNA viruses rely on the host cell for translation, these structural differences suggest derogations to the general rules of eukaryotic translation.
(+) ssRNA viruses code for their own RNA-dependent RNA polymerase (RdRp) but also need host factors for formation of the replicase complex. Replication is initiated by copying the (+) strand to a complementary (−) strand intermediate that then serves as template for the production of (+) strand genomic RNAs through asymmetrical replication. These progeny molecules are then used for translation, replication, or as genomic RNA in new virions. Translation and replication of the same RNA templates must be regulated since the 5′ to 3′ movement of ribosomes on the RNA conflicts with the 3′ to 5′ activity of the RdRp. This is achieved by the interaction of host and viral factors with each other at both ends and sometimes along the viral RNA (Barry and Miller, 2002; Walter et al., 2002).
Like host mRNAs, there is evidence that viral RNAs also circularize; viral genomes that lack either 5′ cap, 3′ poly(A), or both display different mechanisms for formation of 5′ and 3′ end interactions.
Viral RNAs without 5′ Cap or 3′ Poly(A)
Bacteriophage Qβ uses RNA-RNA interactions between the 5′ and 3′ ends of its RNA. Such long-distance RNA interactions are required for replication since mutations that destabilize them abolish replication (Klovins and van Duin, 1999). In the case of Flaviviruses, the genome is also circularized through RNA-RNA interactions: dengue virus initiation of (−) strand RNA synthesis depends upon complementary sequences at the 5′ and 3′ ends (You and Padmanabhan, 1999).
The RNA of barley yellow dwarf virus (BYDV; Fig. 1B) base-pairs stem loops in the 3′ and 5′ UTRs. These interactions between ends are thought to be responsible for positioning the replication complex at the site of initiation for (−) strand RNA synthesis. Disruption of the base-pairing between the ends abolishes viral RNA replication. It is proposed that BYDV uses this base-pairing to allow the replicase working from the 3′ end to shut off translation of viral RNA and free the RNA of ribosomes to allow replication (Barry and Miller, 2002). Cellular proteins may also participate in joining the 5′ and 3′ ends.
Other (+) ssRNA plant viruses recruit host factors through features in their genomic RNA. An interaction between the 5′ UTR and 3′ translation enhancer (TED) of satellite tobacco necrosis virus (STNV; Tombusviridae) facilitates its cap-independent translation. Translation of STNV RNA lacking a functional TED can be restored in vitro by capping the 5′ end. This, combined with the capacity of the STNV TED RNA fragment to bind to eIF4F and eIF(iso)4F complexes, suggests that the 3′ UTR TED is a functional mimic of a 5′ cap group (Gazo et al., 2004).
The genomic RNA of turnip yellow mosaic virus (TYMV; Tymoviridae) has a tRNA-like structure at its 3′ end that can complex with host translation elongation factor eEF1A. For TYMV, Matsuda et al. (2004) have shown that (−) strand synthesis by the viral RdRp is repressed upon binding of eEF1A. This repression could occur early during infection to help coordinate template use between translation and replication. In the case of cricket paralysis virus (Dicistroviridae), the 5′ IRES contains a tRNAmet mimic to recruit eEF1A and 2 (Jan et al., 2003).
Viral RNA with a 5′ Cap But No 3′ Poly(A)
Viruses with a 5′ cap and a nonpolyadenylated 3′ end use different mechanisms to achieve similar results. The 3′ end of alfalfa mosaic virus (AMV; Bromoviridae) RNA has a tRNA-like structure instead of a poly(A) tail and features a series of stem-loops in that 3′ region. The AMV coat protein (CP) binds to these hairpins to stimulate translation perhaps by mimicking the binding of PABP to the poly(A) tail of mRNAs. The binding of one or more molecules of CP to the 3′ end of AMV RNAs is required for efficient translation in vivo. It is proposed that CP promotes circularization of viral RNAs by binding simultaneously to the 3′ UTR and to translation initiation factors assembled at the 5′ end (Neeleman et al., 2004). The dissociation of CP from the complex may allow the shift from translation to replication by allowing access of the 3′ UTR region to the replicase complex. The 3′ UTRs of brome mosaic virus (BMV; Bromoviridae) also contain a tRNA-like structure that stimulates translation (Zeenko et al., 2002; Barends et al., 2004). The 3′ tRNA-like of tobacco mosaic virus (TMV; Tobamovirus) is preceded by a pseudoknot domain to which binds eIF1A; the involvement of eIF1A is important as mutations that reduce the binding also reduce (−) strand synthesis and virus accumulation. The exact mechanism is not clear but eIF1A might stimulate replication in association with other host factors (see Zeenko et al., 2002). eIF1A is also found enriched in replication bodies in infected cells, along with ribosomes, further supporting the hypothesis that it is a required factor for efficient TMV translation and replication.
Mutational screens for BMV replication in yeast (Saccharomyces cerevisiae; used as a model BMV replication system) have allowed the identification of several host proteins that are implicated in translation and replication of viral RNA (Nouiery et al., 2003). Some are also involved in the replication of a double-stranded RNA virus from the yeast itself (Chong et al., 2004), raising the possibility that common mechanisms might be at play in these very different systems. The yeast factors (part of the Sm-like family of proteins) are involved in RNA processing (e.g. mRNA turnover, tRNA processing, decapping) in a broad range of organisms. The complex of yeast factors encoded by LSM1/LSM7 is required for translation of viral genomic RNAs, but not for translation of viral subgenomic mRNAs or most cellular mRNAs. Viral genomic RNAs destined for replication are distinguished from other viral and cell RNAs early in infection through interaction with the Lsm1–7 complex (Nouiery et al., 2003). Lsm1 may be part of the molecular switch that recruits genomic RNAs from translation to replication.
Outside the (+) ssRNA viruses sensu stricto, rotaviruses (Reoviridae, infecting animals) have double-stranded RNA genomes that are exposed to the same functional requirements as (+) ssRNA viruses at the single-stranded stage of their multiplication cycle (Piron et al., 1998). The rotavirus RNA is circularized via virus protein NSP3 that bridges the 3′ end of viral RNA and eIF4G (Vende et al., 2000). The 3′ sequence of rotaviral RNAs and NSP3 fulfil roles similar to the mRNA poly(A) and to PABP, respectively (Piron et al., 1998; Fig. 1C). Furthermore, NSP3 competes with PABP for eIF4G and interferes with circularization of cellular mRNAs leading to inhibition of host protein synthesis (Padilla-Noriega et al., 2002).
Capped and Polyadenylated Viral RNAs
The genome of some RNA viruses resembles cellular mRNAs with a 5′ cap and a 3′ poly(A). Circularization of these genomic RNAs (e.g. the coronaviruses and plant potexviruses) likely follows the canonical cellular mechanism: viral RNA circularization is mediated by a cap-eIF4E-eIF4G-PABP-poly(A) tail interaction. Using coronavirus replicons, it was shown that a poly(A) tail long enough for efficient PABP binding is essential for virus replication such as seen with cellular RNAs (Fig. 1A). The circularization of coronaviruses is suggested to be important for translation and for assembly of the replicase complex (Spagnolo and Hogue, 2000).
Viral Genomes with a 5′ VPg and a 3′ Poly(A)
(+) ssRNA viruses of the families Picornaviridae, Potyviridae, Comoviridae, and Caliciviridae lack a 5′ cap, but their RNA is covalently linked at the 5′ end to the VPg protein. End-to-end RNA interactions are found in several picornaviruses including poliovirus (PV), hepatitis A virus, and encephalomyocarditis virus (EMCV; Bergamini et al., 2000; Michel et al., 2000). Circularization of PV RNA is mediated by PABP; interactions between 3CD (the viral polymerase precursor), poly(C) binding protein (PCBP; a host protein regulating the stability and expression of several cellular mRNAs; Ostareck-Lederer et al., 1998), and PABP hold both ends of the PV RNA together (Herold and Andino, 2001; Fig. 1D). These ribonucleoprotein (RNP) complexes at the ends of the viral genome are necessary for replication and stimulate viral RNA translation. The same synergetic effect was shown for EMCV and hepatitis A virus (Bergamini et al., 2000).
Host proteins interacting with PABPs of both animal (Khaleghpour et al., 2001) and plant (Wang et al., 2004) inhibit translation in vitro. Such inhibition of cap-dependent translation could lead to preferential translation of viral RNAs. In animal cells, cleavage of PABP by viral proteases from Picornaviridae and Caliciviridae interferes with cap-dependent translation. Ribosome-associated PABP is preferentially cleaved resulting in inactivation of ribosome complexes attached to the poly(A) tail (Kerekatte et al., 1999; Kuyumcu-Martinez et al., 2004). The implications for viral translation are unclear as both caliciviruses and enteroviruses have poly(A) tails and a bypass mechanism is probably needed to selectively inhibit host translation without affecting viral translation. It is also possible that cleavage of PABP is required for switching between translation and RNA replication.
The presence of a virus-encoded protein covalently attached to the 5′ end of the virus genome is unique to (+) ssRNA viruses and is a departure from the canonical cellular mRNA structure. Removal of VPg from the feline calicivirus (Caliciviridae) RNA leads to a decrease in viral RNA translation (Herbert et al., 1997). The speculation is that the 5′ VPg acts as a surrogate for the cap and functions as a recruiter of the translation initiation complex in a cap-independent mechanism (Herbert et al., 1997). This is further supported by the observations of Daughenbaugh et al. (2003) that eIF3 binds to VPg in Norwalk virus (Caliciviridae). It is possible that a direct interaction between VPg and eIF3 bound to 40S ribosomal subunits attracts the initiation complex to the viral RNA, resulting in depletion of factors for host mRNA translation.
The RNA of plant potyviruses (Potyviridae, pircorna-like viruses) have a 5′ VPg and a 3′ poly(A). The RNA polymerase of zucchini yellow mosaic virus and the VPg-Pro of turnip mosaic virus (TuMV) interact with PABP (Wang et al., 2000; Leonard et al., 2004). The interaction between the viral RdRp and PABP might facilitate RNA replication by promoting an interaction with the poly(A) tail or facilitate the removal of PABP from the poly(A) tail to allow RdRp to initiate RNA replication. In addition, the potyvirus VPg interacts with different isoforms of the cap-binding eIF4E (Wittmann et al., 1997; Schaad et al., 2000) and the interaction is a determinant of infection (Leonard et al., 2000). In Arabidopsis (Arabidopsis thaliana), mutational inactivation of the gene encoding eIF(iso)4E results in the loss of susceptibility to several potyviruses (Duprat et al., 2002; Lellis et al., 2002). VPg and cap bind with similar affinities to different sites of eIF4E (Leonard et al., 2000; T. Michon and O. Le Gall, unpublished data), suggesting either a competition between VPg-bound (viral) and capped (cellular) mRNAs or a sequestration of translation factors by VPg in infected cells, or both. Circularization of the potyvirus RNA could occur through a noncanonical process where the cap structure is replaced by the 5′ VPg; VPg could bind eIF4E that binds eIF4G and PABP, therefore bringing together the 3′ poly(A) and the 5′-bound VPg (Fig. 1E). Alternatively, a more direct binding resulting from the interaction between VPg-Pro and PABP could achieve the same result. These two mechanisms may reflect different mechanistic approaches used at different stages of the virus life cycle allowing for differential regulation of template use by modification of the molecular components of the RNP complex.
CONCLUDING REMARKS
In eukaryotes, translation is temporally, spatially, and functionally uncoupled from RNA synthesis. Unlike cellular mRNAs, synthesis of viral RNAs follows, rather than precedes, translation; and unlike cellular mRNAs, (+) ssRNA viruses display a variety of structures at the 5′ and 3′ ends. Despite this structural diversity, similar host functions are recruited by cellular and viral RNAs alike. (+) ssRNA viruses make efficient use of resources; many host and viral proteins involved in translation also appear to be involved in replication. The fact that host factors involved in virus replication are part of cellular translation machineries could simply be the consequence of the coupling of translation and replication in (+) ssRNA viruses (White et al., 1992; Novak and Kirkegaard, 1994; Taylor and Carr, 2000).
Conservation of circularization in (+) ssRNA viruses, despite structural differences, provides these viruses with access to the host cell translation machinery and leads to enhanced translation of the genomic RNA (Le et al., 1997; Gallie, 1998; Wei et al., 1998; Borman et al., 2000). Viral RNA circularization could also coordinate translation and RNA synthesis through the localization of the viral RdRp at the appropriate start site (Herold and Andino, 2001; Barry and Miller, 2002). Deviation from the canonical model provides opportunities for differential regulation of translation and template use at different stages of the virus cycle.
The identity and role(s) played by host factors in the virus cycle remains a major unknown in plant virology. Improved knowledge in this area will likely provide plant biology with novel views on fundamental cell processes (e.g. translation), as was the case for bacteriophage research on the understanding of basic prokaryote molecular biology in the second half of the twentieth century. In addition, such knowledge might form the foundation for directed plant breeding efforts to develop virus resistance in crops. For example, the discovery of the eIF(iso)4E interaction with TuMV VPg protein was instrumental in the identification of eIF4E as a recessive resistance gene to potyviruses, which has been used for many years by breeders to protect crops (Ruffel et al., 2002; Nicaise et al., 2003; Gao et al., 2004). Such applications, and the basic understanding of the composition and roles of these large RNP complexes in virus translation and replication, will drive many research efforts.
This work was supported by the Natural Science and Engineering Research Council of Canada, by the Fonds Québécois pour la Recherche-Nature et Technologie, by Valorisation Recherche Québec, by the Québec Department of International Relations, by the EU coordinated action ResistVir, by the French Consulate in Québec, and by the Association de Recherches sur les Nicotianées.
References
- Barends S, Rudinger-Thirion J, Florentz C, Giege R, Pleij CW, Kraal B (2004) tRNA-like structure regulates translation of Brome mosaic virus RNA. J Virol 78: 4003–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JK, Miller WA (2002) A −1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc Natl Acad Sci USA 99: 11133–11138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamini G, Preiss T, Hentze MW (2000) Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6: 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman AM, Michel YM, Kean KM (2000) Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′-end. Nucleic Acids Res 28: 4068–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning KS (2004) Plant translation initiation factors: it is not easy to be green. Biochem Soc Trans 32: 589–591 [DOI] [PubMed] [Google Scholar]
- Chong JL, Chuang RY, Tung L, Chang TH (2004) Ded1p, a conserved DExD/H-box translation factor, can promote yeast L-A virus negative-strand RNA synthesis in vitro. Nucleic Acids Res 32: 2031–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh KF, Fraser CS, Hershey JW, Hardy ME (2003) The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J 22: 2852–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J 32: 927–934 [DOI] [PubMed] [Google Scholar]
- Gallie DR (1998) A tail of two termini: a functional interaction between a termini of a mRNA is a prerequisite for efficient translation initiation. Gene 216: 1–11 [DOI] [PubMed] [Google Scholar]
- Gao Z, Johansen E, Eyers S, Thomas CL, Noel Ellis TH, Maule AJ (2004) The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J 40: 376–385 [DOI] [PubMed] [Google Scholar]
- Gazo BM, Murphy P, Gatchel JR, Browning KS (2004) A novel interaction of cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J Biol Chem 79: 13584–13592 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963 [DOI] [PubMed] [Google Scholar]
- Herbert TP, Brierley I, Brown TD (1997) Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J Gen Virol 78: 1033–1040 [DOI] [PubMed] [Google Scholar]
- Herold J, Andino R (2001) Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol Cell 7: 581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Kinzy TG, Sarnow P (2003) Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc Natl Acad Sci USA 100: 15410–15415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerekatte V, Keiper BD, Badorff C, Cai A, Knowlton KU, Rhoads RE (1999) Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J Virol 73: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N (2001) Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell 7: 205–216 [DOI] [PubMed] [Google Scholar]
- Klovins J, van Duin J (1999) A long-range pseudoknot in Qbeta RNA is essential for replication. J Mol Biol 294: 875–884 [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE (2004) Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol Cell Biol 24: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H, Tanguay RL, Balasta ML, Wei CC, Browning KS, Metz AM, Goss DJ, Gallie DR (1997) Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem 272: 16247–16255 [DOI] [PubMed] [Google Scholar]
- Lellis AD, Kasschau KD, Whitham SA, Carrington JC (2002) Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr Biol 12: 1046–1051 [DOI] [PubMed] [Google Scholar]
- Leonard S, Plante D, Wittmann S, Daigneault N, Fortin MG, Laliberté JF (2000) Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J Virol 74: 7730–7737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard S, Viel C, Beauchemin C, Daigneault N, Fortin MG, Laliberte JF (2004) Interaction of VPg-Pro of Turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J Gen Virol 85: 1055–1063 [DOI] [PubMed] [Google Scholar]
- Matsuda D, Yoshinari S, Dreher TW (2004) eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology 321: 47–56 [DOI] [PubMed] [Google Scholar]
- Michel YM, Poncet D, Piron M, Kean KM, Borman MA (2000) Cap-poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J Biol Chem 41: 32268–32276 [DOI] [PubMed] [Google Scholar]
- Neeleman L, Linthorst HJ, Bol JF (2004) Efficient translation of alfamovirus RNAs requires the binding of coat protein dimers to the 3′ termini of the viral RNAs. J Gen Virol 85: 231–240 [DOI] [PubMed] [Google Scholar]
- Nicaise V, German-Retana S, Sanjuan R, Dubrana MP, Mazier M, Maisonneuve B, Candresse T, Caranta C, Le Gall O (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol 132: 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouiery AO, Diez J, Falk SP, Chen J, Ahlquist P (2003) Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for Brome mosaic virus genomic RNA translation. Mol Cell Biol 23: 4094–4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JE, Kirkegaard K (1994) Coupling between genome translation and replication in an RNA virus. Genes Dev 8: 1726–1737 [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Hentze MW (1998) Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem Sci 23: 409–411 [DOI] [PubMed] [Google Scholar]
- Padilla-Noriega L, Paniagua O, Guzman-Leon S (2002) Rotavirus protein NSP3 shuts off host cell protein synthesis. Virology 298: 1–7 [DOI] [PubMed] [Google Scholar]
- Piron M, Vende P, Cohen J, Poncet D (1998) Rotavirus RNA-binding protein NSP3 interacts with eIF4G1 and evicts the poly(A)-binding protein from eIF4F. EMBO J 17: 5811–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Dussault MH, Palloix A, Moury B, Bendahmane A, Robaglia C, Caranta C (2002) A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J 32: 1067–1075 [DOI] [PubMed] [Google Scholar]
- Schaad MC, Anderberg RJ, Carrington JC (2000) Strain-specific interaction of the Tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology 273: 300–306 [DOI] [PubMed] [Google Scholar]
- Spagnolo JF, Hogue BG (2000) Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J Virol 74: 5053–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DN, Carr JP (2000) The GCD10 subunit of yeast eIF-3 binds the methyltransferase-like domain of the 126 and 183 kDa replicase proteins of Tobacco mosaic virus in the yeast two-hybrid system. J Gen Virol 81: 1587–1591 [DOI] [PubMed] [Google Scholar]
- Vende P, Piron M, Castagne N, Poncet D (2000) Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J Virol 74: 7064–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter BL, Parsley TB, Ehrenfeld E, Semler BL (2002) Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J Virol 76: 12008–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ullah Z, Grumet R (2000) Interaction between zucchini yellow mosaic potyvirus RNA-dependent RNA polymerase and host poly-(A) binding protein. Virology 275: 433–443 [DOI] [PubMed] [Google Scholar]
- Wang X, Ullah Z, Grumet R (2004) Identification and characterization of proteins that interact with the carboxy terminus of poly(A)-binding protein and inhibit translation in vitro. Plant Mol Biol 54: 85–98 [DOI] [PubMed] [Google Scholar]
- Wei CC, Balasta ML, Ren J, Goss DJ (1998) Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry 37: 1910–1916 [DOI] [PubMed] [Google Scholar]
- Welch EM, Wang W, Peltz SW (2000) Translation termination: it's not the end of the story. In N Sonenberg, JWB Hershey, MB Mathews, eds, Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 467–486
- White KA, Bancroft JB, Mackie GA (1992) Coding capacity determines in vivo accumulation of a defective RNA of Clover yellow mosaic virus. J Virol 66: 3069–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann S, Chatel H, Fortin MG, Laliberté JF (1997) Interaction of the viral protein genome linked of Turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso)4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology 234: 84–92 [DOI] [PubMed] [Google Scholar]
- You S, Padmanabhan R (1999) A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′ end of exogenous viral RNA templates requires 5′-and 3′ terminal complementary sequence motifs of the viral RNA. J Biol Chem 274: 33714–33722 [DOI] [PubMed] [Google Scholar]
- Zeenko VV, Ryabova LA, Spirin AS, Rothnie HM, Hess D, Browning KS, Hohn T (2002) Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J Virol 76: 5678–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]